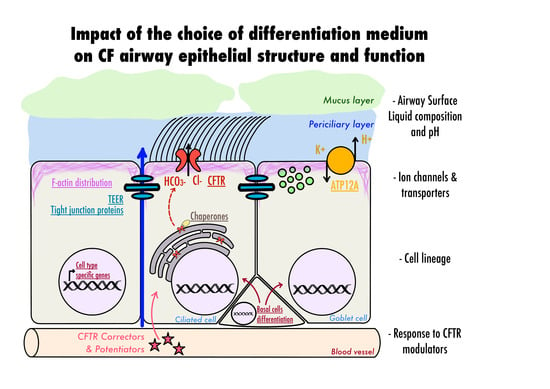
Choice of Differentiation Media Significantly Impacts Cell Lineage and Response to CFTR Modulators in Fully Differentiated Primary Cultures of Cystic Fibrosis Human Airway Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
Solutions
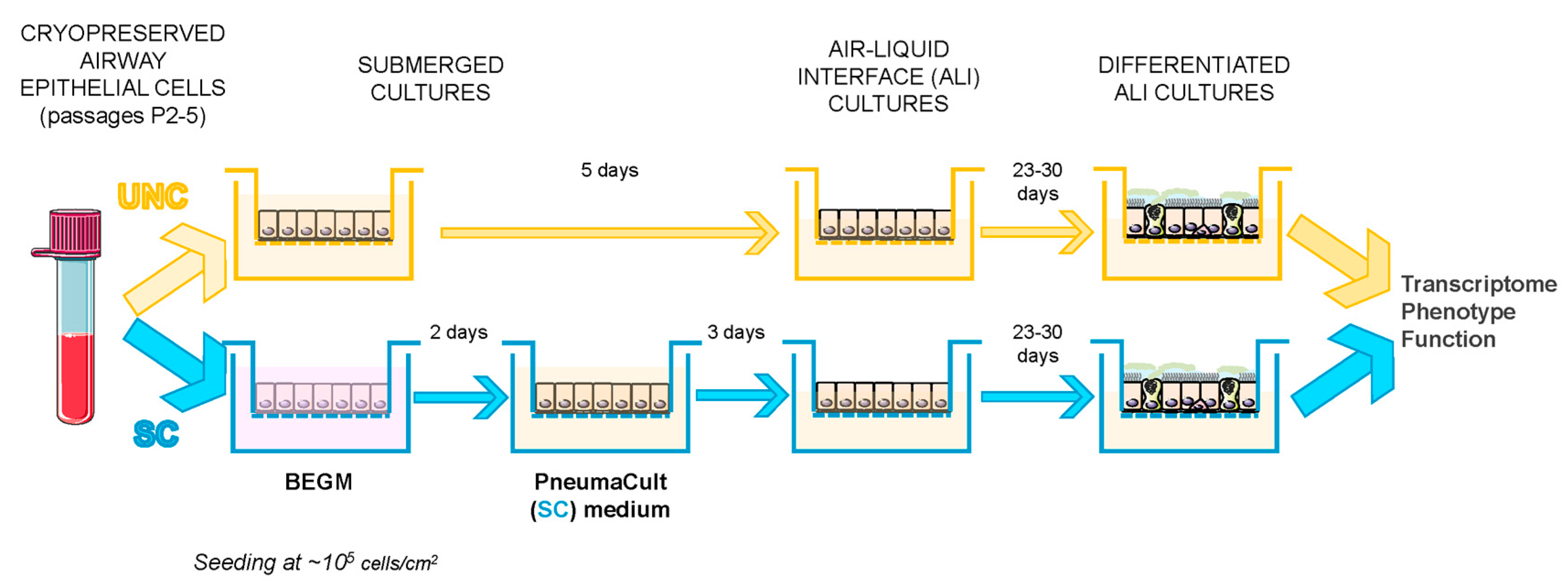
2.2. Cell Growth and Differentiation Protocol
2.3. RNA Extraction, RNA-Sequencing, and Real-Time Quantitative PCR Analysis
2.4. Protein Extraction, Quantification, and Western Blot (WB)
2.5. Histological and Immunofluorescence Staining
2.5.1. Histology
2.5.2. Immunofluorescence Staining
2.6. Intracellular pH
2.7. Short-Circuit Current Measurements in Ussing Chamber
2.8. ASL pH
2.9. Data Analysis
2.9.1. RNA-Sequencing Analysis
2.9.2. Airway Epithelial Cell Subtype Identification
2.9.3. Patterns of Gene Expression and Signaling Pathways Using NetworkAnalyst
2.9.4. Statistical Analysis of Other Data
3. Results
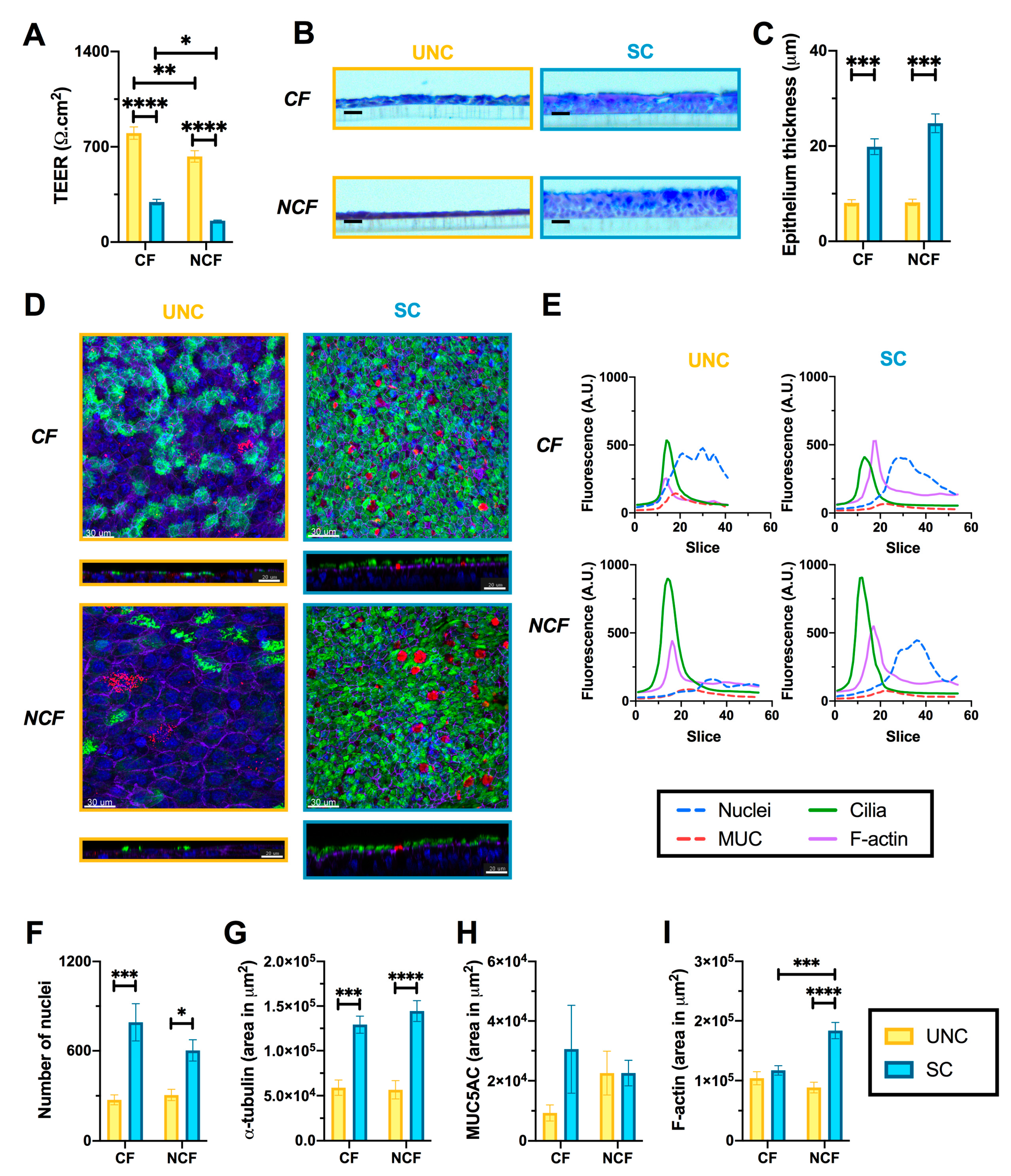
3.1. UNC and SC Media Induce Distinct Epithelial Phenotypes in CF and NCF Epithelia
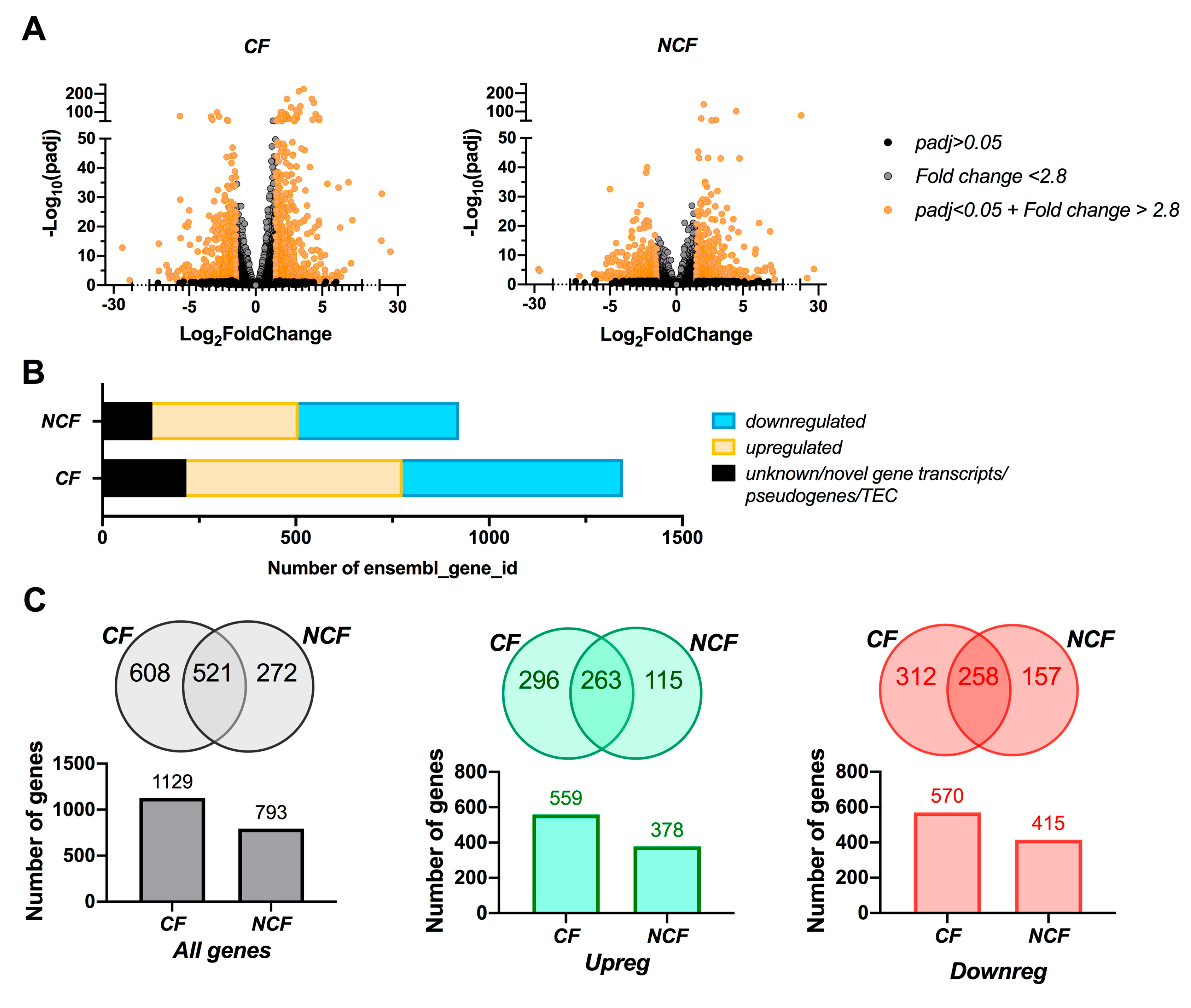
3.2. CF Cells are More Susceptible to Changes in Growth Media than NCF Cells
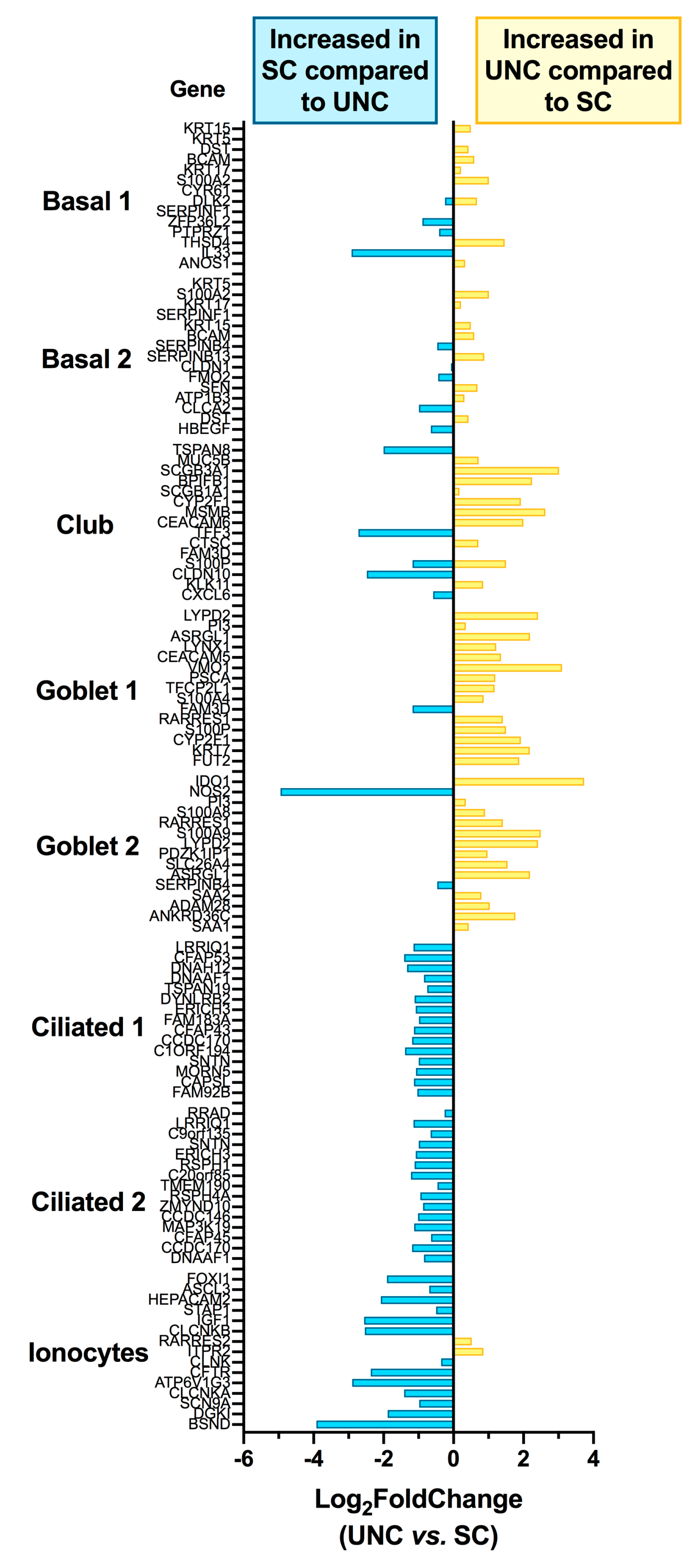
3.3. Growth Conditions Dictate Airway Epithelial Cell Identity
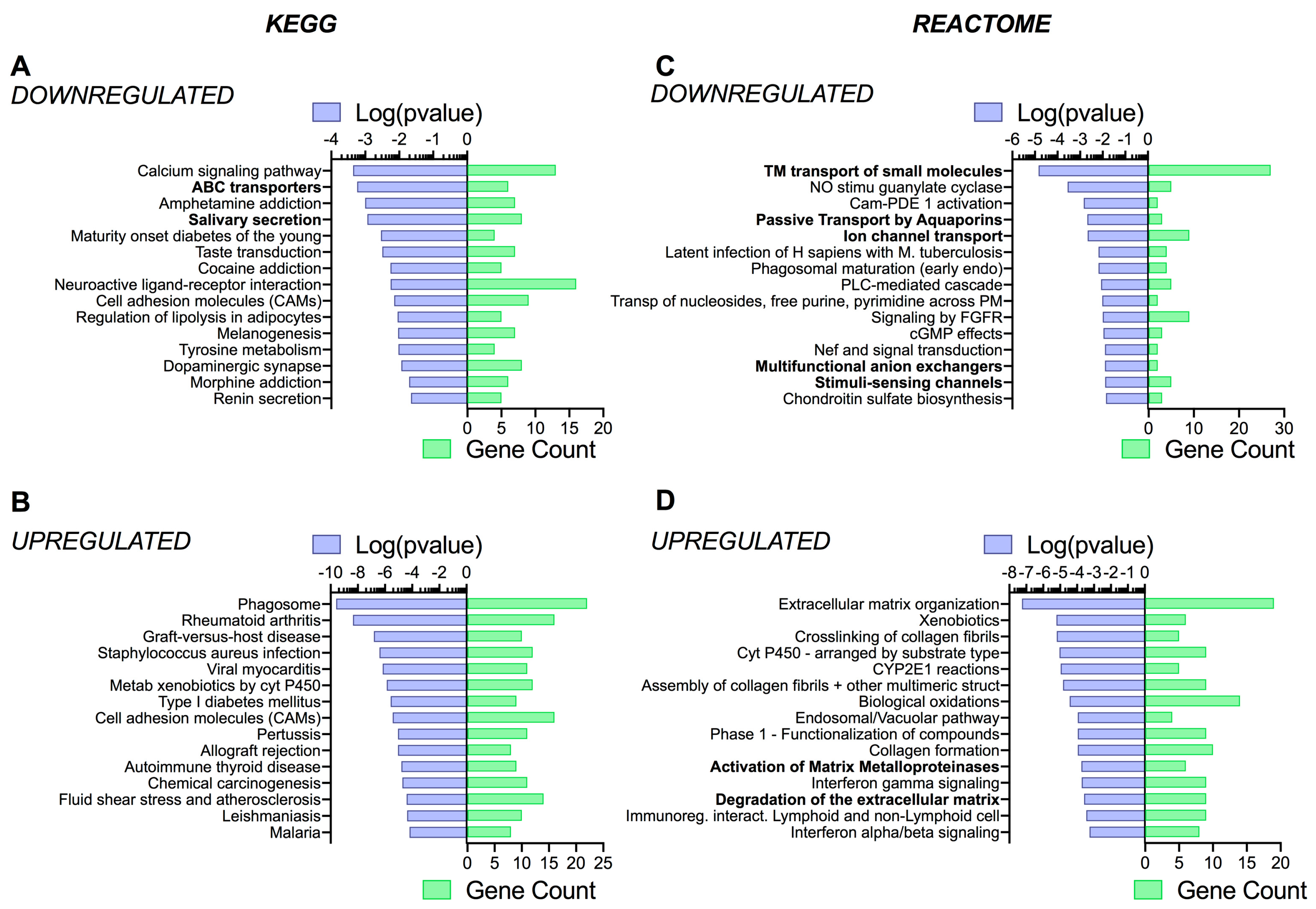
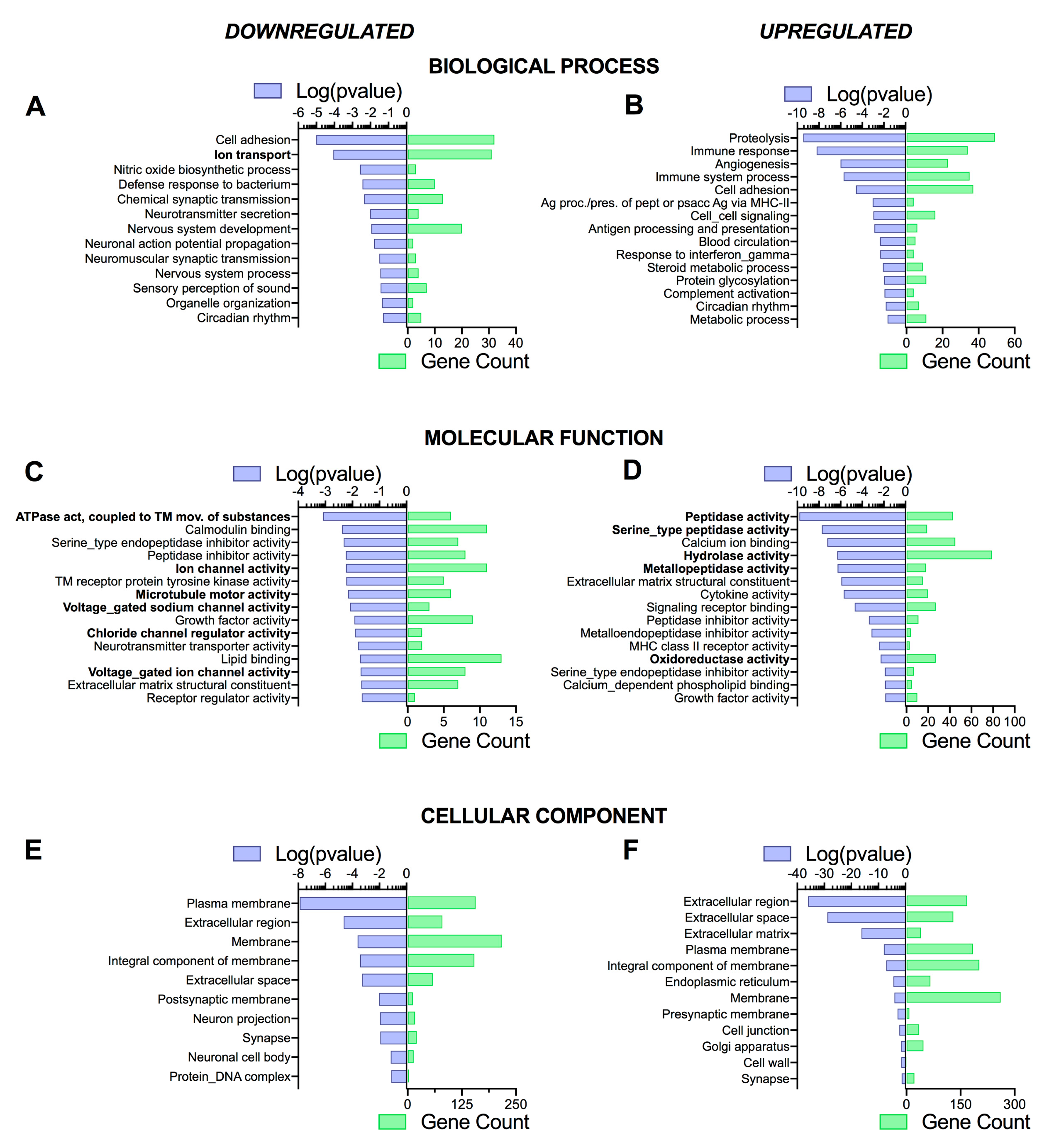
3.4. Pathway Analysis Reveals a Differentiation Medium Dependent Shift in Expression of Genes Involved in Ion and Fluid Transport Homeostasis
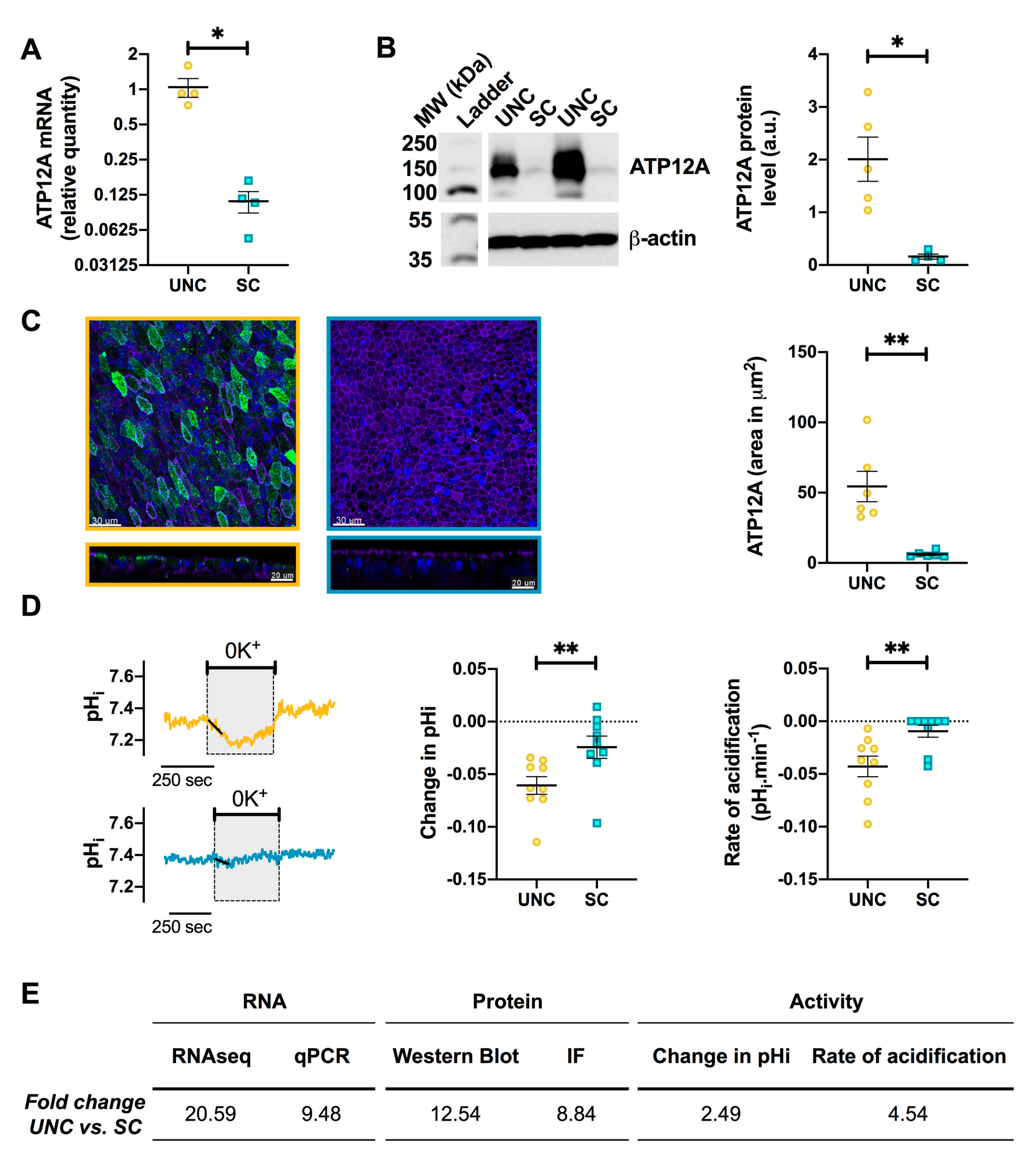
3.5. UNC-Grown CF Cells Express Higher Levels of ATP12A
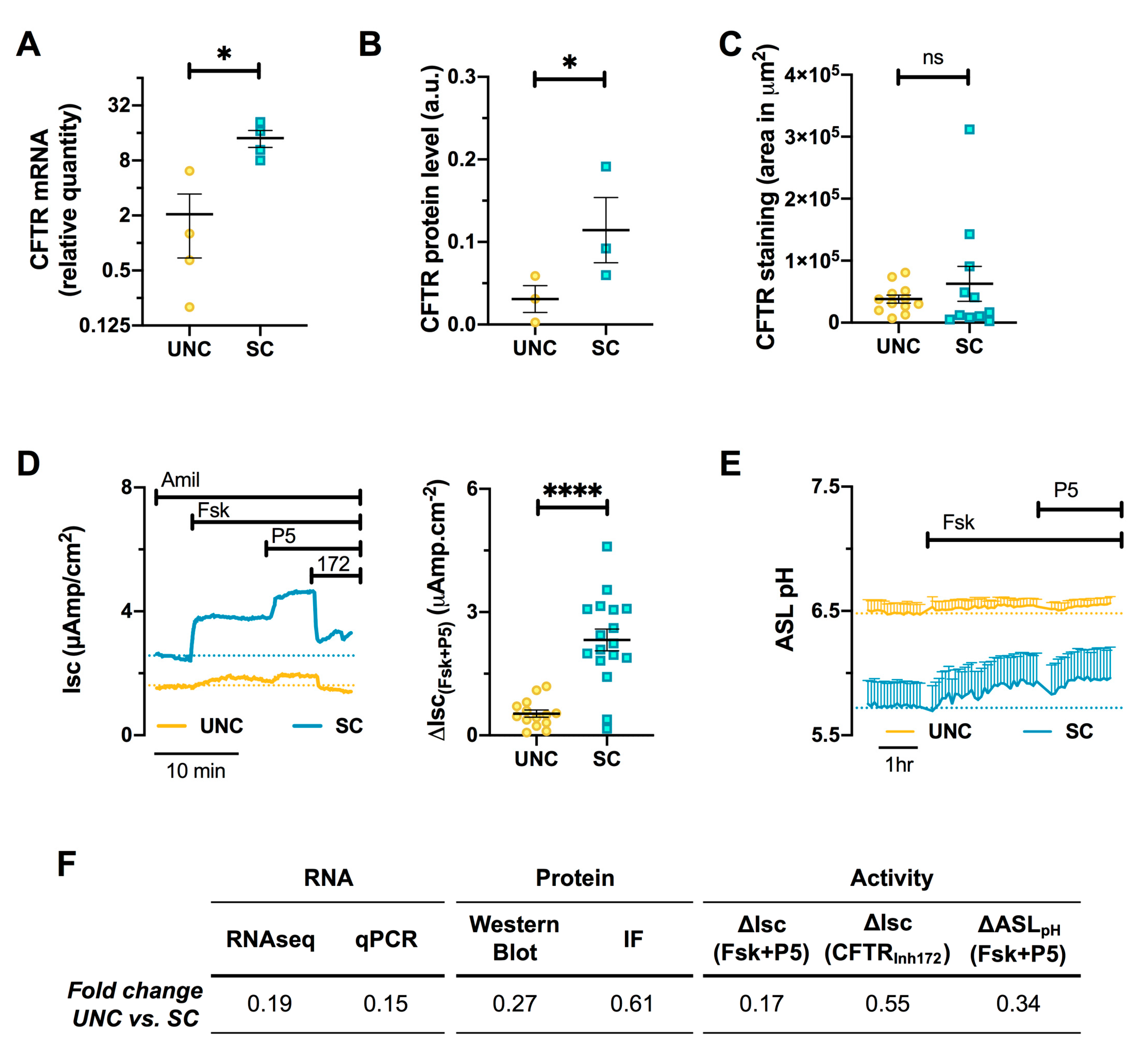
3.6. CFTR Expression and Function are Altered by Differentiation Media in CF Cells
3.7. Differentiation Media Modulate CF Cells Response to CFTR Modulators
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Franks, T.J.; Colby, T.V.; Travis, W.D.; Tuder, R.M.; Reynolds, H.Y.; Brody, A.R.; Cardoso, W.V.; Crystal, R.G.; Drake, C.J.; Engelhardt, J.; et al. Resident cellular components of the human lung: Current knowledge and goals for research on cell phenotyping and function. Proc. Am. Thorac. Soc. 2008, 5, 763–766. [Google Scholar] [CrossRef]
- Vieira Braga, F.A.; Kar, G.; Berg, M.; Carpaij, O.A.; Polanski, K.; Simon, L.M.; Brouwer, S.; Gomes, T.; Hesse, L.; Jiang, J.; et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019, 25, 1153–1163. [Google Scholar] [CrossRef] [Green Version]
- Whitsett, J.A. Airway Epithelial Differentiation and Mucociliary Clearance. Ann. Am. Thorac. Soc. 2018, 15, S143–S148. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.R.; Hogan, B.L.M. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 2011, 27, 493–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plasschaert, L.W.; Žilionis, R.; Choo-Wing, R.; Savova, V.; Knehr, J.; Roma, G.; Klein, A.M.; Jaffe, A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018, 560, 377–381. [Google Scholar] [CrossRef]
- Brewington, J.J.; Filbrandt, E.T.; LaRosa, F.J.; Moncivaiz, J.D.; Ostmann, A.J.; Strecker, L.M.; Clancy, J.P. Brushed nasal epithelial cells are a surrogate for bronchial epithelial CFTR studies. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyay, S.; Palmberg, L. Air-Liquid Interface: Relevant In Vitro Models for Investigating Air Pollutant-Induced Pulmonary Toxicity. Toxicol. Sci. 2018, 164, 21–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiemstra, P.S.; Tetley, T.D.; Janes, S.M. Airway and alveolar epithelial cells in culture. Eur. Respir. J. 2019, 54. [Google Scholar] [CrossRef]
