1. Introduction
According to the National Cancer Registry (NCR), more than 100,000 South Africans are diagnosed with cancer each year, of which 21% is attributed to breast cancer [
1]. Development of breast cancer is characterized by the deregulation of cell cycle control and resistance to apoptosis [
2]. Tumour cells can acquire resistance to apoptosis by expressing a number of anti-apoptotic proteins such as the Inhibitors of Apoptosis Protein (IAP) family of proteins that include survivin [
3]. These proteins inhibit apoptosis by hindering caspase cascade and therefore, inhibiting both intrinsic and extrinsic pathways [
4]. The resistance to apoptosis is multifactorial and there are several role players that can be implicated in caspase inhibition or independent. As reviewed in Mohammad et al. [
5], Bcl-2 family of proteins, autophagy, proteasome pathway and epigenetics all play a role in resistance to apoptosis.
Although several efforts have been made to reduce the death rates associated with cancer, to date, there has been no ultimate chemotherapeutic drug available to successfully overcome this disease. Targeting biomolecules that regulate molecular mechanisms has become popular and survivin has attracted a lot of research interest across the globe due to the discovery of a wide range of splice variants with diverse biological functions [
4,
6]. Survivin, encoded by the
Baculoviral IAP repeat containing 5 (BIRC5) gene, is the smallest and best studied member of the IAPs family, which is involved in the regulation of cell division, promoting angiogenesis, inhibition of both caspase-dependent and caspase-independent apoptosis in cancer cells [
7,
8]. Survivin wild-type is strongly expressed during embryonic and foetal development but rarely found in normal adult tissues [
4]. It is, however, up-regulated in transformed cancer cell lines and most common types of human cancers [
9,
10], where it promotes tumour survival by reducing apoptosis as well as favouring endothelial cell proliferation and migration.
Alternative splicing of the
BIRC5 gene produces six survivin splice variants, namely, wild type survivin, survivin 2B, survivin 2α, survivin 3B, survivin ∆Ex3 and survivin 3α [
6]. Survivin has been acknowledged as an essential molecular marker and target in a range of cancer diagnosis and therapeutics [
11].
As
2O
3 has been shown to exert anticancer activities against solid cancers, including breast cancer [
12,
13]. As
2O
3 has also been demonstrated to inhibit lung adenocarcinoma cell line (H1355) growth by down-regulating survivin expression and through the activation of p38 and c-Jun N-terminal kinases (JNK) pathways [
14]. Inhibition of Phosphoinositide 3-Kinase (PI3K) or extracellular signal-regulated kinases (ERK) signalling led to clear inhibition of survivin expression. However, pre-treatment with p38 Mitogen-Activated Protein Kinase (MAPK) inhibitor led to up-regulated survivin levels.
The role and the expression of the survivin splice variants are not fully understood and there is no study which had proven that As2O3 has any effect on the splicing machinery of survivin and its splice variants. This study focused on analysing the expression pattern of the different survivin splice variants during both As2O3-induced apoptosis and cell cycle arrest in breast cancer MCF-7 cells.
2. Methods and Materials
2.1. Materials
The MCF-7 cells were kindly donated by Prof Mervin Meyer from the University of the Western Cape, South Africa. Dulbecco’s Modified Eagle Medium (DMEM) and foetal bovine serum (FBS) were purchased from Hyclone (Hyclone, South Logan, UT, USA). Antibiotic mixture containing penicillin and streptomycin (Pen-Strep), phosphate buffered saline (PBS) the MTT [3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide], 4′, 6-diamidino-2-phenylindole (DAPI), Trizol reagent were obtained from ThermoFisher Scientific (ThermoFischer Scientific, Waltham, MA, USA) while Dimethyl Sulfoxide (DMSO) was purchased from (Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). The AMV II Reverse transcription system was purchased from Promega (Promega, Madison, WI, USA) while the EmeraldAmp® GT-PCR Kit was procured from Takara Bio (Takara Bio, Kasatsu, Shiga Prefecture, Japan). The Muse® Assay Kits (Muse® Count and Viability Assay, Muse® Cell Cycle Assay, Muse® Annexin V and Dead Cell Assay, Muse® MitoPotential Assay, Muse® Multi-Caspase Assay, Muse® MAPK Assay and Muse® PI3k Assay) were all purchased from Merck-Millipore (Merck-Millipore, Darmstadt, Germany). All the reagents were used without further purification or alterations.
2.2. Cell Culture
MCF-7 cells were cultured in DMEM supplemented with 10% FBS and 1% antibiotic mixture of Penicillin and Streptomycin and maintained in culture flasks at 37 °C in a humidified chamber containing 5% CO2.
2.3. Cytotoxicity Assay
The MCF-7 cells viability was tested by the MTT assay to evaluate the cytotoxicity of the As
2O
3. Briefly, the MCF-7 cells were diluted into a single cell suspension and 2 × 10
3 cells/well were seeded in 96-well culture plates and allowed to attach, overnight. The cells were washed with 1 × PBS, then treated with different concentrations of As
2O
3, cobalt chloride and curcumin for 24 h. After 24 h, the treatment was discarded and wells were washed with 1 × PBS. Then, 10 µL of MTT reagent (5 mg/mL) was added to each well and the plates were incubated for 4 h in the CO
2 incubator. Following incubation, 100 µL DMSO was added to dissolve formazan crystals and the absorbance readings were taken at 560 nm using a microplate reader (Promega). The cell viability was assessed using the formula:
The IC50s were extrapolated from the graph of cell viability versus the concentration of the compounds and confirmed using the MUSE® Cell Viability Assay. Cobalt chloride was used as a positive control for cell cycle assays only as it is known to induce the G2/M cell cycle arrest while curcumin was used as a positive control for apoptosis. These two positive controls were therefore used for the relevant experiments.
2.4. Muse® Count and Viability Assay
In order to confirm the cell cycle arrest and apoptotic inducing concentrations, MCF-7 cells (2 × 105 cells/well) were cultured in six well plates and treated with 11 and 32 µM As2O3, 100 µM cobalt chloride and 100 µM curcumin for 24 h. Following, treatment, the cells were trypsinized and centrifuged at 300× g for 5 min. The cells were then resuspended in 20 µL cell culture medium and 380 µL Muse® Count and Viability Reagent (Merck-Millipore) was added to each sample. The samples were incubated for 5 min in the dark at room temperature and then analysed using the Muse® Cell Analyser (Merck-Millipore).
2.5. Light and Fluorescence Microscopic Analysis
MCF-7 cells (5 × 104 cells/well) were seeded in 24 well plates, overnight. After incubation, the cells were then exposed to various concentrations of As2O3 (0, 4, 11, 16 and 32 µM), cobalt chloride (50 and 100 µM) and curcumin (50 and 100 µM) for 24 h. After treatment, the cells were washed with sterile 1 × PBS and fixed with 3.7% paraformaldehyde for 10 min at room temperature. The cells were then washed twice with sterile 1 × PBS and stained with DAPI (5 µg/mL) for 15 min in the dark, at room temperature. Following incubation, the cells were washed twice with sterile 1 × PBS. Morphological changes of MCF-7 cells in each group were observed under the Olympus CKX53 Inverted Microscope, (Olympus, Shinjuku, Tokyo, Japan) and Eclipse Ti-U fluorescence microscope (Nikon Instruments Inc., Melville, NY, USA), for light and fluorescence microscopy, respectively. The images were captured with DSRI-1 camera and an LC-30 camera set, respectively.
2.6. Cell Cycle Assay
To confirm whether As2O3 induces cell cycle arrest in MCF-7 cells, Muse® Cell Cycle Kit was used. MCF-7 cells (2 × 105 cells/well) were cultured in six well plates and allowed to settle, overnight. The cells were then treated with 11 µM of As2O3 and 100 µM of cobalt chloride and incubated for 24 h in appropriate cell culture conditions mentioned above. Following incubation, the cells were mildly trypsinized and centrifuged at 300× g for 5 min. The cells were then washed with sterile 1 × PBS and fixed for 3 h in 70% ethanol at −20 °C. Following incubation, the cells were centrifuged, washed with sterile 1 × PBS and 200 µL of the Muse® Cell Cycle reagent was added and incubated in the dark for 30 min. The samples were analysed using the Muse® Cell Analyser (Merck-Millipore).
