Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria monocytogenes
Abstract
:1. Introduction
2. Essential Yet Potentially Toxic Metals
3. Cadmium and Arsenic: Non-Essential Toxic Metals
4. Arsenic Resistance
5. Arsenic Resistance Determinants
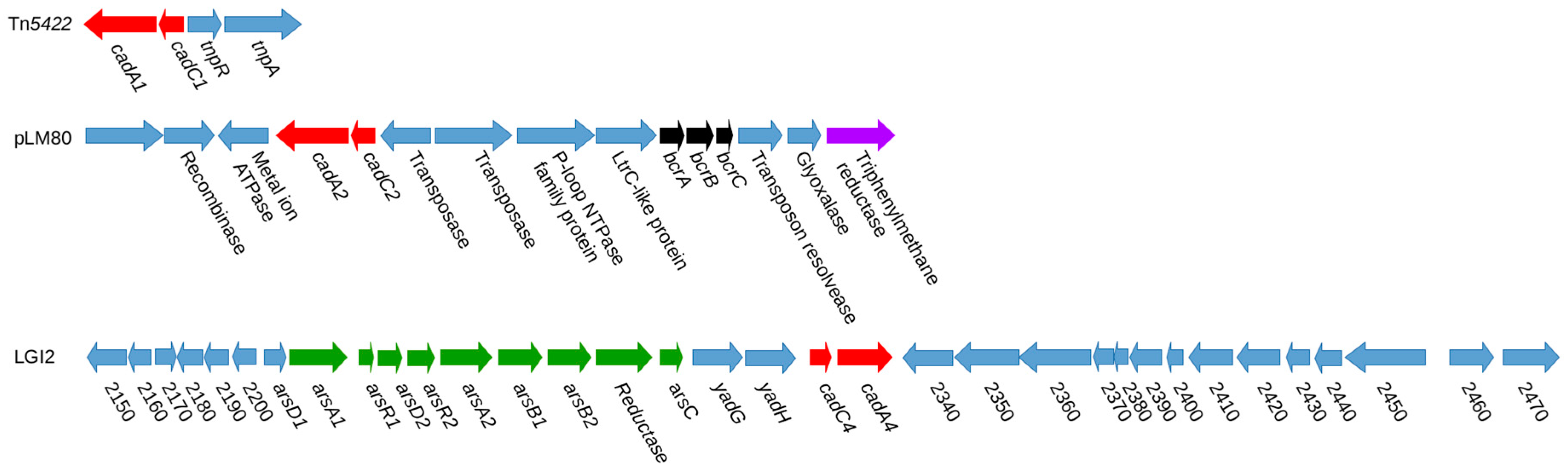
6. Cadmium Resistance
7. Plasmid-Associated Cadmium Resistance Determinants
8. Chromosomal Cadmium Resistance Determinants
9. Impacts of Heavy Metal Resistance Determinants on other Adaptations, Including Virulence
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Painter, J.; Slutsker, L. Listeriosis in humans. In Listeria, Listeriosis and Food Safety; CRC Press: Boca Raton, FL, USA, 2007; Volume 30, pp. 85–109. ISBN 978-0-8247-5750-2. [Google Scholar]
- Kathariou, S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Perrodeau, É.; Leclercq, A.; Cazenave, B.; Pilmis, B.; Henry, B.; Lopes, A.; Maury, M.M.; Moura, A.; Goffinet, F.; et al. Clinical features and prognostic factors of listeriosis: The MONALISA national prospective cohort study. Lancet Infect. Dis. 2017. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Chikindas, M.L. Listeria: A foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 2007, 113, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Freitag, N.E.; Boor, K.J. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 2006, 74, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes—From saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Jesse, H.E.; Roberts, I.S.; Cavet, J.S. Metal ion homeostasis in Listeria monocytogenes and importance in host-pathogen interactions. Adv. Microb. Physiol. 2014, 65, 83–123. [Google Scholar] [CrossRef]
- Alloway, B.J. Heavy Metals in Soils; Springer: Dordrecht, The Netherlands, 2013; p. 614. [Google Scholar] [CrossRef]
- Nunes, I.; Jacquiod, S.; Brejnrod, A.; Holm, P.E.; Brandt, K.K.; Priemé, A.; Sørensen, S.J. Coping with copper: Legacy effect of copper on potential activity of soil bacteria following a century of exposure. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef]
- Lindh, S.; Razmara, P.; Bogart, S.; Pyle, G. Comparative tissue distribution and depuration characteristics of copper nanoparticles and soluble copper in rainbow trout (Oncorhynchus mykiss). Environ. Toxicol. Chem. 2018. [Google Scholar] [CrossRef]
- Jordanova, M.; Hristovski, S.; Musai, M.; Boškovska, V.; Rebok, K.; Dinevska-Ќovkarovska, S.; Melovski, L. Accumulation of heavy metals in some organs in barbel and chub from Crn Drim River in the Republic of Macedonia. Bull. Environ. Contam. Toxicol. 2018, 101, 392–397. [Google Scholar] [CrossRef]
- Li, P.; Du, B.; Chan, H.M.; Feng, X.; Li, B. Mercury bioaccumulation and its toxic effects in rats fed with methylmercury polluted rice. Sci. Total Environ. 2018, 633, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Zalewski, P.; Truong-Tran, A.; Lincoln, S.; Ward, D.; Shankar, A.; Coyle, P.; Jayaram, L.; Copley, A.; Grosser, D.; Murgia, C.; et al. Use of a zinc fluorophore to measure labile pools of zinc in body fluids and cell-conditioned media. Biotechniques 2006, 40, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Carrigan, P.E.; Hentz, J.G.; Gordon, G.; Morgan, J.L.; Raimondo, M.; Anbar, A.D.; Miller, L.J. Distinctive heavy metal composition of pancreatic juice in patients with pancreatic carcinoma. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2656–2663. [Google Scholar] [CrossRef] [PubMed]