- Lechner, J.F.; Haugen, A.; Autrup, H.; McClendon, I.A.; Trump, B.F.; Harris, C.C. Clonal growth of epithelial cells from normal adult human bronchus. Cancer Res. 1981, 41, 2294–2304. [Google Scholar]
- Whitcutt, M.J.; Adler, K.B.; Wu, R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell. Dev. Biol. 1988, 24, 420–428. [Google Scholar] [CrossRef]
- Gruenert, D.C.; Finkbeiner, W.E.; Widdicombe, J.H. Culture and transformation of human airway epithelial cells. Am. J. Physiol. 1995, 268, L347–L360. [Google Scholar] [CrossRef]
- Pezzulo, A.A.; Starner, T.D.; Scheetz, T.E.; Traver, G.L.; Tilley, A.E.; Harvey, B.-G.; Crystal, R.G.; McCray, P.B.; Zabner, J. The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. Am. J. Physiol. Lung Cell Mol. Physiol. 2011, 300, L25–L31. [Google Scholar] [CrossRef] [Green Version]
- Dvorak, A.; Tilley, A.E.; Shaykhiev, R.; Wang, R.; Crystal, R.G. Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am. J. Respir. Cell Mol. Biol. 2011, 44, 465–473. [Google Scholar] [CrossRef] [Green Version]
- de Jong, P.M.; van Sterkenburg, M.A.; Hesseling, S.C.; Kempenaar, J.A.; Mulder, A.A.; Mommaas, A.M.; Dijkman, J.H.; Ponec, M. Ciliogenesis in human bronchial epithelial cells cultured at the air-liquid interface. Am. J. Respir. Cell Mol. Biol. 1994, 10, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Suprynowicz, F.A.; Upadhyay, G.; Krawczyk, E.; Kramer, S.C.; Hebert, J.D.; Liu, X.; Yuan, H.; Cheluvaraju, C.; Clapp, P.W.; Boucher, R.C.; et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20035–20040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentzsch, M.; Boyles, S.E.; Cheluvaraju, C.; Chaudhry, I.G.; Quinney, N.L.; Cho, C.; Dang, H.; Liu, X.; Schlegel, R.; Randell, S.H. Pharmacological Rescue of Conditionally Reprogrammed Cystic Fibrosis Bronchial Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2017, 56, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Barkauskas, C.E.; Chung, M.-I.; Fioret, B.; Gao, X.; Katsura, H.; Hogan, B.L.M. Lung organoids: Current uses and future promise. Development 2017, 144, 986–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellani, S.; Di Gioia, S.; di Toma, L.; Conese, M. Human Cellular Models for the Investigation of Lung Inflammation and Mucus Production in Cystic Fibrosis. Anal. Cell. Pathol. (Amst.) 2018, 2018, 3839803. [Google Scholar] [CrossRef] [Green Version]
- Peters-Hall, J.R.; Coquelin, M.L.; Torres, M.J.; LaRanger, R.; Alabi, B.R.; Sho, S.; Calva-Moreno, J.F.; Thomas, P.J.; Shay, J.W. Long-term culture and cloning of primary human bronchial basal cells that maintain multipotent differentiation capacity and CFTR channel function. Am. J. Physiol. Lung Cell Mol. Physiol. 2018, 315, L313–L327. [Google Scholar] [CrossRef]
- Fulcher, M.L.; Randell, S.H. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 2013, 945, 109–121. [Google Scholar] [CrossRef]
- Gianotti, A.; Delpiano, L.; Caci, E. In vitro Methods for the Development and Analysis of Human Primary Airway Epithelia. Front. Pharmacol. 2018, 9, 1176. [Google Scholar] [CrossRef] [Green Version]
- Müller, L.; Brighton, L.E.; Carson, J.L.; Fischer, W.A.; Jaspers, I. Culturing of human nasal epithelial cells at the air liquid interface. J. Vis. Exp. 2013. [Google Scholar] [CrossRef] [PubMed]