2.7. Annexin V and Dead Cell Assay
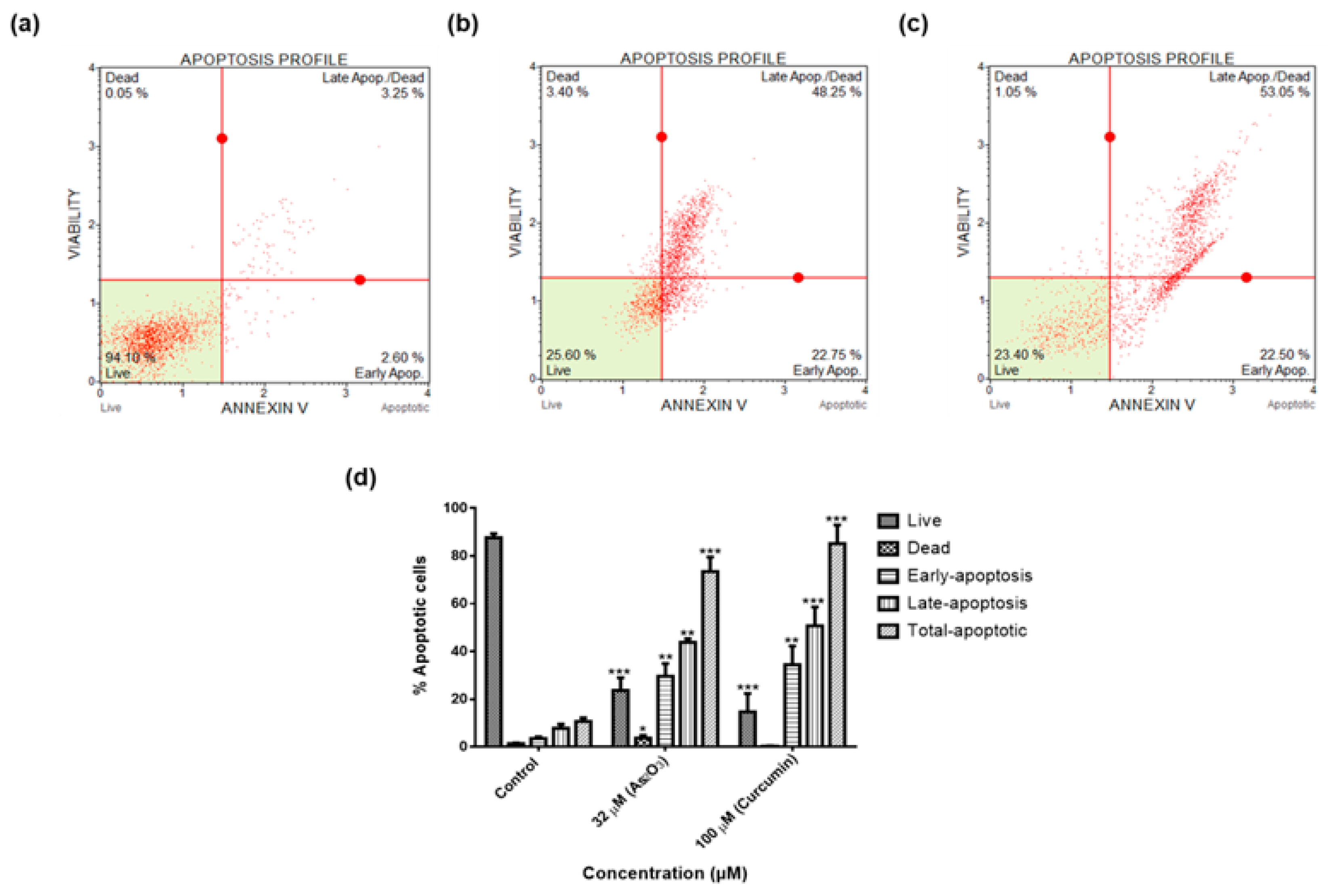
To confirm whether As2O3 induces apoptosis in MCF-7 cells, the Annexin V and Dead Cell Kit (MerkMillipore, Darmstadt, Germany) was used. MCF-7 Cells (2 × 105 cells/well) were seeded and allowed to adhere in six well plates, overnight. The cells were treated with 32 µM of As2O3 and 100 µM curcumin, which was used as a positive control, for 24 h. After the incubation period, the cells were trypsinized and centrifuged at 300× g for 5 min. The cells were re-suspended in 100 µL of DMEM with 1% FBS and 100 µL of Annexin V and Dead Cell Reagent was added to each tube. Each sample was vortexed for 5 s and incubated in the dark at room temperature for 20 min. Samples were then analysed using the Muse® Cell Analyser (Merck-Millipore).
2.8. Mitochondrial Membrane Potential Assay
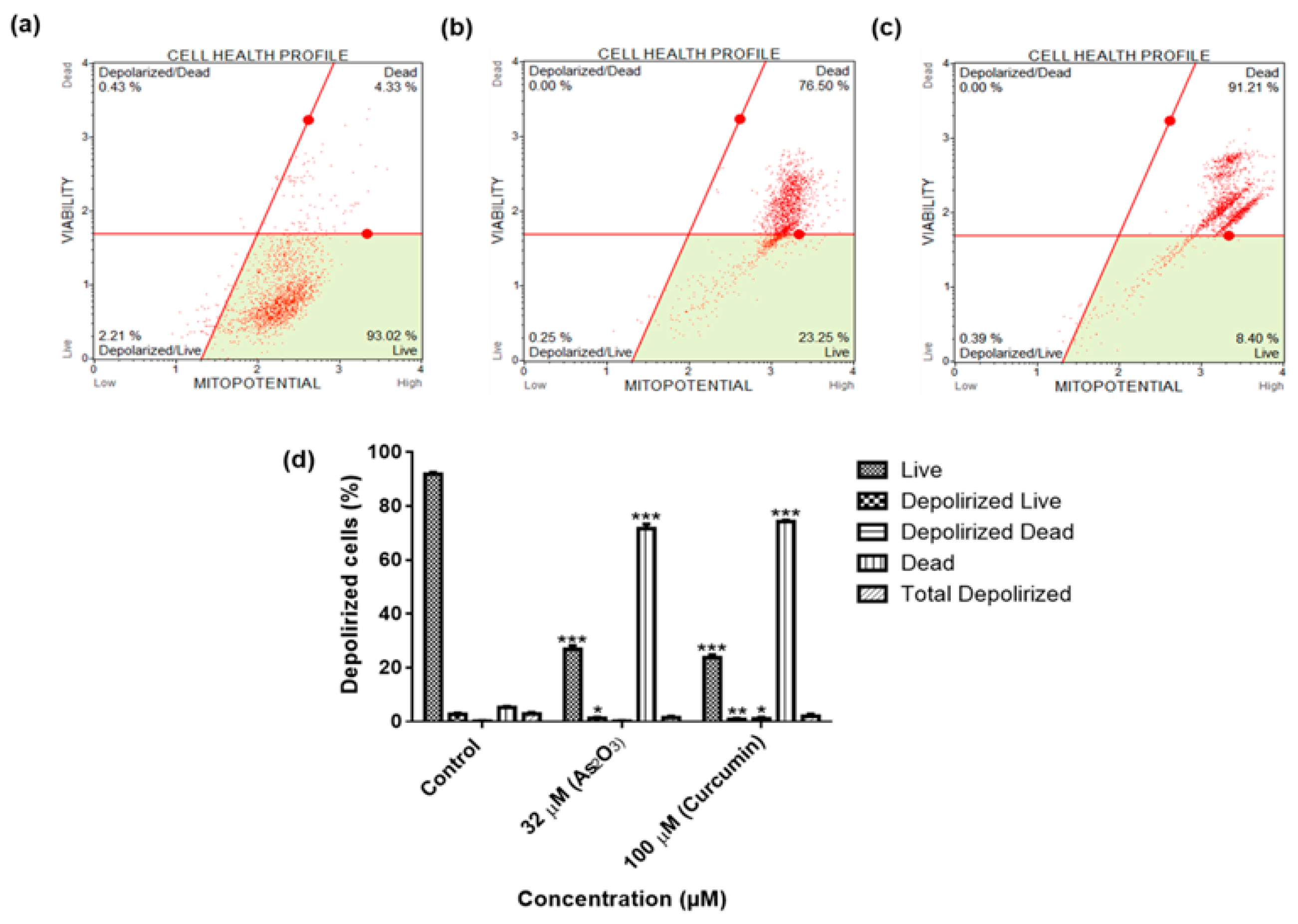
Mitochondrial membrane depolarization, a marker of apoptosis, was detected using the Muse® MitoPotential Assay kit according to the manufacturer’s protocol (Merck-Millipore). Briefly, 2 × 105 cells/well were cultured in six well plates and allowed to adhere to the bottom of the plate, overnight. Following incubation, the cells were treated with 32 µM As2O3 and 100 µM curcumin. After 24 h treatment, the cells were trypsinized and centrifuged at 300× g for 5 min. The cell suspension for each sample was mixed with 95 µL MitoPotential dye and incubated for 20 min at 37 °C. After incubation, 5 µL MitoPotential 7-AAD was and placed in the dark for 5 min. The samples were then analysed using the Muse® Cell Analyzer (Merck-Millipore).