- White, C.; Lee, J.; Kambe, T.; Fritsche, K.; Petris, M.J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 2009, 284, 33949–33956. [Google Scholar] [CrossRef]
- Argüello, J.M.; Raimunda, D.; Padilla-Benavides, T. Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 2013, 3, 73. [Google Scholar] [CrossRef]
- McLaughlin, H.P.; Hill, C.; Gahan, C.G.M. The impact of iron on Listeria monocytogenes; inside and outside the host. Curr. Opin. Biotechnol. 2011, 22, 194–199. [Google Scholar] [CrossRef]
- Lechowicz, J.; Krawczyk-Balska, A. An update on the transport and metabolism of iron in Listeria monocytogenes: The role of proteins involved in pathogenicity. Biometals 2015, 28, 587–603. [Google Scholar] [CrossRef]
- Latorre, M.; Olivares, F.; Reyes-Jara, A.; López, G.; González, M. CutC is induced late during copper exposure and can modify intracellular copper content in Enterococcus faecalis. Biochem. Biophys. Res. Commun. 2011, 406, 633–637. [Google Scholar] [CrossRef]
- Pi, H.; Patel, S.J.; Argüello, J.M.; Helmann, J.D. The Listeria monocytogenes Fur-regulated virulence protein FrvA is an Fe(II) efflux P1B4-type ATPase. Mol. Microbiol. 2016, 100, 1066–1079. [Google Scholar] [CrossRef]
- McLaughlin, H.P.; Xiao, Q.; Rea, R.B.; Pi, H.; Casey, P.G.; Darby, T.; Charbit, A.; Sleator, R.D.; Joyce, S.A.; Cowart, R.E.; et al. A putative P-type ATPase required for virulence and resistance to haem toxicity in Listeria monocytogenes. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Santo, C.E.; Quaranta, D.; Grass, G. Antimicrobial metallic copper surfaces kill Staphylococcus haemolyticus via membrane damage. Microbiologyopen 2012, 1, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Senadheera, D.B.; Lévesque, C.M.; Cvitkovitch, D.G. The copYAZ operon functions in copper efflux, biofilm formation, genetic transformation, and stress tolerance in Streptococcus mutans. J. Bacteriol. 2015, 197, 2545–2557. [Google Scholar] [CrossRef]
- Yousuf, B.; Ahire, J.J.; Dicks, L.M.T. Understanding the antimicrobial activity behind thin- and thick-rolled copper plates. Appl. Microbiol. Biotechnol. 2016, 100, 5569–5580. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Costolo, B.; Brown, P.; Kathariou, S. Penicillin-binding protein encoded by pbp4 is involved in mediating copper stress in Listeria monocytogenes. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.L.; Klawitter, L.A.; Bhaduri, S.; Stahl, H.G. Arsenite resistance in Listeria monocytogenes. Food Microbiol. 1991, 8, 161–166. [Google Scholar] [CrossRef]
- Lebrun, M.; Loulergue, J.; Chaslus-Dancla, E.; Audurier, A. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl. Environ. Microbiol. 1992, 58, 3183–3186. [Google Scholar]
- McLauchlin, J.; Hampton, M.D.; Shah, S.; Threlfall, E.J.; Wieneke, A.A.; Curtis, G.D. Subtyping of Listeria monocytogenes on the basis of plasmid profiles and arsenic and cadmium susceptibility. J. Appl. Microbiol. 1997, 83, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ward, T.J.; Graves, L.M.; Tarr, C.L.; Siletzky, R.M.; Kathariou, S. Population structure of Listeria monocytogenes serotype 4b isolates from sporadic human listeriosis in the United States, 2003–2008. Appl. Environ. Microbiol. 