IL-1β and TNF-α Modulation of Proliferated and Committed Myoblasts: IL-6 and COX-2-Derived Prostaglandins as Key Actors in the Mechanisms Involved
Abstract
:1. Introduction
2. Materials and Methods
2.1. C2C12 Culture and Pro-Inflammatory Stimulus
2.2. Assessment of Cell Proliferation by BrdU Incorporation
Treatment with Inhibitors
2.3. Measurements of Prostaglandin Release by ELISA
2.4. Cyclooxygenase-2 Protein Expression by Western Blotting
2.5. Cyclooxygenase-2 Gene Expression by Quantitative PCR (qPCR)
2.6. Expression of Myogenic Regulatory Factors
2.7. Myokines Release by Multiplex
2.8. Statistical Analysis
3. Results
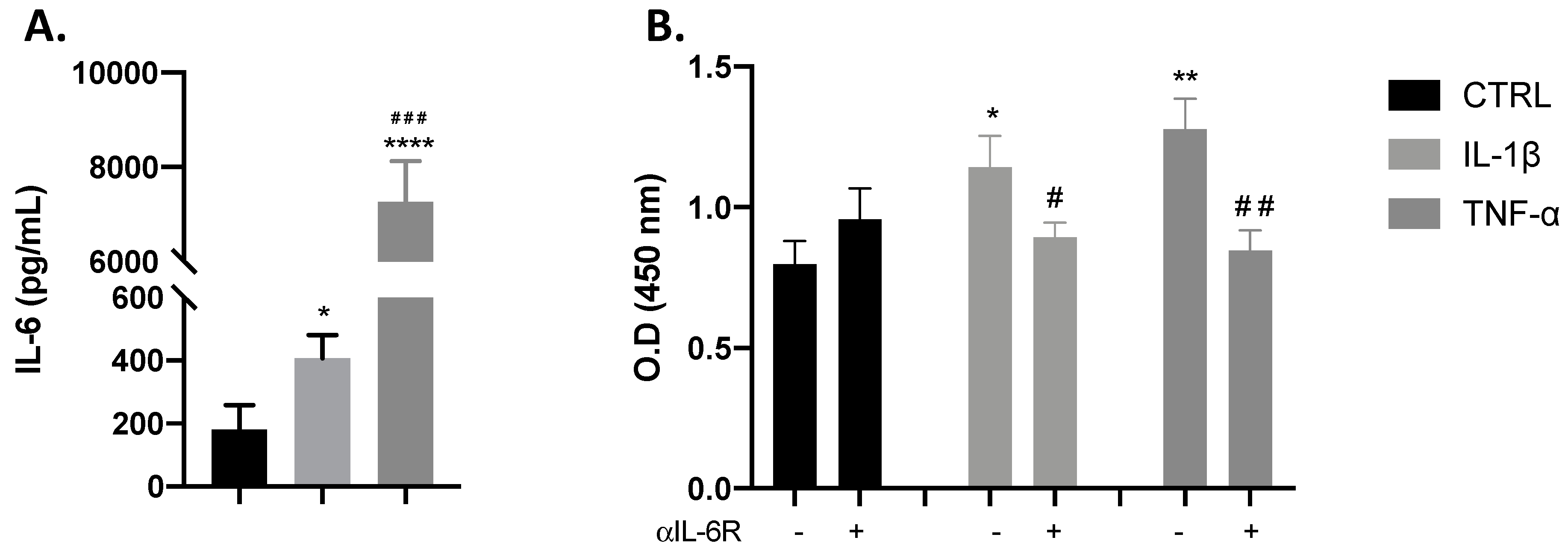
3.1. Proliferative Effects of IL-1β and TNF-α Occur through the IL-6 Receptor
3.2. COX-2 Pathway Is Involved in IL-1β-Induced Proliferation
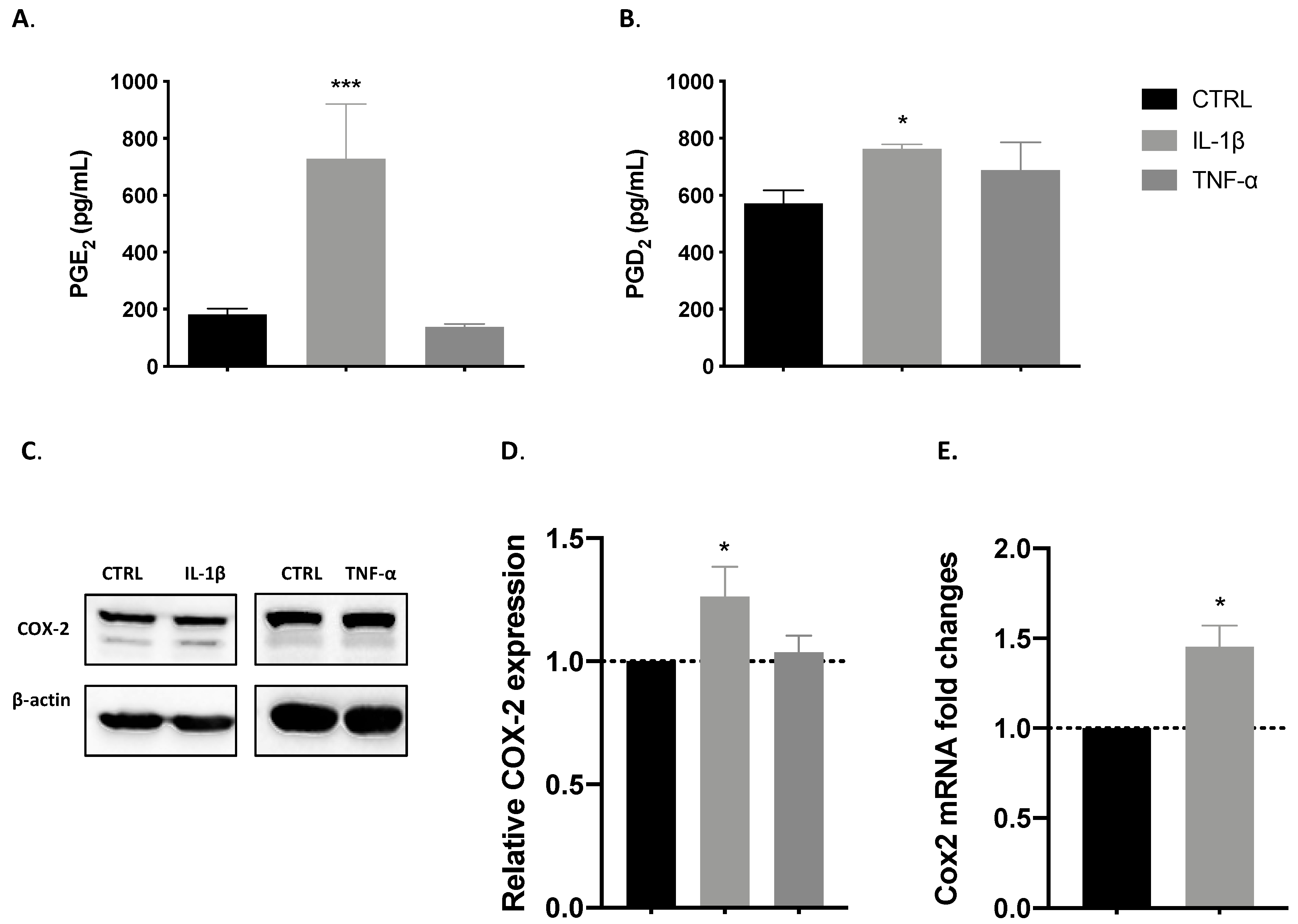
3.2.1. IL-1β but Not TNF-α Induced the Release of COX-Derived Mediators
3.2.2. IL-1β but Not TNF-α Induces COX-2 Expression
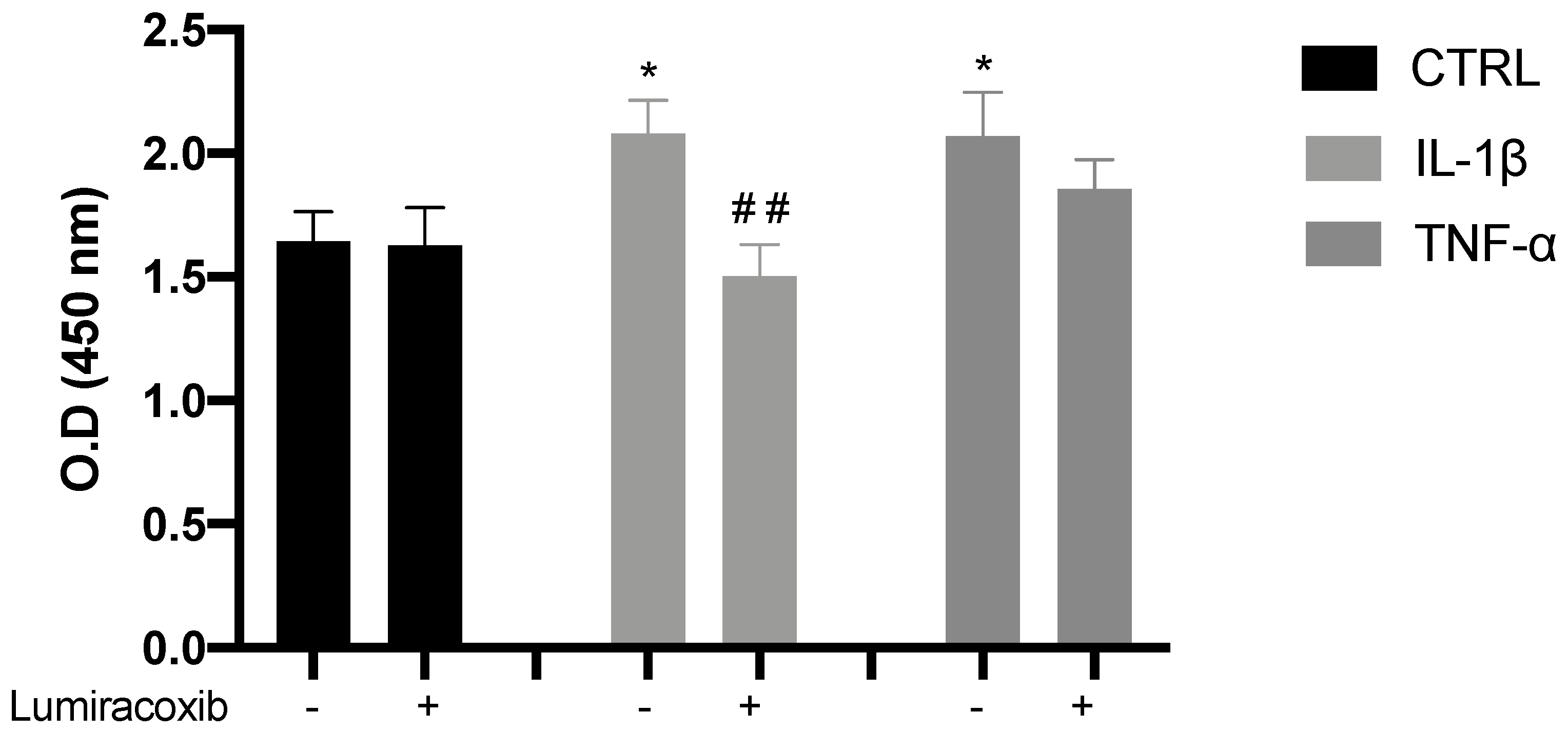
3.2.3. IL-1β Induces Proliferation through the Synthesis of COX-2-Derived Mediators
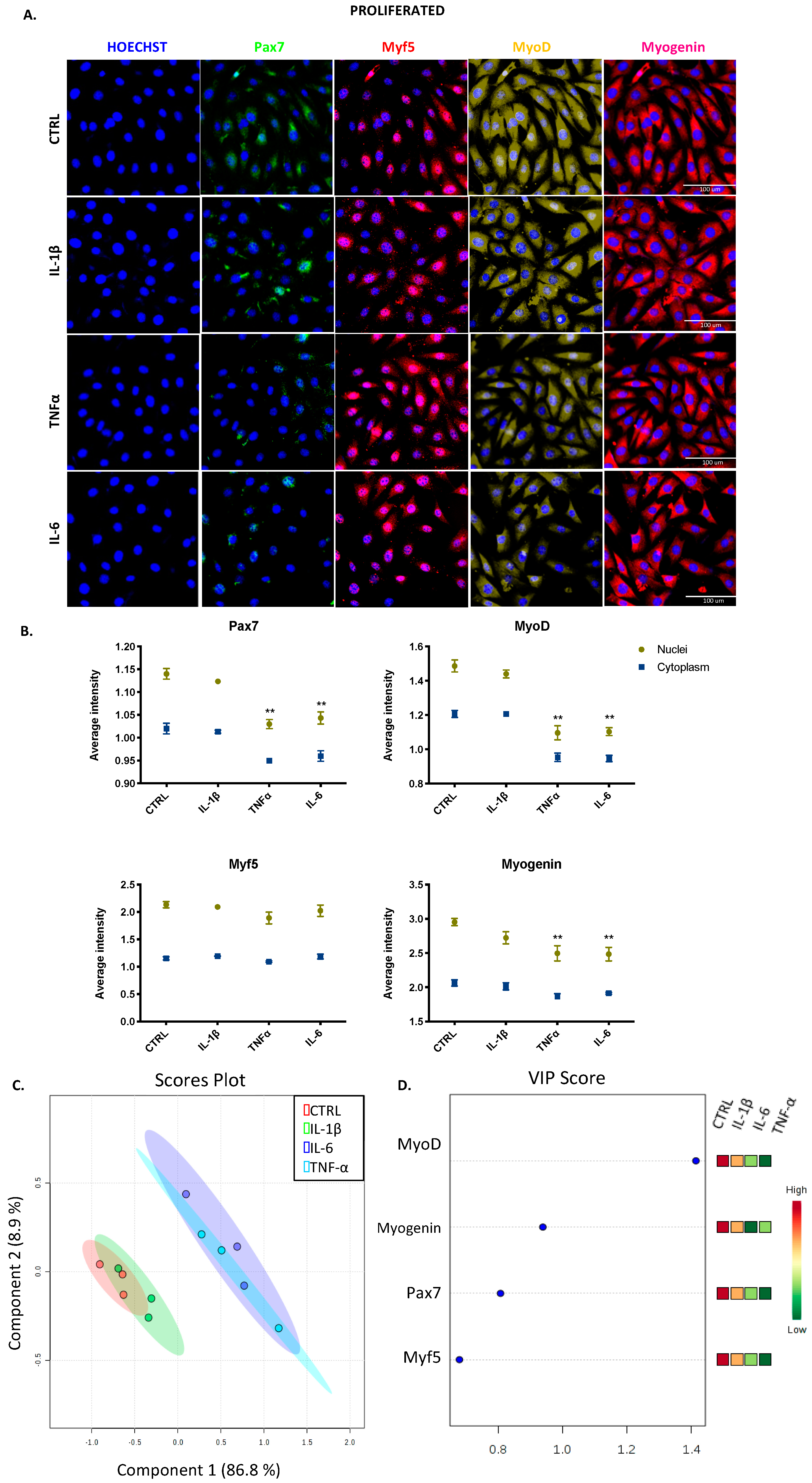
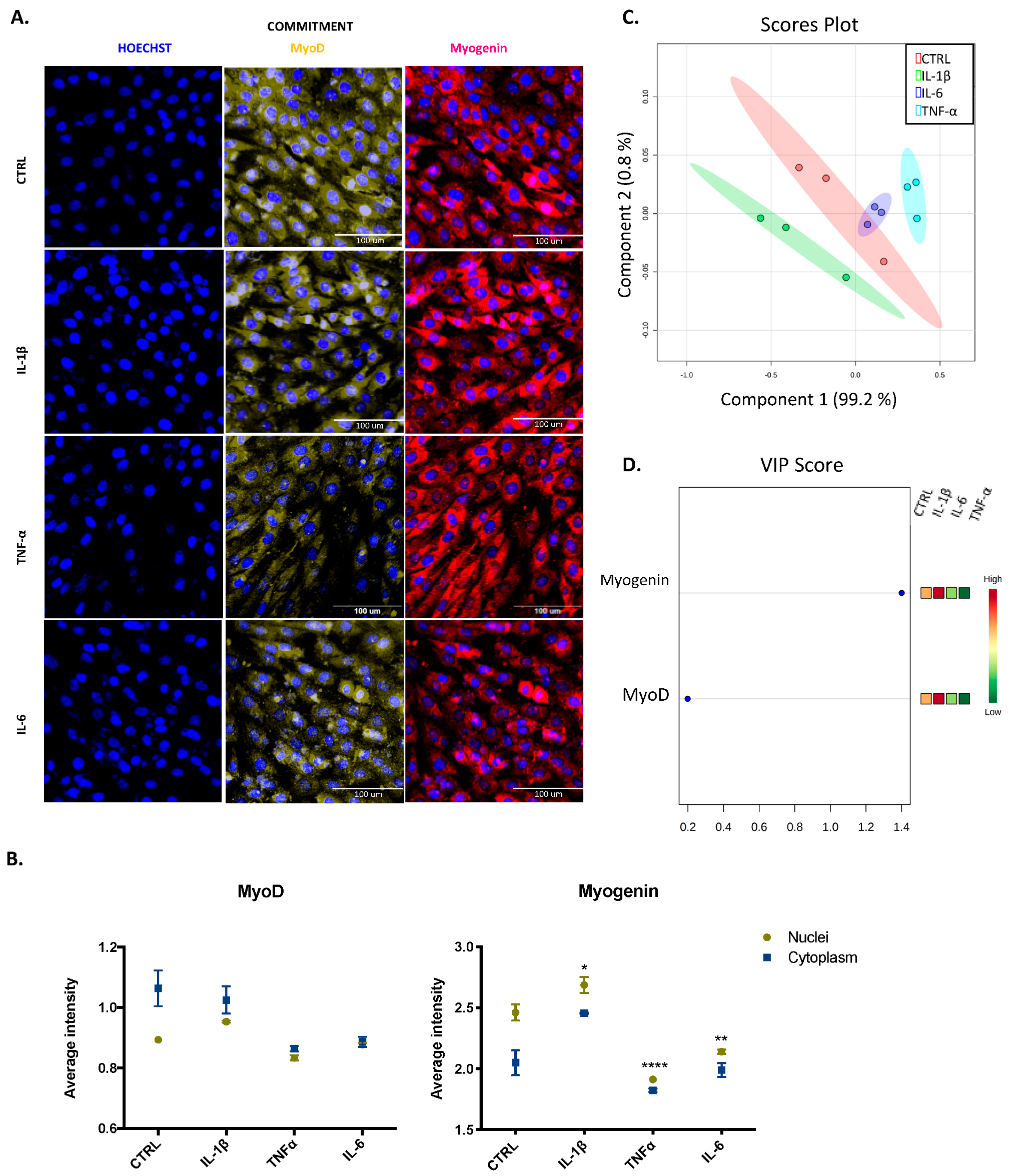
3.3. IL-1β, TNF-α, and IL-6 Can Modulate the Expression of Myogenic Regulatory Factors in Both Proliferated and Committed Cells Displaying Distinct Effects
3.4. IL-1β and TNF-α Positively Modulate the Production of Myokines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, N.C.; Rudnicki, M.A. Satellite cells: The architects of skeletal muscle. Curr. Top. Dev. Biol. 2014, 107, 161–181. [Google Scholar] [CrossRef] [PubMed]