2.9. Multi-Caspase Assay
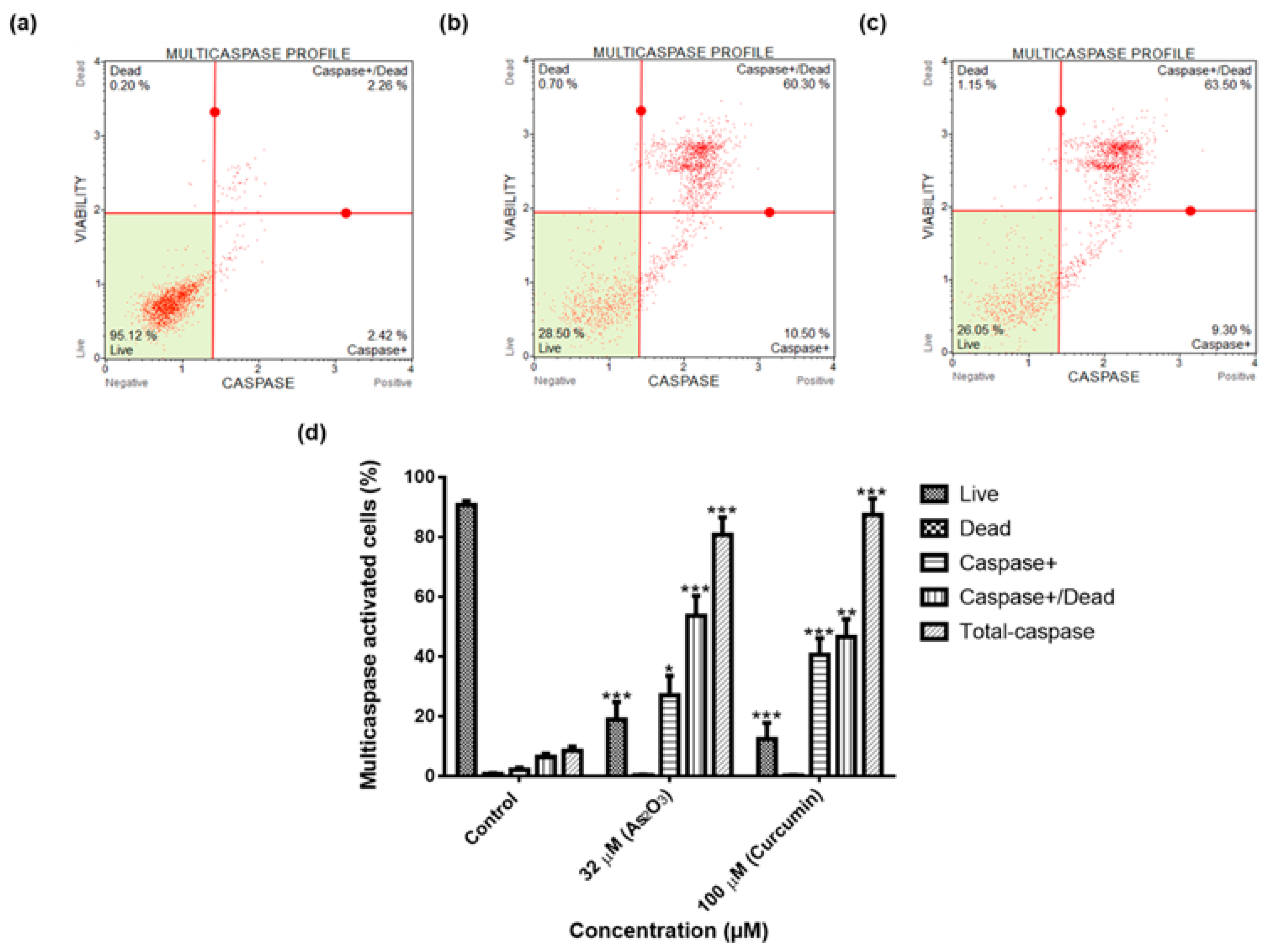
To classify apoptosis induced by As2O3, the Multi-Caspase Assay was performed as instructed by the Muse® Multi-Caspase Kit protocol (Merck Millipore). MFC-7 cells (2 × 105 cells/well) were cultured in six well plates and allowed to adhere to the bottom of the plate, overnight. Following the incubation overnight, the cells were treated with 32 µM As2O3 and 100 µM curcumin for 24 h, in appropriate cell culture conditions. Following treatment, the cells were trypsinized and centrifuged at 300× g for 5 min. Multi-Caspase buffer was used to re-suspend the cells and 50 µL of each sample was transferred to a new tube and 5 µL of Multi-Caspase reagent was added. Samples were mixed and incubated at 37 °C in CO2 incubator for 30 min, followed by addition of 150 µL 7-AAD and placed in the dark for 5 min at room temperature. The samples were then analysed using the Muse® Cell Analyser (Merck-Millipore).
2.10. MAPK Dual Pathway Activation Assay
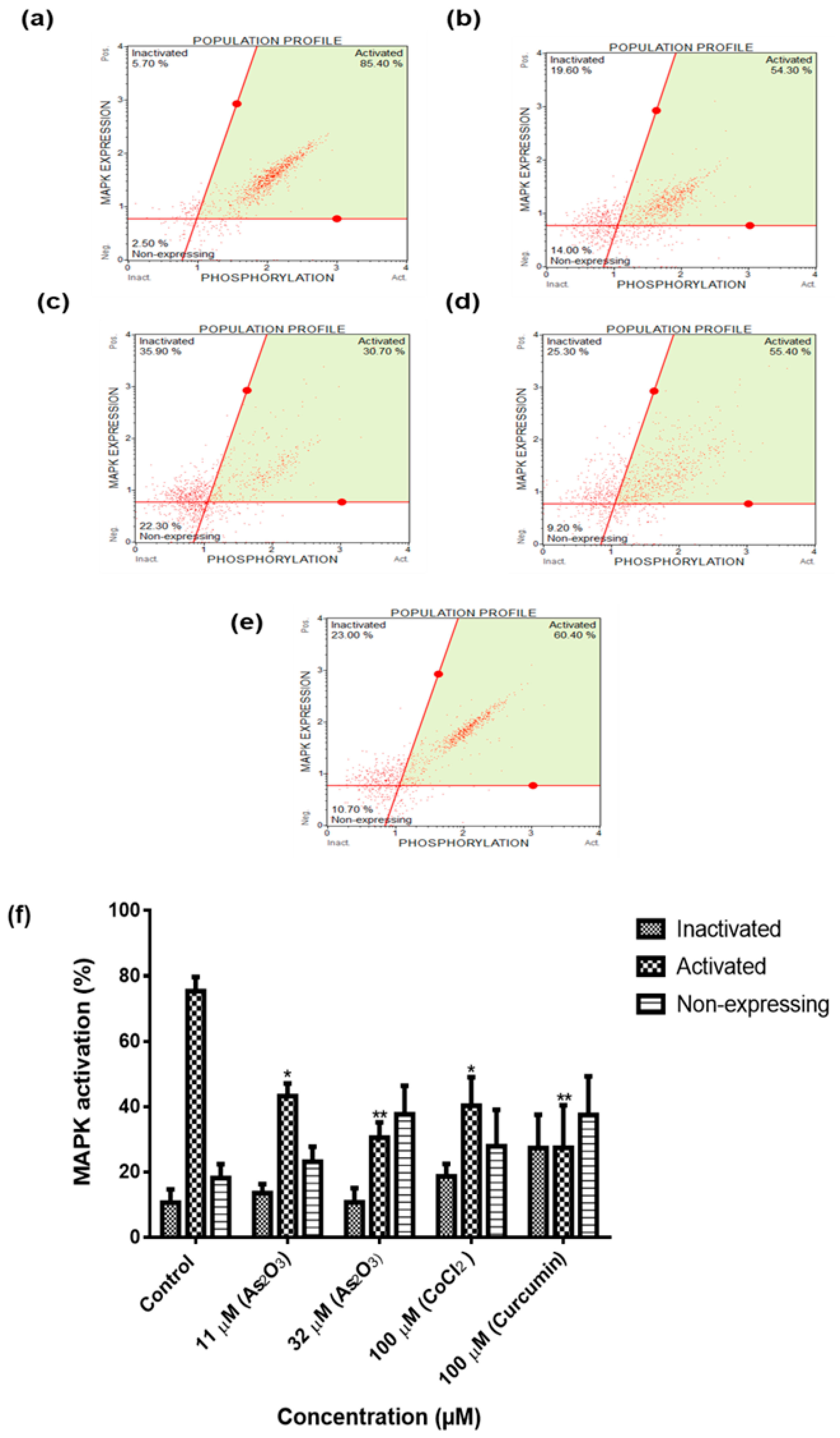
The Muse® MAPK Assay was employed to detect the total protein expression and to measure the phosphotransferase activity of the mitogen-activated protein kinase according to the manufacturer (Merck-Millipore). The cells were seeded (1 × 105 cells/well) in six well plates and allowed to adhere overnight in a 37 °C humidified incubator containing 5% CO2. The cells were then treated with 11 and 32 µM As2O3, 100 µM cobalt chloride and 100 µM curcumin for 24 h. Following treatment, the cells were trypsinized and centrifuged at 300× g for 5 min. The cells were resuspended in 500 µL of 1 × MAPK buffer and equal amounts of fixation buffer were added. The samples were placed on ice for 5 min. The samples were centrifuged at 300× g for 5 min, permeabilized by adding 1 mL of ice-cold permeabilization buffer and placed on ice for 5 min. Following incubation, the cells were centrifuged at 300× g for 5 min, resuspended in 450 µL of 1 × MAPK buffer and 10 µL of antibody working cocktail (5 µL of anti-phospho-ERK1/2 (Thr202/Thr204, Thr185/Thr187), phycoerythrin and 5 µL of anti-ERK1/2 PECy5 conjugated antibodies) and incubated for 30 min in the dark at room temperature. Following incubation step, 100 µL of 1 × MAPK buffer was added to each sample and centrifuged at 300× g for 5 min. The cells were resuspended in 200 µL of 1 × MAPK buffer and analysed using Muse® Cell Analyser (Merck-Millipore).