2014, 80, AEM-00454. [Google Scholar] [CrossRef] [PubMed]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative genomics of Listeria species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Elhanafi, D.; Dutta, V.; Kathariou, S. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998–1999 outbreak. Appl. Environ. Microbiol. 2010, 76, 8231–8238. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.E.; Fouts, D.E.; Mongodin, E.F.; Ravel, J.; DeBoy, R.T.; Kolonay, J.F.; Rasko, D.A.; Angiuoli, S.V.; Gill, S.R.; Paulsen, I.T.; et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004, 32, 2386–2395. [Google Scholar] [CrossRef] [PubMed]
- Kuenne, C.; Voget, S.; Pischimarov, J.; Oehm, S.; Goesmann, A.; Daniel, R.; Hain, T.; Chakraborty, T. Comparative analysis of plasmids in the genus Listeria. PLoS ONE 2010, 5, e12511. [Google Scholar] [CrossRef] [PubMed]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Rakic-Martinez, M.; Graves, L.M.; Ward, T.J.; Siletzky, R.M.; Kathariou, S. Genetic determinants for cadmium and arsenic resistance among Listeria monocytogenes serotype 4b isolates from sporadic human listeriosis patients. Appl. Environ. Microbiol. 2013, 79, 2471–2476. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212. [Google Scholar] [CrossRef]
- Lebrun, M.; Audurier, A.; Cossart, P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J. Bacteriol. 1994, 176, 3040–3048. [Google Scholar] [CrossRef]
- Lee, S.; Ward, T.J.; Jima, D.D.; Parsons, C.; Kathariou, S. The arsenic resistance-associated Listeria genomic island LGI2 exhibits sequence and integration site diversity and a propensity for three Listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Cheng, Y.; Siletzky, R.M.; Kathariou, S. Genomic divisions/lineages, epidemic clones, and population structure. In Handbook of Listeria monocytogenes; CRC Press: Boca Raton, FL, USA, 2008; pp. 337–358. [Google Scholar]
- Pasquali, F.; Palma, F.; Guillier, L.; Lucchi, A.; De Cesare, A.; Manfreda, G. Listeria monocytogenes sequence types 121 and 14 repeatedly isolated within one year of sampling in a rabbit meat processing plant: Persistence and ecophysiology. Front. Microbiol. 2018, 9, 596. [Google Scholar] [CrossRef]
- Kaur, S.; Kamli, M.R.; Ali, A. Role of arsenic and its resistance in nature. Can. J. Microbiol. 2011, 57, 769–774. [Google Scholar] [CrossRef]
- Ordóñez, E.; Letek, M.; Valbuena, N.; Gil, J.A.; Mateos, L.M. Analysis of genes involved in arsenic resistance in Corynebacterium glutamicum ATCC 13032. Appl. Environ. Microbiol. 2005, 71, 6206–6215. [Google Scholar] [CrossRef]
- Tisa, L.S.; Rosen, B.P. Molecular characterization of an anion pump. The ArsB protein is the membrane anchor for the ArsA protein. J. Biol. Chem. 1990, 265, 190–194. [Google Scholar] [PubMed]
- López-Maury, L.; Florencio, F.J.; Reyes, J.C. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2003, 185, 5363–5371. [Google Scholar] [CrossRef] [PubMed]
- Gladysheva, T.B.; Oden, K.L.; Rosen, B.P. Properties of the arsenate reductase of plasmid R773. Biochemistry 1994, 33, 7288–7293. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Rosen, B.P. The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol. Microbiol. 1993, 8, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Klumpp, J.; Schuppler, M.; Loessner, M.J. Genome sequence of Listeria monocytogenes Scott A, a clinical isolate from a food-borne listeriosis outbreak. J. Bacteriol. 2011, 193, 4284–4285. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Lee, S.; Jayeola, V.; Kathariou, S. Novel cadmium resistance determinant in Listeria monocytogenes. Appl. Environ. Microbiol. 2017, 83, e02580-16. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.-H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016. [Google Scholar] [CrossRef]