- Becsky, D.; Gyulai-Nagy, S.; Balind, A.; Horvath, P.; Dux, L.; Keller-Pinter, A. Myoblast Migration and Directional Persistence Affected by Syndecan-4-Mediated Tiam-1 Expression and Distribution. Int. J. Mol. Sci. 2020, 21, 823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domingues-Faria, C.; Vasson, M.P.; Goncalves-Mendes, N.; Boirie, Y.; Walrand, S. Skeletal muscle regeneration and impact of aging and nutrition. Ageing Res. Rev. 2016, 26, 22–36. [Google Scholar] [CrossRef]
- Zammit, P.S.; Partridge, T.A.; Yablonka-Reuveni, Z. The skeletal muscle satellite cell: The stem cell that came in from the cold. J. Histochem. Cytochem. 2006, 54, 1177–1191. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Schuler, S.C.; Huttner, S.S.; von Eyss, B.; von Maltzahn, J. Adult stem cells at work: Regenerating skeletal muscle. Cell Mol. Life Sci. 2019, 76, 2559–2570. [Google Scholar] [CrossRef] [Green Version]
- Forcina, L.; Cosentino, M.; Musaro, A. Mechanisms Regulating Muscle Regeneration: Insights into the Interrelated and Time-Dependent Phases of Tissue Healing. Cells 2020, 9, 1297. [Google Scholar] [CrossRef]
- Philippou, A.; Maridaki, M.; Theos, A.; Koutsilieris, M. Cytokines in muscle damage. Adv. Clin. Chem. 2012, 58, 49–87. [Google Scholar] [CrossRef]
- Howard, E.E.; Pasiakos, S.M.; Blesso, C.N.; Fussell, M.A.; Rodriguez, N.R. Divergent Roles of Inflammation in Skeletal Muscle Recovery From Injury. Front. Physiol. 2020, 11, 87. [Google Scholar] [CrossRef] [Green Version]