2.11. PI3K Dual Pathway Activation Assay
The Muse® phosphoinositide 3-kinase (PI3K) Assay was performed to detect the extent of Akt phosphorylation relative to the total Akt expression in given cell population according to the manufacturer (Merck-Millipore). Similar to MAPK Assay, the cells (2 × 105 cells/well) were seeded in six well plates and allowed to attach, overnight in a 37 °C humidified incubator containing 5% CO2. The cells were then treated with 11 and 32 µM As2O3, 100 µM cobalt chloride and 100 µM curcumin for 24 h. Following treatment, the cells were trypsinized and centrifuged at 300× g for 5 min. The cells were resuspended in 500 µL of 1 × PI3K buffer and equal amounts of fixation buffer were added. The samples were placed on ice for 5 min. The samples were centrifuged at 300× g for 5 min, permeabilized by adding 1 mL of ice-cold permeabilization buffer and placed on ice for 5 min. Following incubation, the cells were centrifuged at 300× g for 5 min, resuspended in 450 µL of 1 × PI3K buffer and 10 µL of antibody working cocktail (5 µL of anti-phospho-Akt [Ser473], Alexa, Fluor®555 and 5 µL of anti-Akt/PKB, PECy5 conjugated antibodies) and incubated for 30 min in the dark at room temperature. Following incubation step, 100 µL of 1 × PI3K buffer was added to each sample and centrifuged at 300× g for 5 min. The cells were resuspended in 200 µL of 1 × PI3K buffer and analysed using Muse® Cell Analyser (Merck-Millipore).
2.12. Polymerase Chain Reaction
Total RNA was isolated from treated and untreated MCF-7 cells using TRIzol reagent from ThermoFisher Scientific, USA. Complementary deoxyribonucleic acid (cDNA) was synthesized from the total RNA using the AMV II Reverse transcription System (Promega, USA). The cDNA samples were normalized to a consistent concentration of 200 ng. The cDNA samples were then subjected to PCR using primers which amplify the different survivin splice variants (
Table 1).
The EmeraldAmp® GT PCR Kit (Takara Bio, USA) was used and the manufacturer’s instructions were followed. Briefly, the 25 µL reaction mix containing 12.5 µL EmeraldAmp®, GT PCR Master Mix (Takara Bio), 1 µl of each primer, 9.5 µl of water and 1 µl of DNA template. The PCR products were visualised on a 1.3% agarose gels. The gels were then viewed using a Chemidoc XRS image analyser (BioRad, Hercules, CA, USA).
2.13. Immunocytochemistry
To view the localization and expression of survivin proteins after As2O3 treatment, the cells were subjected to immunofluorescence staining. The MCF-7 cells were treated with 11 µM and 32 µM As2O3, 100 µM cobalt chloride and 100 µM curcumin for 24 h at 37 °C in a humidified atmosphere. The cells were washed with sterile 1 X PBS and fixed with 4% of paraformaldehyde for 15 min. After fixing, the cells were permeabilized with 0.25% Triton™ × -100 for 10 min and antibody non-specific binding was blocked using 5% BSA for 1 h at room temperature. The cells were labelled with 5 µg/mL survivin recombinant rabbit monoclonal antibody (Product # 700387) [ThermoFischer Scientific, USA] for 1 h at room temperature. The cells were then washed with sterile 1 × PBS, then labelled with 4 µg/mL Alexa Flour® 488 goat anti-rabbit IgG secondary antibody (Product # A-11008) [ThermoFischer Scientific, US] for 30 min in the dark. The cell nuclei were identified by counterstaining with DAPI for 5 min. The samples were examined using an Eclipse Ti-U fluorescence microscope (Nickon Instruments Inc.) at 20 × magnification and images were captured with DSRI-1 camera.
2.14. Statistical Significance
All data was analysed using Graph pad prism version 6.0 statistical software and the values are expressed as mean ± standard error of mean (SEM). Statistical significance was considered at p < 0.05 using the Turkey Cramer multiple comparison test. The asterisk (*) (**) and (***) show p < 0.05, p < 0.01 and p < 0.001, respectively.
4. Discussion
The purpose of this investigation was to evaluate the potential effect of As
2O
3 on the expression of the different survivin splice variants during cell cycle progression and apoptosis of breast cancer cell line, MCF-7. To date, there is a limited number of studies that have directly or indirectly investigated the expression of survivin variants in breast cancer cells. The focus has been on the wild-type only, where its expression has been shown to favour the carcinogenesis process [
3]. Additionally, As
2O
3 has been used for ages to treat proteolytic leukaemia [
15]. In order to understand the expression of survivin splice variant during As
2O
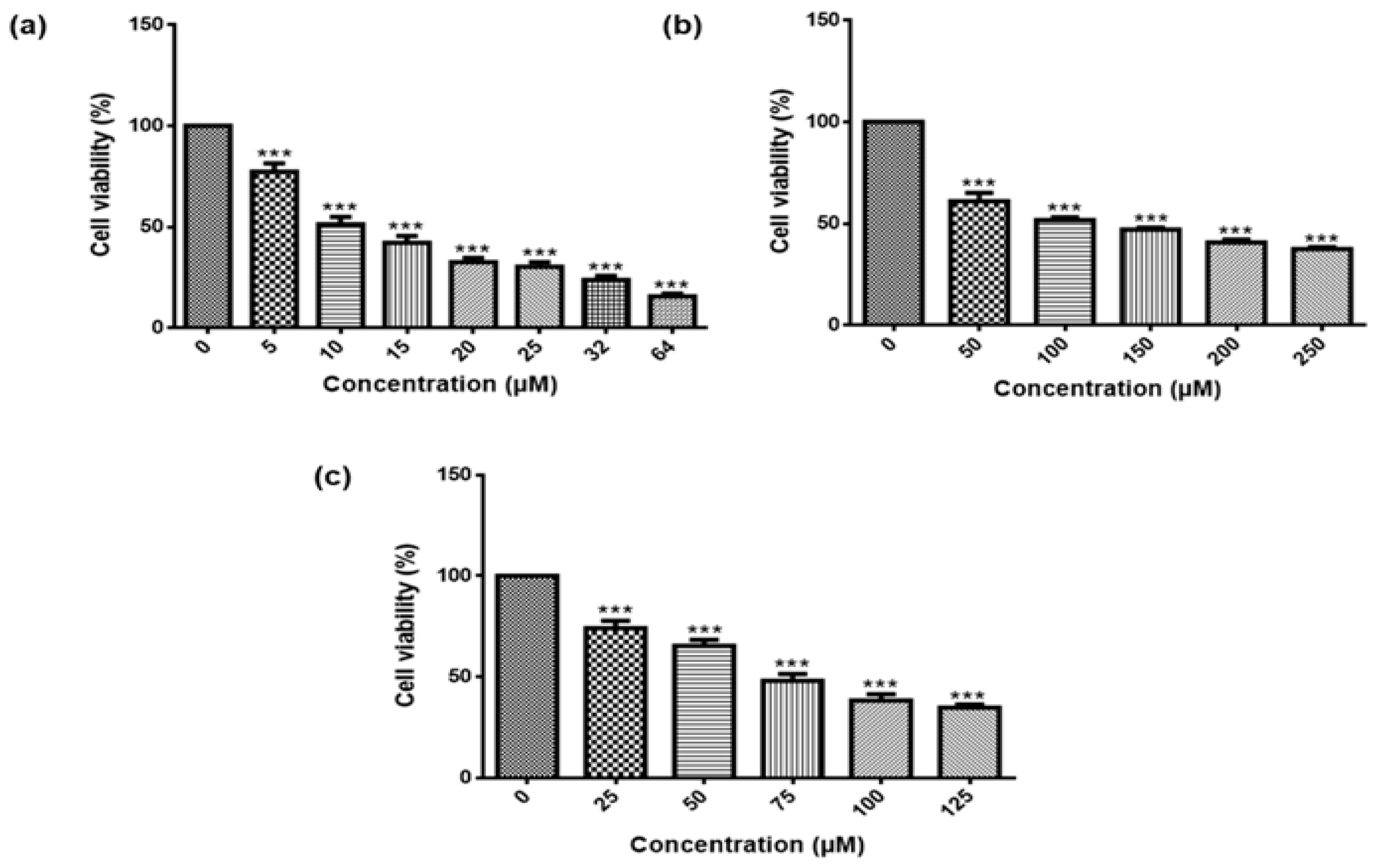
3-induced cell cycle arrest and apoptosis, the viability of MCF-7 cells was investigated using MTT Assay and the Muse
® Count and Viability Assay. Both confirmed that As
2O
3 reduced the viability of the MCF-7 cells in a concentration-dependent manner (
Figure 1a and
Figure 2f). This is in line with the previous reports [
13,
16], which showed that As
2O
3 suppresses the growth of MCF-7 cells. Kasukabe et al. [
16] also showed that Cotylenin A enhanced As
2O
3-induced growth inhibition of MCF-7 and MDA-MB-231 cells. The 11 µM As
2O
3 suppressed 50% cell growth inhibition while 32 µM of As
2O
3 yielded the lowest cell viability against the MCF-7 cells and was then used to investigate apoptosis in this study while 11 µM was used to investigate cell cycle arrest potential of As
2O
3 in MCF-7 cells.