- Neuberger, T.; Burton, B.; Clark, H.; Van Goor, F. Use of primary cultures of human bronchial epithelial cells isolated from cystic fibrosis patients for the pre-clinical testing of CFTR modulators. Methods Mol. Biol. 2011, 741, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Schaefer, N.; Chu, H.W. Air-Liquid Interface Culture of Human and Mouse Airway Epithelial Cells. Methods Mol. Biol. 2018, 1809, 91–109. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, M.L.; Gabriel, S.; Burns, K.A.; Yankaskas, J.R.; Randell, S.H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005, 107, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.E.; Torr, E.E.; Mohd Jamili, N.H.; Bosquillon, C.; Sayers, I. Evaluation of differentiated human bronchial epithelial cell culture systems for asthma research. J. Allergy (Cairo) 2012, 2012, 943982. [Google Scholar] [CrossRef] [Green Version]
- Yonker, L.M.; Mou, H.; Chu, K.K.; Pazos, M.A.; Leung, H.; Cui, D.; Ryu, J.; Hibbler, R.M.; Eaton, A.D.; Ford, T.N.; et al. Development of a Primary Human Co-Culture Model of Inflamed Airway Mucosa. Sci. Rep. 2017, 7, 8182. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.-K.; Yoo, J.-W.; Lin, H.; Kim, Y.-S.; Kim, D.-D.; Choi, Y.-M.; Park, S.-K.; Lee, C.-H.; Roh, H.-J. Air-liquid interface culture of serially passaged human nasal epithelial cell monolayer for in vitro drug transport studies. Drug Deliv. 2005, 12, 305–311. [Google Scholar] [CrossRef] [Green Version]
- Mathis, C.; Poussin, C.; Weisensee, D.; Gebel, S.; Hengstermann, A.; Sewer, A.; Belcastro, V.; Xiang, Y.; Ansari, S.; Wagner, S.; et al. Human bronchial epithelial cells exposed in vitro to cigarette smoke at the air-liquid interface resemble bronchial epithelium from human smokers. Am. J. Physiol. Lung Cell Mol. Physiol. 2013, 304, L489–L503. [Google Scholar] [CrossRef]
- Azzopardi, D.; Haswell, L.E.; Foss-Smith, G.; Hewitt, K.; Asquith, N.; Corke, S.; Phillips, G. Evaluation of an air-liquid interface cell culture model for studies on the inflammatory and cytotoxic responses to tobacco smoke aerosols. Toxicol. In Vitro 2015, 29, 1720–1728. [Google Scholar] [CrossRef] [Green Version]
- Gianotti, A.; Capurro, V.; Delpiano, L.; Mielczarek, M.; García-Valverde, M.; Carreira-Barral, I.; Ludovico, A.; Fiore, M.; Baroni, D.; Moran, O.; et al. Small Molecule Anion Carriers Correct Abnormal Airway Surface Liquid Properties in Cystic Fibrosis Airway Epithelia. Int. J. Mol. Sci. 2020, 21, 1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Bukreyev, A.; Thompson, C.I.; Watson, B.; Peeples, M.E.; Collins, P.L.; Pickles, R.J. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 2005, 79, 1113–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caves, E.A.; Cook, S.A.; Lee, N.; Stoltz, D.; Watkins, S.; Shair, K.H.Y. Air-Liquid Interface Method to Study Epstein-Barr Virus Pathogenesis in Nasopharyngeal Epithelial Cells. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgener, E.B.; Moss, R.B. Cystic fibrosis transmembrane conductance regulator modulators: Precision medicine in cystic fibrosis. Curr. Opin. Pediatr. 2018, 30, 372–377. [Google Scholar] [CrossRef]
- Lin, H.; Li, H.; Cho, H.-J.; Bian, S.; Roh, H.-J.; Lee, M.-K.; Kim, J.S.; Chung, S.-J.; Shim, C.-K.; Kim, D.-D. Air-liquid interface (ALI) culture of human bronchial epithelial cell monolayers as an in vitro model for airway drug transport studies. J. Pharm. Sci. 2007, 96, 341–350. [Google Scholar] [CrossRef]
- Wainwright, C.E.; Elborn, J.S.; Ramsey, B.W.; Marigowda, G.; Huang, X.; Cipolli, M.; Colombo, C.; Davies, J.C.; De Boeck, K.; Flume, P.A.; et al. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N. Engl. J. Med. 2015, 373, 220–231. [Google Scholar] [CrossRef] [Green Version]