- Otis, J.S.; Niccoli, S.; Hawdon, N.; Sarvas, J.L.; Frye, M.A.; Chicco, A.J.; Lees, S.J. Pro-inflammatory mediation of myoblast proliferation. PLoS ONE 2014, 9, e92363. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.; Hershko, D.D.; Robb, B.W.; Wray, C.J.; Hasselgren, P.O. IL-1beta stimulates IL-6 production in cultured skeletal muscle cells through activation of MAP kinase signaling pathway and NF-kappa B. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 284, 1249–1254. [Google Scholar] [CrossRef]
- Chaweewannakorn, C.; Tsuchiya, M.; Koide, M.; Hatakeyama, H.; Tanaka, Y.; Yoshida, S.; Sugawara, S.; Hagiwara, Y.; Sasaki, K.; Kanzaki, M. Roles of IL-1alpha/beta in regeneration of cardiotoxin-injured muscle and satellite cell function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2018, 315, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Tews, D.S.; Goebel, H.H. Cytokine expression profile in idiopathic inflammatory myopathies. J. Neuropathol. Exp. Neurol. 1996, 55, 342–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bleecker, J.L.; Meire, V.I.; Declercq, W.; Van Aken, E.H. Immunolocalization of tumor necrosis factor-alpha and its receptors in inflammatory myopathies. Neuromuscul. Disord. 1999, 9, 239–246. [Google Scholar] [CrossRef]
- Zador, E.; Mendler, L.; Takacs, V.; de Bleecker, J.; Wuytack, F. Regenerating soleus and extensor digitorum longus muscles of the rat show elevated levels of TNF-alpha and its receptors, TNFR-60 and TNFR-80. Muscle Nerve 2001, 24, 1058–1067. [Google Scholar] [CrossRef]
- Warren, G.L.; Hulderman, T.; Jensen, N.; McKinstry, M.; Mishra, M.; Luster, M.I.; Simeonova, P.P. Physiological role of tumor necrosis factor alpha in traumatic muscle injury. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002, 16, 1630–1632. [Google Scholar] [CrossRef] [Green Version]