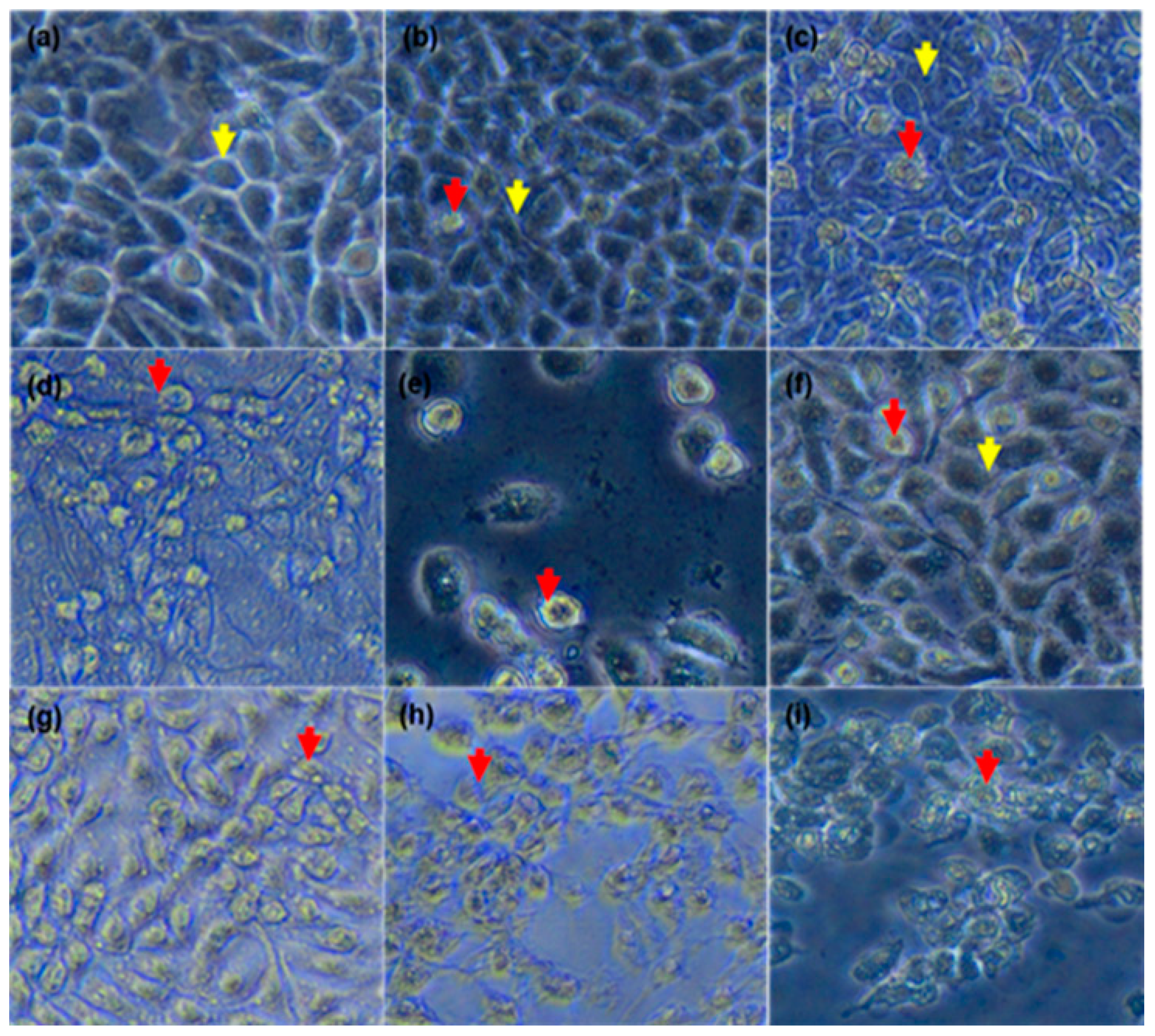
As demonstrated in
Figure 3a, the untreated cells maintained their epithelial morphological shape. However, the cells treated with As
2O
3 exhibited morphological changes typical to apoptosis (
Figure 3b–e). As
2O
3 reduced the MCF-7 cell numbers in a concentration dependent manner, which can be attributed to characteristics observed from the treated cells. The treated cells displayed increased cellular volumes with the formation of condensed chromatin, raptured nuclear membrane and cytoplasmic blebs. These are characteristics of cells undergoing apoptosis. This data concurred what was demonstrated by Liu et al. (2015) where it was shown that As
2O
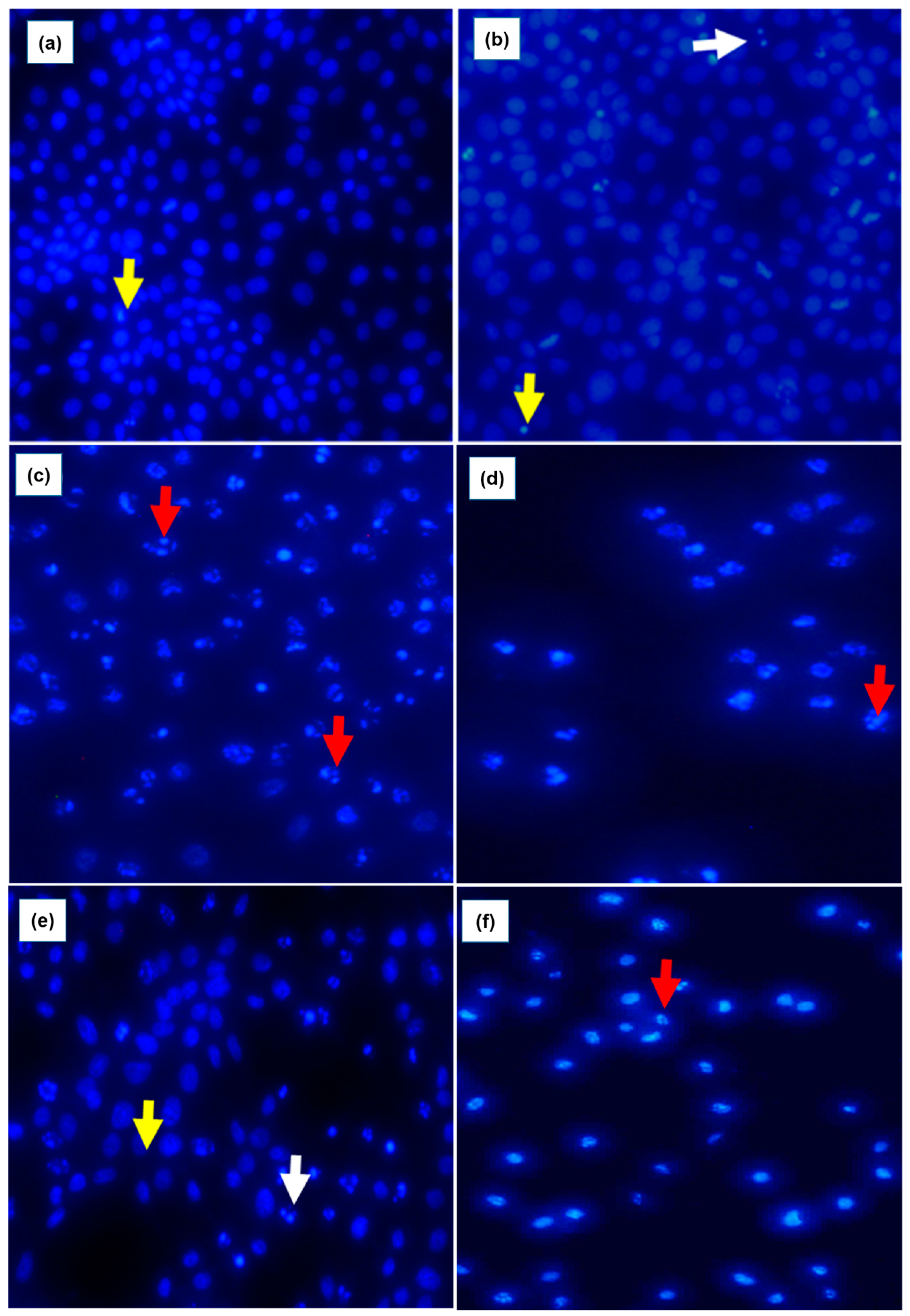
3 treated cells changed from polygonal to round shape, displaying partly condensed chromatin and the appearance of vacuoles. Fluorescence microscopy further confirmed apoptotic features induced by As
2O
3 (
Figure 4). When the untreated control cells were stained with DAPI, they showed normal intact nuclei, whereas the treated MCF-7 cells with 11 µM As
2O
3 displayed mostly mitotic morphology, suggesting that the cells may be arrested during the G2/M cell cycle arrest checkpoint. As the concentration of As
2O
3 increased to 16 µM (
Figure 4d). Cell numbers decreased with several apoptotic features such membrane blebbing, DNA fragmentation and nuclear condensation, appearing. The cells that were treated with 32 µM, showed more membrane blebbing and nuclei fragmentation (
Figure 4d). This concurred with other studies that reported that As
2O
3 induces apoptosis in breast cancer MCF-7 cells [
17,
18]. As the concentration of As
2O
3 increased, the number of apoptotic cells also increased, which confirmed that this compound induces cellular apoptosis in a concentration-dependent manner. As
2O
3 has been previously shown to reduce the activity of IKKβ, which is a catalytic subunit of the IKK complex, which was shown to be reduced by As
2O
3 [
13]. As
2O
3 has also been shown to be a Notch-1 inhibitor, inactivating the Notch Signalling pathway in breast cancer [
19]. Notch inactivation has been shown to cause down-regulation of antiapoptotic genes such as Bcl-2 and NF-κB, resulting in the inhibition of cell growth, invasion and induction of apoptosis [
20].
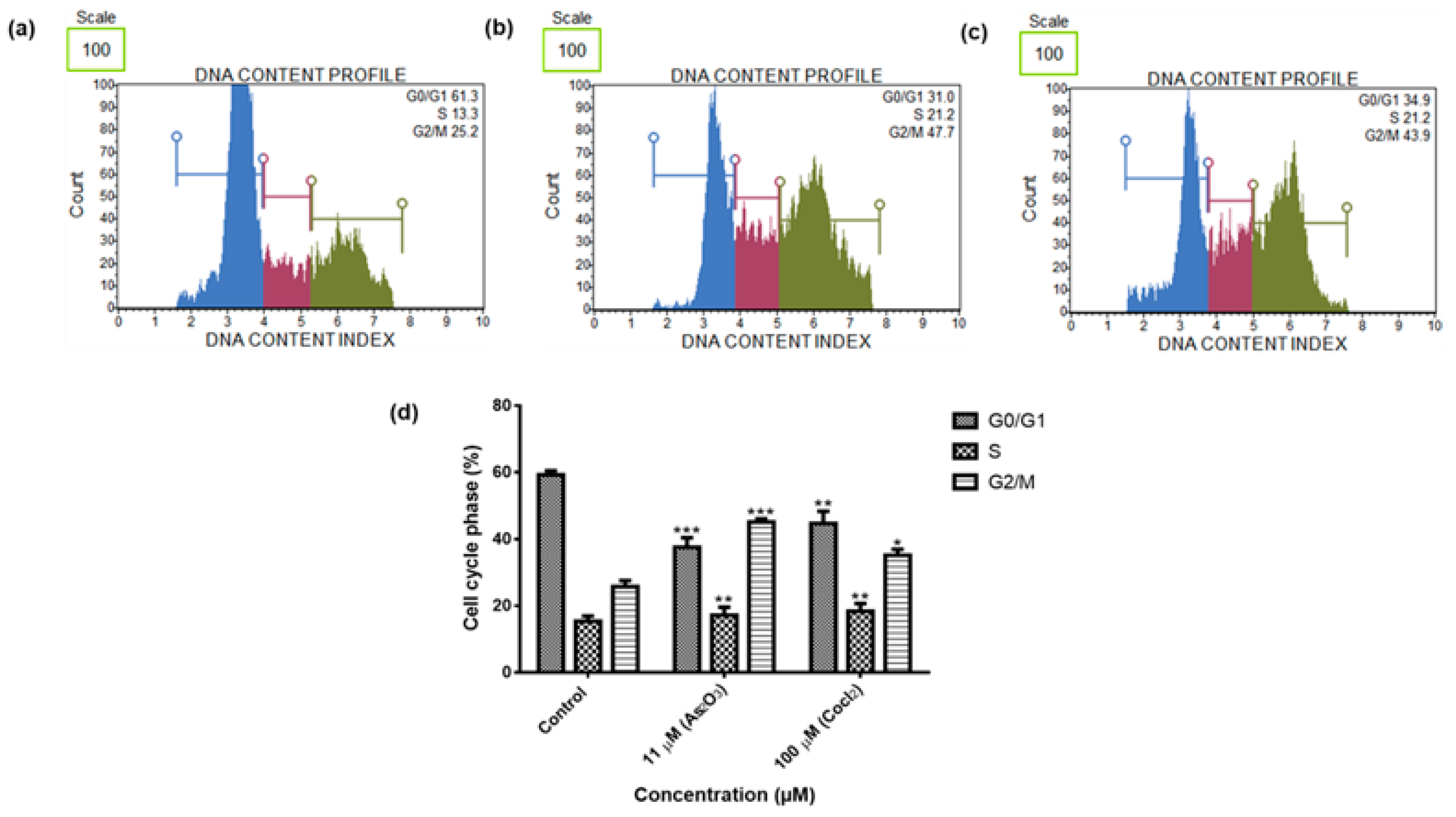