- Awatade, N.T.; Uliyakina, I.; Farinha, C.M.; Clarke, L.A.; Mendes, K.; Solé, A.; Pastor, J.; Ramos, M.M.; Amaral, M.D. Measurements of Functional Responses in Human Primary Lung Cells as a Basis for Personalized Therapy for Cystic Fibrosis. EBioMedicine 2015, 2, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Cao, D.; Neuberger, T.; Turnbull, A.; Singh, A.; Joubran, J.; Hazlewood, A.; et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. USA 2009, 106, 18825–18830. [Google Scholar] [CrossRef] [Green Version]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.J.; Burton, B.; Stack, J.H.; Straley, K.S.; Decker, C.J.; Miller, M.; McCartney, J.; Olson, E.R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 2011, 108, 18843–18848. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.L.; Saint-Criq, V.; Hwang, T.-C.; Csanády, L. Ion channels as targets to treat cystic fibrosis lung disease. J. Cyst. Fibros. 2018, 17, S22–S27. [Google Scholar] [CrossRef] [Green Version]
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- Saint-Criq, V.; Gray, M.A. Role of CFTR in epithelial physiology. Cell. Mol. Life Sci. 2017, 74, 93–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haq, I.J.; Gray, M.A.; Garnett, J.P.; Ward, C.; Brodlie, M. Airway surface liquid homeostasis in cystic fibrosis: Pathophysiology and therapeutic targets. Thorax 2016, 71, 284–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.X.; Ostedgaard, L.S.; Hoegger, M.J.; Moninger, T.O.; Karp, P.H.; McMenimen, J.D.; Choudhury, B.; Varki, A.; Stoltz, D.A.; Welsh, M.J. Acidic pH increases airway surface liquid viscosity in cystic fibrosis. J. Clin. Investig. 2016, 126, 879–891. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, G.; Koch, W.; Ritter, D.; Gutleb, A.C.; Larsen, S.T.; Loret, T.; Zanetti, F.; Constant, S.; Chortarea, S.; Rothen-Rutishauser, B.; et al. Air–Liquid Interface In Vitro Models for Respiratory Toxicology Research: Consensus Workshop and Recommendations. Appl. In Vitro Toxicol. 2018, 4, 91–106. [Google Scholar] [CrossRef] [Green Version]
- Ruiz García, S.; Deprez, M.; Lebrigand, K.; Cavard, A.; Paquet, A.; Arguel, M.-J.; Magnone, V.; Truchi, M.; Caballero, I.; Leroy, S.; et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Rayner, R.E.; Makena, P.; Prasad, G.L.; Cormet-Boyaka, E. Optimization of Normal Human Bronchial Epithelial (NHBE) Cell 3D Cultures for in vitro Lung Model Studies. Sci. Rep. 2019, 9, 500. [Google Scholar] [CrossRef]
- Lee, D.D.H.; Petris, A.; Hynds, R.E.; O’Callaghan, C. Ciliated Epithelial Cell Differentiation at Air-Liquid Interface Using Commercially Available Culture Media. Methods Mol. Biol. 2020, 2109, 275–291. [Google Scholar] [CrossRef]
- Randell, S.H.; Fulcher, M.L.; O’Neal, W.; Olsen, J.C. Primary epithelial cell models for cystic fibrosis research. Methods Mol. Biol. 2011, 742, 285–310. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Turner, M.J.; Saint-Criq, V.; Patel, W.; Ibrahim, S.H.; Verdon, B.; Ward, C.; Garnett, J.P.; Tarran, R.; Cann, M.J.; Gray, M.A. Hypercapnia modulates cAMP signalling and cystic fibrosis transmembrane conductance regulator-dependent anion and fluid secretion in airway epithelia. J. Physiol. (Lond.) 2016, 594, 1643–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saint-Criq, V.; Kim, S.H.; Katzenellenbogen, J.A.; Harvey, B.J. Non-genomic estrogen regulation of ion transport and airway surface liquid dynamics in cystic fibrosis bronchial epithelium. PLoS ONE 2013, 8, e78593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saint-Criq, V.; Haq, I.J.; Gardner, A.I.; Garnett, J.P.; Ward, C.; Brodlie, M.; Gray, M.A. Real-Time, Semi-Automated Fluorescent Measurement of the Airway Surface Liquid pH of Primary Human Airway Epithelial Cells. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Benner, M.J.; Hancock, R.E.W. NetworkAnalyst--integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, W167–W174. [Google Scholar] [CrossRef] [Green Version]