- Levitt, D.E.; Yeh, A.Y.; Prendergast, M.J.; Budnar, R.G., Jr.; Adler, K.A.; Cook, G.; Molina, P.E.; Simon, L. Chronic Alcohol Dysregulates Skeletal Muscle Myogenic Gene Expression after Hind Limb Immobilization in Female Rats. Biomolecules 2020, 10, 441. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, M.E.; Londino, J.; McGinnis, M.; Weathington, N.; Adair, J.; Suber, T.; Kagan, V.; Chen, K.; Zou, C.; Chen, B.; et al. Tumor Necrosis Factor Alpha Regulates Skeletal Myogenesis by Inhibiting SP1 Interaction with cis-Acting Regulatory Elements within the Fbxl2 Gene Promoter. Mol. Cell. Biol. 2020, 40, e00040-20. [Google Scholar] [CrossRef]
- Li, Y.P.; Schwartz, R.J. TNF-alpha regulates early differentiation of C2C12 myoblasts in an autocrine fashion. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2001, 15, 1413–1415. [Google Scholar] [CrossRef]
- Li, Y.P.; Niu, A.; Wen, Y. Regulation of myogenic activation of p38 MAPK by TACE-mediated TNFalpha release. Front. Cell Dev. Biol. 2014, 2, 21. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.E.; Jin, B.; Li, Y.P. TNF-alpha regulates myogenesis and muscle regeneration by activating p38 MAPK. Am. J. Physiol. Cell Physiol. 2007, 292, 1660–1671. [Google Scholar] [CrossRef]
- Zhan, M.; Jin, B.; Chen, S.E.; Reecy, J.M.; Li, Y.P. TACE release of TNF-alpha mediates mechanotransduction-induced activation of p38 MAPK and myogenesis. J. Cell Sci. 2007, 120, 692–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacios, D.; Mozzetta, C.; Consalvi, S.; Caretti, G.; Saccone, V.; Proserpio, V.; Marquez, V.E.; Valente, S.; Mai, A.; Forcales, S.V.; et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 