To further elucidate the cell cycle arrest potential of As
2O
3, we examined DNA content using Muse
® Cell Cycle Kit. In our study, as it can be seen in
Figure 5d, the G2/M percentage of cells after treatment with 11 µM As
2O
3 (45.13 ± 0.942) was much higher than the complement of G2/M cells in the control untreated cells (25.73 ± 1.824). Our cell cycle results showed that As
2O
3 induced the G2/M cell cycle arrest of MCF-7 cells. Several studies also have reported As
2O
3 as a therapeutic agent which can induce cell cycle arrest and expression of certain cell cycle-specific genes in malignant tumours [
21,
22]. Santarelli et al. [
23] reviewed the function of survivin and survivin based treatment strategies and highlighted that survivin is involved in cell cycle regulation, involved in spindle formation. This is in line with our finding that showed that survivin and survivin 2B is upregulated during arsenic trioxide-induced G2M cell cycle arrest, which suggest that this variant be part of the chromosomal passenger complex (CPC). Our study further suggest that the upregulation of survivin 3α may be involved in cellular survival in unfavourable condition. Our study also show that survivin 2B may be an antagonist to both survivin and survivin 3α during As
2O
3-induced apoptosis. Additionally, our data showed that As
2O
3 induced late apoptosis rather than early apoptosis (
Figure 6d). The same trend was previously observed in the breast cancer cell line MCF-7 [
24] and other cell lines which include T80, HEY and SKOV3 ovarian cancer cells [
25]. Our data suggest that As
2O
3 induced the extrinsic pathway as As
2O
3 was shown to have no effect on the mitochondrial-mediated apoptotic pathway (
Figure 7d). This is in line with another study that showed the same results where HepG2 cells were exposed to As
2O
3 for a period of 7 days [
26]. The caspases (cysteine-directed aspartate-specific proteases) are important in directing the process of programmed cell death in response to apoptotic signals [
27]. In our study, Multi-Caspase Assay revealed that 32 µM As
2O
3 lead to activation of several caspases including (1, 3, 4, 5, 6, 7, 8 and 9) and this shows that As
2O
3 induces caspase-dependent apoptosis because all the activated caspases include the initiator caspases (caspase 8 and 9) and execution caspases (3 and 7) (
Figure 8d).