- Shah, V.S.; Meyerholz, D.K.; Tang, X.X.; Reznikov, L.; Abou Alaiwa, M.; Ernst, S.E.; Karp, P.H.; Wohlford-Lenane, C.L.; Heilmann, K.P.; Leidinger, M.R.; et al. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science 2016, 351, 503–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delpiano, L.; Thomas, J.J.; Yates, A.R.; Rice, S.J.; Gray, M.A.; Saint-Criq, V. Esomeprazole Increases Airway Surface Liquid pH in Primary Cystic Fibrosis Epithelial Cells. Front. Pharmacol. 2018, 9, 1462. [Google Scholar] [CrossRef]
- Tarran, R.; Button, B.; Picher, M.; Paradiso, A.M.; Ribeiro, C.M.; Lazarowski, E.R.; Zhang, L.; Collins, P.L.; Pickles, R.J.; Fredberg, J.J.; et al. Normal and cystic fibrosis airway surface liquid homeostasis. The effects of phasic shear stress and viral infections. J. Biol. Chem. 2005, 280, 35751–35759. [Google Scholar] [CrossRef] [Green Version]
- Ferrera, L.; Baroni, D.; Moran, O. Lumacaftor-rescued F508del-CFTR has a modified bicarbonate permeability. J. Cyst. Fibros. 2019, 18, 602–605. [Google Scholar] [CrossRef]
- Fiore, M.; Picco, C.; Moran, O. Correctors modify the bicarbonate permeability of F508del-CFTR. Sci. Rep. 2020, 10, 8440. [Google Scholar] [CrossRef]
- Schuldiner, M.; Yanuka, O.; Itskovitz-Eldor, J.; Melton, D.A.; Benvenisty, N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2000, 97, 11307–11312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, T.J.; Cardoso, W.V. Growth factors in lung development and disease: Friends or foe? Respir. Res. 2002, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Zhao, Y.H.; Chang, M.M. Growth and differentiation of conducting airway epithelial cells in culture. Eur. Respir. J. 1997, 10, 2398–2403. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Sarkar, R.R. Comparison of human cell signaling pathway databases—evolution, drawbacks and challenges. Database (Oxford) 2015, 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krug, S.M.; Günzel, D.; Conrad, M.P.; Lee, I.-F.M.; Amasheh, S.; Fromm, M.; Yu, A.S.L. Charge-selective claudin channels. Ann. N. Y. Acad. Sci. 2012, 1257, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Soini, Y. Claudins in lung diseases. Respir. Res. 2011, 12, 70. [Google Scholar] [CrossRef] [Green Version]
- Flynn, A.N.; Itani, O.A.; Moninger, T.O.; Welsh, M.J. Acute regulation of tight junction ion selectivity in human airway epithelia. Proc. Natl. Acad. Sci. USA 2009, 106, 3591–3596. [Google Scholar] [CrossRef] [Green Version]
- Shah, V.S.; Ernst, S.; Tang, X.X.; Karp, P.H.; Parker, C.P.; Ostedgaard, L.S.; Welsh, M.J. Relationships among CFTR expression, HCO3- secretion, and host defense may inform gene- and cell-based cystic fibrosis therapies. Proc. Natl. Acad. Sci. USA 2016, 113, 5382–5387. [Google Scholar] [CrossRef] [Green Version]
- Simonin, J.; Bille, E.; Crambert, G.; Noel, S.; Dreano, E.; Edwards, A.; Hatton, A.; Pranke, I.; Villeret, B.; Cottart, C.-H.; et al. Airway surface liquid acidification initiates host defense abnormalities in Cystic Fibrosis. Sci. Rep. 2019, 9, 6516. [Google Scholar] [CrossRef]