2010, 7, 455–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belizario, J.E.; Fontes-Oliveira, C.C.; Borges, J.P.; Kashiabara, J.A.; Vannier, E. Skeletal muscle wasting and renewal: A pivotal role of myokine IL-6. Springerplus 2016, 5, 619. [Google Scholar] [CrossRef] [Green Version]
- Loell, I.; Lundberg, I.E. Can muscle regeneration fail in chronic inflammation: A weakness in inflammatory myopathies? J. Intern. Med. 2011, 269, 243–257. [Google Scholar] [CrossRef]
- Podbregar, M.; Lainscak, M.; Prelovsek, O.; Mars, T. Cytokine response of cultured skeletal muscle cells stimulated with proinflammatory factors depends on differentiation stage. Sci. World J. 2013, 2013, 617170. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Panguluri, S.K.; Gupta, S.K.; Dahiya, S.; Lundy, R.F.; Kumar, A. Tumor necrosis factor-alpha regulates distinct molecular pathways and gene networks in cultured skeletal muscle cells. PLoS ONE 2010, 5, e13262. [Google Scholar] [CrossRef]
- Peake, J.M.; Della Gatta, P.; Suzuki, K.; Nieman, D.C. Cytokine expression and secretion by skeletal muscle cells: Regulatory mechanisms and exercise effects. Exerc. Immunol. Rev. 2015, 21, 8–25. [Google Scholar]
- Little, H.C.; Tan, S.Y.; Cali, F.M.; Rodriguez, S.; Lei, X.; Wolfe, A.; Hug, C.; Wong, G.W. Multiplex Quantification Identifies Novel Exercise-regulated Myokines/Cytokines in Plasma and in Glycolytic and Oxidative Skeletal Muscle. Mol. Cell. Proteom. 2018, 17, 1546–1563. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.H.; Kim, J.; Song, P.; Lee, T.G.; Suh, P.G.; Ryu, S.H. Secretomics for skeletal muscle cells: A discovery of novel regulators? Adv. Biol. Regul. 2012, 52, 340–350. [Google Scholar] [CrossRef]