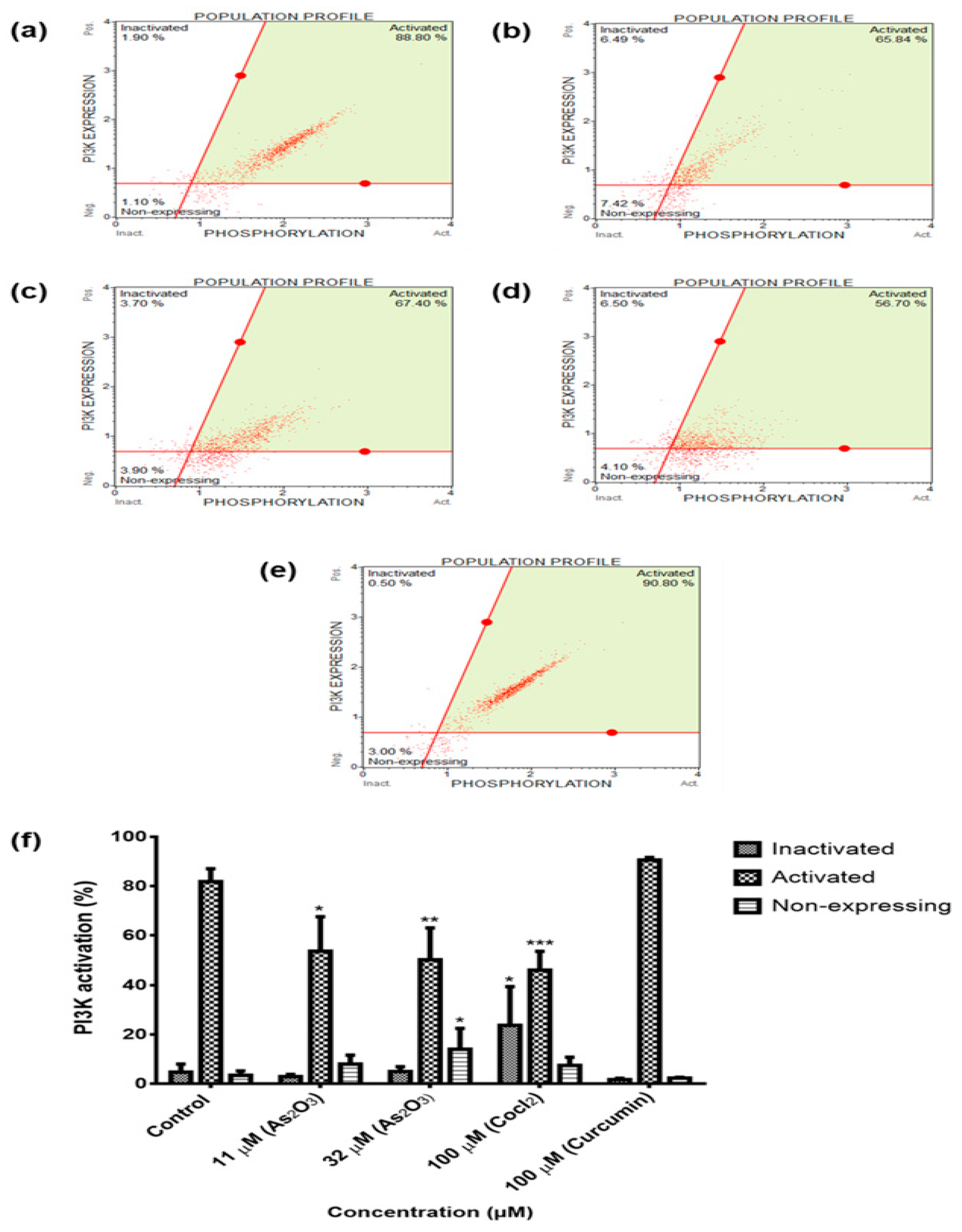
To further characterize the mechanism by which As
2O
3 suppress cell growth of MCF-7 cells, intracellular pathways known to be activated during proliferation of cancer cells were analysed. Many of these signal transduction pathways involve activation of protein serine/threonine kinase of extracellular signal-regulated kinases (ERK) family by oestrogen and involved in cellular growth [
28]. To determine the effect of As
2O
3 on these cell survival pathways, the MCF-7 cells were exposed to 11and 32 µM As
2O
3 for 24 h and the levels of MAPK and PI3K activation were determine using the Muse
® Cell Analyser. The MAPK (
Figure 9f) and PI3K (
Figure 10f) were significantly deactivated during As
2O
3-induced cell cycle arrest and apoptosis. Previously, As
2O
3 was shown to inhibit the activation of MAPK and PI3K pathways in U118-MG cells and gastric cancer cells [
29,
30]. As
2O
3 has also been demonstrated to inhibit lung adenocarcinoma cell line (H1355) growth by down-regulating survivin expression through the activation of p38 and c-Jun N-terminal kinases (JNK) pathways [
14]. Inhibition of PI3K or ERK signalling led to explicit inhibition of survivin expression. However, pre-treatment with p38 MAPK inhibitor led to up-regulated survivin levels. PI3K, ERK and p38 MAPK are all involved in regulating survivin expression and up-regulation of survivin may provide a potential cancer therapy [
31]. For the first time, this study has demonstrated the involvement of the novel survivin splice, especially, survivin 2B in the inactivation of the two cell survival pathways that are involved in breast cancer development.
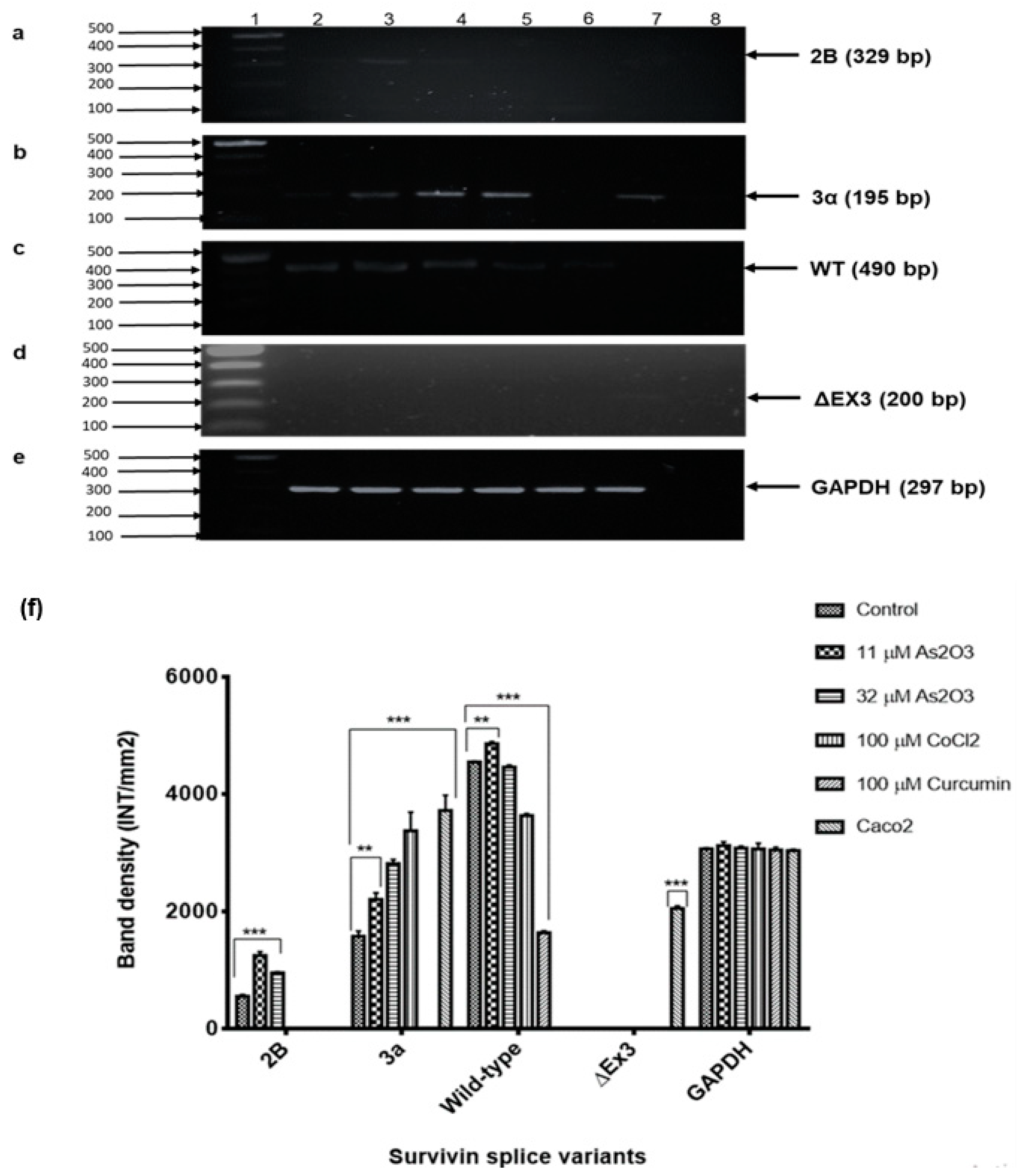
Reverse-Transcription Polymerase Chain Reaction (RT-PCR) was used to analyse the differential expression of survivin spliced variants during As
2O
3-induced cell cycle arrest and apoptosis in the breast MCF-7 cancer cells. Although expression of survivin during cell cycle progression has been studied, little is known about the expression of spliced variants, especially in As
2O
3-mediated cell cycle arrest. Survivin 2B is a variant that is pro-apoptotic or anti-apoptotic depending on the type and stage of cancer. In this study, the expression of variant 2B was undetectable in the untreated MCF-7 cells but upregulated during As
2O
3-induced cell cycle arrest (
Figure 11a) and the expression was downregulated during As
2O
3-induced apoptosis. This data suggests that survivin 2B has a role in cell cycle regulation than apoptosis. Previously, a more variable expression of survivin 2B level was found at different breast cancer stages [
9]. Survivin wild-type was found to be highly expressed in the MCF-7 cells and the expression was decreased during As
2O
3-induced G2/M cell cycle arrest and apoptosis. Our data revealed low expression of the anti-apoptotic survivin variant 3α in MCF-7 cells (
Figure 11b). Its expression was found to be upregulated during As
2O
3-induced cell cycle arrest and apoptosis.