- Lee, S.; Chen, Y.; Gorski, L.; Ward, T.J.; Osborne, J.; Kathariou, S. Listeria monocytogenes source distribution analysis indicates regional heterogeneity and ecological niche preference among serotype 4b clones. MBio 2018, 9. [Google Scholar] [CrossRef]
- Mullapudi, S.; Siletzky, R.M.; Kathariou, S. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 2008, 74, 1464–1468. [Google Scholar] [CrossRef]
- Ratani, S.S.; Siletzky, R.M.; Dutta, V.; Yildirim, S.; Osborne, J.A.; Lin, W.; Hitchins, A.D.; Ward, T.J.; Kathariou, S. Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl. Environ. Microbiol. 2012, 78, 6938–6945. [Google Scholar] [CrossRef]
- Xu, D.; Li, Y.; Zahid, M.S.H.; Yamasaki, S.; Shi, L.; Li, J.; Yan, H. Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int. J. Food Microbiol. 2014, 190, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.; Barbosa, J.; Stasiewicz, M.; Vongkamjan, K.; Moreno Switt, A.; Hogg, T.; Gibbs, P.; Teixeira, P.; Wiedmann, M. Diverse geno-and phenotypes of persistent Listeria monocytogenes isolates from fermented meat sausage production facilities in Portugal. Appl. Environ. Microbiol. 2011, 77, 2701–2715. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.; Gilmour, A. Characterization of recurrent and sporadic Listeria monocytogenes isolates from raw milk and nondairy foods by pulsed-field gel electrophoresis, monocin typing, plasmid profiling, and cadmium and antibiotic resistance determination. Appl. Environ. Microbiol. 2001, 67, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, M.; Audurier, A.; Cossart, P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J. Bacteriol. 1994, 176, 3049–3061. [Google Scholar] [CrossRef] [PubMed]
- Canchaya, C.; Giubellini, V.; Ventura, M.; De Los Reyes-Gavilán, C.G.; Margolles, A. Mosaic-Like sequences containing transposon, phage, and plasmid elements among Listeria monocytogenes plasmids. Appl. Environ. Microbiol. 2010, 76, 4851–4857. [Google Scholar] [CrossRef] [PubMed]
- Dutta, V.; Elhanaf, D.; Kathariou, S. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Dutta, V.; Elhanafi, D.; Osborne, J.; Martinez, M.R.; Kathariou, S. Genetic characterization of plasmid-associated triphenylmethane reductase in Listeria monocytogenes. Appl. Environ. Microbiol. 2014, 80, 5379–5385. [Google Scholar] [CrossRef]
- Katharios-Lanwermeyer, S.; Rakic-Martinez, M.; Elhanafi, D.; Ratani, S.; Tiedje, J.M.; Kathariou, S. Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other listeriae. Appl. Environ. Microbiol. 2012, 78, 7549–7556. [Google Scholar] [CrossRef]
- Mullapudi, S.; Siletzky, R.M.; Kathariou, S. Diverse cadmium resistance determinants in Listeria monocytogenes isolates from the turkey processing plant environment. Appl. Environ. Microbiol. 2010, 76, 627–630. [Google Scholar] [CrossRef]
- Lawrence, C. Surface-Active Quaternary Ammonium Germicides; Academic Press Inc. Publishers: New York, NY, USA, 1950. [Google Scholar]
- Qiagen CLC Genomics Workbench. Available online: https://www.qiagenbioinformatics.com (accessed on 17 December 2018).