- Furuichi, Y.; Manabe, Y.; Takagi, M.; Aoki, M.; Fujii, N.L. Evidence for acute contraction-induced myokine secretion by C2C12 myotubes. PLoS ONE 2018, 13, e0206146. [Google Scholar] [CrossRef] [Green Version]
- Eckel, J. Chapter 3: Skeletal Muscle A Novel Secretory Organ. In The Cellular Secretome and Organ Crosstalk; Academic Press: Cambridge, MA, USA, 2018; pp. 65–90. [Google Scholar] [CrossRef]
- Bondesen, B.A.; Mills, S.T.; Kegley, K.M.; Pavlath, G.K. The COX-2 pathway is essential during early stages of skeletal muscle regeneration. Am. J. Physiol. Cell Physiol. 2004, 287, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Bondesen, B.A.; Jones, K.A.; Glasgow, W.C.; Pavlath, G.K. Inhibition of myoblast migration by prostacyclin is associated with enhanced cell fusion. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2007, 21, 3338–3345. [Google Scholar] [CrossRef]
- Novak, M.L.; Billich, W.; Smith, S.M.; Sukhija, K.B.; McLoughlin, T.J.; Hornberger, T.A.; Koh, T.J. COX-2 inhibitor reduces skeletal muscle hypertrophy in mice. Am. J. Physiol. Regul Integr. Comp. Physiol. 2009, 296, 1132–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, A.T.V.; Palla, A.R.; Blake, M.R.; Yucel, N.D.; Wang, Y.X.; Magnusson, K.E.G.; Holbrook, C.A.; Kraft, P.E.; Delp, S.L.; Blau, H.M. Prostaglandin E2 is essential for efficacious skeletal muscle stem-cell function, augmenting regeneration and strength. Proc. Natl. Acad. Sci. USA 2017, 114, 6675–6684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, C.; Zhao, R.; Vallejo, J.; Igwe, O.; Bonewald, L.; Wetmore, L.; Brotto, M. Prostaglandin