The findings of this study agree with the previous studies that have shown that apoptosis induction decreases the expression of survivin wild-type [
32,
33]. This study showed that survivin ΔEx3 is undetectable in MCF-7 cells and during As
2O
3-induced cell cycle arrest and apoptosis but was detected in caco2 colon cancer cells. This supports the previous report that showed little or no expression of ΔEx3 in breast carcinoma [
34]. Vegran et al. [
35], revealed that survivin wild-type, survivin-ΔEx3 and survivin 2B have no prognostic role in breast carcinoma. There is concrete evidence showing that survivin variants are highly expressed in cancer cells including MCF-7 cells and demonstrated a cell-cycle dependent expression with a noticeable increase in the G2/M phase [
36]. Our study, for the first time suggests that survivin splice variants are differentially expressed in MCF-7 during both As
2O
3-induced cell cycle arrest and apoptosis. These results may suggest that the mode of action of As
2O
3 in the inhibition of cell proliferation may be dependent on the express of different survivin splice variants. Furthermore, the role of survivin in breast cancer progression may be boundless to its role in the inhibition of apoptosis, which is a hallmark of cancer. In this study, we have documented for the first time that Survivin 2B is involved in As
2O
3-mediated G2M cell cycle arrest in MCF-7 cells. Previously, G2M cell cycle arrest has been shown to involve the inactivation of PI3K/Akt and ERα pathways [
37]. Arsenic trioxide was found to upregulate survivin 2B during G2M cell cycle arrest which was associated with the deactivation of PI3K/Akt pathway. This suggests that strategies targeting survivin 2B and PI3K/Akt may be beneficial for anticancer drug development against breast cancer. Li et al. (2009) showed that As2O3 induces apoptosis and G2M cell cycle arrest by inhibiting PI3K/Akt signalling by Cbl and p53 activation. Our study suggests that survivin 2B plays a role in As
2O
3-mediated PI3K/Akt inactivation and this variant can be targeted for anticancer drug development against breast cancer.
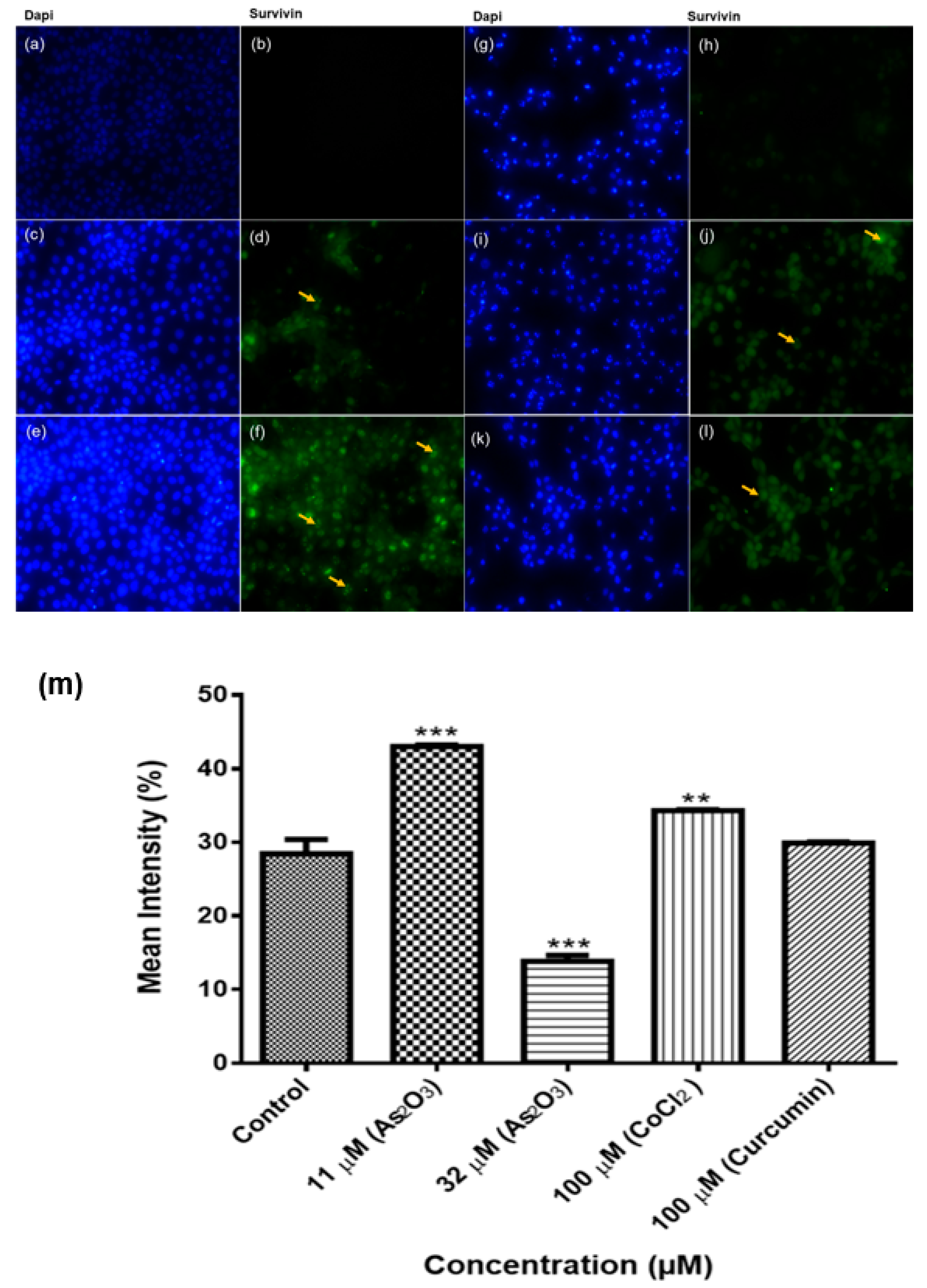
Human survivin is located on chromosome 17q25, approximately 3% of the distance from the telomere and it comprises three introns and four exons, encoding 142 amino acids, including one copy of the BIR, essential for apoptosis inhibition and 16.5 kDa molecular weight [
38]. In our study, we performed an immunocytochemical analysis of the expression of survivin proteins in breast cancer MCF-7 cells. Survivin proteins were expressed in the nucleus and cytoplasm of MCF-7 cells. The expression significantly increased during As
2O
3-induced G2/M cell cycle arrest (
Figure 12f) and low expression was observed in As
2O
3-induced apoptosis (
Figure 12h). These data is consistent with the results from conventional PCR analysis of survivin 2B mRNA expression in MCF-7 cells (
Figure 11a). This known but survivin 2B may have significant role in these processes than initially thought. Survivin wild-type is multifunctional, it has been implicated in cell cycle arrest as well as in the inhibition of apoptosis. In this study only arsenic trioxide-induced G2M cell cycle arrest upregulated wild-type survivin while arsenic trioxide-induced-apoptosis downregulated wild-type survivin. Survivin wild-type upregulation during G2M cell cycle arrest has been reported to favour cancer cell survival [
39] and this may be the case with survivin 3α.