- Abou Alaiwa, M.H.; Beer, A.M.; Pezzulo, A.A.; Launspach, J.L.; Horan, R.A.; Stoltz, D.A.; Starner, T.D.; Welsh, M.J.; Zabner, J. Neonates with cystic fibrosis have a reduced nasal liquid pH; a small pilot study. J. Cyst. Fibros. 2014, 13, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Garland, A.L.; Walton, W.G.; Coakley, R.D.; Tan, C.D.; Gilmore, R.C.; Hobbs, C.A.; Tripathy, A.; Clunes, L.A.; Bencharit, S.; Stutts, M.J.; et al. Molecular basis for pH-dependent mucosal dehydration in cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 2013, 110, 15973–15978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abou Alaiwa, M.H.; Reznikov, L.R.; Gansemer, N.D.; Sheets, K.A.; Horswill, A.R.; Stoltz, D.A.; Zabner, J.; Welsh, M.J. pH modulates the activity and synergism of the airway surface liquid antimicrobials β-defensin-3 and LL-37. Proc. Natl. Acad. Sci. USA 2014, 111, 18703–18708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pezzulo, A.A.; Tang, X.X.; Hoegger, M.J.; Abou Alaiwa, M.H.; Ramachandran, S.; Moninger, T.O.; Karp, P.H.; Wohlford-Lenane, C.L.; Haagsman, H.P.; van Eijk, M.; et al. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature 2012, 487, 109–113. [Google Scholar] [CrossRef] [PubMed]
- McShane, D.; Davies, J.C.; Davies, M.G.; Bush, A.; Geddes, D.M.; Alton, E.W.F.W. Airway surface pH in subjects with cystic fibrosis. Eur. Respir. J. 2003, 21, 37–42. [Google Scholar] [CrossRef]
- Schultz, A.; Puvvadi, R.; Borisov, S.M.; Shaw, N.C.; Klimant, I.; Berry, L.J.; Montgomery, S.T.; Nguyen, T.; Kreda, S.M.; Kicic, A.; et al. Airway surface liquid pH is not acidic in children with cystic fibrosis. Nat. Commun. 2017, 8, 1409. [Google Scholar] [CrossRef] [Green Version]
- Coakley, R.D.; Grubb, B.R.; Paradiso, A.M.; Gatzy, J.T.; Johnson, L.G.; Kreda, S.M.; O’Neal, W.K.; Boucher, R.C. Abnormal surface liquid pH regulation by cultured cystic fibrosis bronchial epithelium. Proc. Natl. Acad. Sci. USA 2003, 100, 16083–16088. [Google Scholar] [CrossRef] [Green Version]
- Scudieri, P.; Musante, I.; Caci, E.; Venturini, A.; Morelli, P.; Walter, C.; Tosi, D.; Palleschi, A.; Martin-Vasallo, P.; Sermet-Gaudelus, I.; et al. Increased expression of ATP12A proton pump in cystic fibrosis airways. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [Green Version]
- Garnett, J.P.; Kalsi, K.K.; Sobotta, M.; Bearham, J.; Carr, G.; Powell, J.; Brodlie, M.; Ward, C.; Tarran, R.; Baines, D.L. Hyperglycaemia and Pseudomonas aeruginosa acidify cystic fibrosis airway surface liquid by elevating epithelial monocarboxylate transporter 2 dependent lactate-H+ secretion. Sci. Rep. 2016, 6, 37955. [Google Scholar] [CrossRef]
- Garnett, J.P.; Hickman, E.; Burrows, R.; Hegyi, P.; Tiszlavicz, L.; Cuthbert, A.W.; Fong, P.; Gray, M.A. Novel role for pendrin in orchestrating bicarbonate secretion in cystic fibrosis transmembrane conductance regulator (CFTR)-expressing airway serous cells. J. Biol. Chem. 2011, 286, 41069–41082. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Huang, J.; Billet, A.; Abu-Arish, A.; Goepp, J.; Matthes, E.; Tewfik, M.A.; Frenkiel, S.; Hanrahan, J.W. Pendrin Mediates Bicarbonate Secretion and Enhances Cystic Fibrosis Transmembrane Conductance Regulator Function in Airway Surface Epithelia. Am. J. Respir. Cell Mol. Biol. 2019, 60, 705–716. [Google Scholar] [CrossRef]
- Thornell, I.M.; Rehman, T.; Pezzulo, A.A.; Welsh, M.J. Paracellular bicarbonate flux across human cystic fibrosis airway epithelia tempers changes in airway surface liquid pH. J. Physiol. (Lond.) 2020. [Google Scholar] [CrossRef] [PubMed]