E2 promotes proliferation of skeletal muscle myoblasts via EP4 receptor activation. Cell Cycle 2015, 14, 1507–1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baracos, V.; Rodemann, H.P.; Dinarello, C.A.; Goldberg, A.L. Stimulation of muscle protein degradation and prostaglandin E2 release by leukocytic pyrogen (interleukin-1). A mechanism for the increased degradation of muscle proteins during fever. N. Engl. J. Med. 1983, 308, 553–558. [Google Scholar] [CrossRef]
- Schafers, M.; Sorkin, L.S.; Sommer, C. Intramuscular injection of tumor necrosis factor-alpha induces muscle hyperalgesia in rats. Pain 2003, 104, 579–588. [Google Scholar] [CrossRef]
- Velica, P.; Bunce, C.M. Prostaglandins in muscle regeneration. J. Muscle Res. Cell Motil. 2008, 29, 163–167. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Monda, M.; Vicidomini, C.; Viggiano, A.; Sampaolo, S.; Di Iorio, G.; Viggiano, A.; Viggiano, E.; De Luca, B. Inhibition of prostaglandin synthesis reduces the induction of MyoD expression in rat soleus muscle. J. Muscle Res. Cell Motil. 2009, 30, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Otto, J.C.; DeWitt, D.L.; Smith, W.L. N-glycosylation of prostaglandin endoperoxide synthases-1 and -2 and their orientations in the endoplasmic reticulum. J. Biol. Chem. 1993, 268, 18234–18242. [Google Scholar] [PubMed]
- Otis, J.S.; Burkholder, T.J.; Pavlath, G.K. Stretch-induced myoblast proliferation is dependent on the COX2 pathway. Exp. Cell Res. 2005, 310, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Langen, R.C.; Schols, A.M.; Kelders, M.C.; Wouters, E.F.; Janssen-Heininger, Y.M. Inflammatory cytokines inhibit myogenic differentiation through activation of nuclear factor-kappaB. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2001, 15, 1169–1180. [Google Scholar] [CrossRef] [Green Version]