- Camejo, A.; Buchrieser, C.; Couvé, E.; Carvalho, F.; Reis, O.; Ferreira, P.; Sousa, S.; Cossart, P.; Cabanes, D. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009, 5, e1000449. [Google Scholar] [CrossRef]
- Pombinho, R.; Camejo, A.; Vieira, A.; Reis, O.; Carvalho, F.; Almeida, M.T.; Pinheiro, J.C.; Sousa, S.; Cabanes, D. Listeria monocytogenes CadC regulates cadmium efflux and fine-tunes lipoprotein localization to escape the host immune response and promote infection. J. Infect. Dis. 2017, 215, 1468–1479. [Google Scholar] [CrossRef] [PubMed]
- Osman, D.; Cavet, J.S. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat. Prod. Rep. 2010, 27, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.H.; Yang, M.; He, Z.G. An ArsR-like transcriptional factor recognizes a conserved sequence motif and positively regulates the expression of phoP in mycobacteria. Biochem. Biophys. Res. Commun. 2011, 411, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Farias, P.; Santo, C.E.; Branco, R.; Francisco, R.; Santos, S.; Hansen, L.; Sorensen, S.; Morais, P.V. Natural hot spots for gain of multiple resistances: Arsenic and antibiotic resistances in heterotrophic, aerobic bacteria from marine hydrothermal vent fields. Appl. Environ. Microbiol. 2015, 81, 2534–2543. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. At the Nexus of Antibiotics and Metals: The Impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 2017, 25, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Quan, Y.; Yang, S.; Guo, L.; Zhang, X.; Liu, S.; Chen, S.; Zhou, K.; He, L.; Li, B.; et al. Antibiotic resistance in Salmonella from retail foods of animal origin and its association with disinfectant and heavy metal resistance. Microb. Drug Resist. 2018, 24, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Billman-Jacobe, H.; Liu, Y.; Haites, R.; Weaver, T.; Robinson, L.; Marenda, M.; Dyall-Smith, M. pSTM6-275, a conjugative IncHI2 plasmid of Salmonella enterica that confers antibiotic and heavy-metal resistance under changing physiological conditions. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Gullberg, E.; Albrecht, L.M.; Karlsson, C.; Sandegren, L.; Andersson, D.I. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio 2014, 5, e01918-14. [Google Scholar] [CrossRef]
- Binepal, G.; Gill, K.; Crowley, P.; Cordova, M.; Brady, L.J.; Senadheera, D.B.; Cvitkovitch, D.G. Trk2 Potassium transport system in Streptococcus mutans and its role in potassium homeostasis, biofilm formation, and stress tolerance. J. Bacteriol. 2016, 198, 1087–1100. [Google Scholar] [CrossRef]

| Metal Resistance-Associated Determinant | Annotation |
|---|---|
| arsA | Arsenic efflux ATPase [37] |
| arsB | Membrane transporter [37] |
| arsC | Arsenate reductase [37] |
| arsD | Transcriptional regulator [37] |
| arsR | Transcriptional regulator [37] |
| cadA1 | Cadmium efflux ATPase [38] |
| cadC1 | Transcriptional regulator [38] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parsons, C.; Lee, S.; Kathariou, S. Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria monocytogenes. Genes 2019, 10, 11. https://doi.org/10.3390/genes10010011
Parsons C, Lee S, Kathariou S. Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria monocytogenes. Genes. 2019; 10(1):11. https://doi.org/10.3390/genes10010011
Chicago/Turabian StyleParsons, Cameron, Sangmi Lee, and Sophia Kathariou. 2019. "Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria monocytogenes" Genes 10, no. 1: 11. https://doi.org/10.3390/genes10010011
APA StyleParsons, C., Lee, S., & Kathariou, S. (2019). Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria monocytogenes. Genes, 10(1), 11. https://doi.org/10.3390/genes10010011