- Dudeja, P.K.; Hafez, N.; Tyagi, S.; Gailey, C.A.; Toofanfard, M.; Alrefai, W.A.; Nazir, T.M.; Ramaswamy, K.; Al-Bazzaz, F.J. Expression of the Na+/H+ and Cl-/HCO-3 exchanger isoforms in proximal and distal human airways. Am. J. Physiol. 1999, 276, L971–L978. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.I.; Tasca, A.; Haas, M.; Baltazar, G.; Harland, R.M.; Finkbeiner, W.E.; Walentek, P. Na+/H+ Exchangers Are Required for the Development and Function of Vertebrate Mucociliary Epithelia. Cells Tissues Organs (Print) 2018, 205, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, H.; Sonnentag, T.; Heinzmann, A.; Seidler, B.; Bachmann, O.; Vieillard-Baron, D.; Gregor, M.; Seidler, U. Differential expression and regulation of Na(+)/H(+) exchanger isoforms in rabbit parietal and mucous cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 281, G447–G458. [Google Scholar] [CrossRef] [Green Version]
- Monterisi, S.; Favia, M.; Guerra, L.; Cardone, R.A.; Marzulli, D.; Reshkin, S.J.; Casavola, V.; Zaccolo, M. CFTR regulation in human airway epithelial cells requires integrity of the actin cytoskeleton and compartmentalized cAMP and PKA activity. J. Cell. Sci. 2012, 125, 1106–1117. [Google Scholar] [CrossRef] [Green Version]
- Arndt, V.; Daniel, C.; Nastainczyk, W.; Alberti, S.; Höhfeld, J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol. Biol. Cell 2005, 16, 5891–5900. [Google Scholar] [CrossRef]
- Noh, S.H.; Gee, H.Y.; Kim, Y.; Piao, H.; Kim, J.; Kang, C.M.; Lee, G.; Mook-Jung, I.; Lee, Y.; Cho, J.W.; et al. Specific autophagy and ESCRT components participate in the unconventional secretion of CFTR. Autophagy 2018, 14, 1761–1778. [Google Scholar] [CrossRef] [Green Version]
- Pranke, I.M.; Sermet-Gaudelus, I. Biosynthesis of cystic fibrosis transmembrane conductance regulator. Int. J. Biochem. Cell Biol. 2014, 52, 26–38. [Google Scholar] [CrossRef]
- Estabrooks, S.; Brodsky, J.L. Regulation of CFTR Biogenesis by the Proteostatic Network and Pharmacological Modulators. Int. J. Mol. Sci. 2020, 21, 452. [Google Scholar] [CrossRef] [Green Version]
- Avella, M.; Loriol, C.; Boulukos, K.; Borgese, F.; Ehrenfeld, J. SLC26A9 stimulates CFTR expression and function in human bronchial cell lines. J. Cell. Physiol. 2011, 226, 212–223. [Google Scholar] [CrossRef]
- Sato, Y.; Thomas, D.Y.; Hanrahan, J.W. The anion transporter SLC26A9 localizes to tight junctions and is degraded by the proteasome when co-expressed with F508del-CFTR. J. Biol. Chem. 2019, 294, 18269–18284. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, C.A.; Mitra, S.; Mishra, S.K.; Wang, X.; Zhao, Y.; Pilewski, J.M.; Madden, D.R.; Frizzell, R.A. The CFTR trafficking mutation F508del inhibits the constitutive activity of SLC26A9. Am. J. Physiol. Lung Cell Mol. Physiol. 2017, 312, L912–L925. [Google Scholar] [CrossRef] [PubMed]
- Pedemonte, N.; Bertozzi, F.; Caci, E.; Sorana, F.; Di Fruscia, P.; Tomati, V.; Ferrera, L.; Rodríguez-Gimeno, A.; Berti, F.; Pesce, E.; et al. Discovery of a picomolar potency pharmacological corrector of the mutant CFTR chloride channel. Sci. Adv. 2020, 6, eaay9669. [Google Scholar] [CrossRef] [Green Version]
- Lopes-Pacheco, M.; Boinot, C.; Sabirzhanova, I.; Morales, M.M.; Guggino, W.B.; Cebotaru, L. Combination of Correctors Rescue ΔF508-CFTR by Reducing Its Association with Hsp40 and Hsp27. J. Biol. Chem. 2015, 290, 25636–25645. [Google Scholar] [CrossRef] [Green Version]
- Laselva, O.; Eckford, P.D.; Bartlett, C.; Ouyang, H.; Gunawardena, T.N.; Gonska, T.; Moraes, T.J.; Bear, C.E. Functional rescue of c.3846G>A (W1282X) in patient-derived nasal cultures achieved by inhibition of nonsense mediated decay and protein modulators with complementary mechanisms of action. J. Cyst. Fibros. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montoro, D.T.; Haber, A.L.; Biton, M.; Vinarsky, V.; Lin, B.; Birket, S.E.; Yuan, F.; Chen, S.; Leung, H.M.; Villoria, J.; et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018, 560, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Rock, J.R.; Gao, X.; Xue, Y.; Randell, S.H.; Kong, Y.-Y.; Hogan, B.L.M. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 2011, 8, 639–648. [Google Scholar] [CrossRef] [Green Version]
- Vermeer, P.D.; Panko, L.; Karp, P.; Lee, J.H.; Zabner, J. Differentiation of human airway epithelia is dependent on erbB2. Am. J. Physiol. Lung Cell Mol. Physiol. 2006, 291, L175–L180. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Target Gene | Primer | Sequence | Product Length |
|---|---|---|---|
| 18S rRNA | Forward | 5′-CTCTAGATAACCTCGGGCCG-3′ | 209 |
| Reverse | 5′-GTCGGGAGTGGGTAATTTGC-3′ | ||
| ATP12A | Forward | 5′-GGGGCACACTTGTTCATCTTCTGA-3′ | 128 |
| Reverse | 5′-GCAAAACATCAGTGAGCATCCTG-3′ | ||
| CFTR | Forward | 5′-AGGAGGCAGTCTGTCCTGAA-3′ | 237 |
| Reverse | 5′-CACTGCTGGTATGCTCTCCA-3′ |
| Protein Target | Manufacturer (Reference) | Species Raised in | Dilution/Concentration (WB/IF) |
|---|---|---|---|
| Primary Antibodies | |||
| β-actin | Sigma-Aldrich (A5441) | Mouse | 1:5000 (WB) |
| CFTR (Cystic Fibrosis transmembrane conductance regulator | CFF Therapeutics (596) | Mouse | 1:3000 (WB) |
| 1:250 (IF) | |||
| ATP12A | Sigma-Aldrich (HPA039526) | Rabbit | 1:1000 (WB) |
| 1:400 (IF) | |||
| MUC5AC | Abcam (ab3649, Cambridge, UK) | Mouse | 1:67 (IF) |
| Acetyl-α-Tubulin (Lys40) (D20G3) | Cell Signaling Technology (5335, London, UK) | Rabbit | 1:800 (IF) |
| Phalloidin (Alexa Fluor 647) | Thermo Fisher Scientific (A30107) | NA | 1.25 µg/mL |
| DAPI (4′,6-diamidino-2-phenylindole) | Sigma-Aldrich (D9542) | NA | 1:1000 |
| Secondary Antibodies | |||
| Anti-mouse Immunoglobulin (Ig)G, HorseRadish Peroxidase (HRP)-linked | Cell Signaling Technology (7076) | Horse | 1:5000 (WB) |
| Anti-rabbit IgG, HRP-linked | Cell Signaling Technology (7074) | Goat | 1:5000 (WB) |
| Precision Protein™ StrepTactin-HRP Conjugate | Bio-Rad (161038) | NA | 1:5000 (WB) |
| Anti-Rabbit IgG (Alexa Fluor 488) | Thermo Fisher Scientific (A11034) | Goat | 5 µg/mL (IF) |
| Anti-Mouse IgG (Alexa Fluor 594) | Thermo Fisher Scientific (A11032) | Goat | 2 µg/mL (IF) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saint-Criq, V.; Delpiano, L.; Casement, J.; Onuora, J.C.; Lin, J.; Gray, M.A. Choice of Differentiation Media Significantly Impacts Cell Lineage and Response to CFTR Modulators in Fully Differentiated Primary Cultures of Cystic Fibrosis Human Airway Epithelial Cells. Cells 2020, 9, 2137. https://doi.org/10.3390/cells9092137
Saint-Criq V, Delpiano L, Casement J, Onuora JC, Lin J, Gray MA. Choice of Differentiation Media Significantly Impacts Cell Lineage and Response to CFTR Modulators in Fully Differentiated Primary Cultures of Cystic Fibrosis Human Airway Epithelial Cells. Cells. 2020; 9(9):2137. https://doi.org/10.3390/cells9092137
Chicago/Turabian StyleSaint-Criq, Vinciane, Livia Delpiano, John Casement, Jennifer C. Onuora, JinHeng Lin, and Michael A. Gray. 2020. "Choice of Differentiation Media Significantly Impacts Cell Lineage and Response to CFTR Modulators in Fully Differentiated Primary Cultures of Cystic Fibrosis Human Airway Epithelial Cells" Cells 9, no. 9: 2137. https://doi.org/10.3390/cells9092137