- Prelovsek, O.; Mars, T.; Jevsek, M.; Podbregar, M.; Grubic, Z. High dexamethasone concentration prevents stimulatory effects of TNF-alpha and LPS on IL-6 secretion from the precursors of human muscle regeneration. Am. J. Physiol Regul Integr Comp. Physiol 2006, 291, 1651–1656. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Canoves, P.; Scheele, C.; Pedersen, B.K.; Serrano, A.L. Interleukin-6 myokine signaling in skeletal muscle: A double-edged sword? FEBS J. 2013, 280, 4131–4148. [Google Scholar] [CrossRef]
- Daou, H.N. Exercise as an anti-inflammatory therapy for cancer cachexia: A focus on interleukin-6 regulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2020, 318, 296–310. [Google Scholar] [CrossRef]
- Maurer, M.; Bougoin, S.; Feferman, T.; Frenkian, M.; Bismuth, J.; Mouly, V.; Clairac, G.; Tzartos, S.; Fadel, E.; Eymard, B.; et al. IL-6 and Akt are involved in muscular pathogenesis in myasthenia gravis. Acta Neuropathol. Commun. 2015, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Steyn, P.J.; Dzobo, K.; Smith, R.I.; Myburgh, K.H. Interleukin-6 Induces Myogenic Differentiation via JAK2-STAT3 Signaling in Mouse C2C12 Myoblast Cell Line and Primary Human Myoblasts. Int. J. Mol. Sci 2019, 20, 5273. [Google Scholar] [CrossRef] [Green Version]
- Al-Shanti, N.; Saini, A.; Faulkner, S.H.; Stewart, C.E. Beneficial synergistic interactions of TNF-alpha and IL-6 in C2 skeletal myoblasts--potential cross-talk with IGF system. Growth Factors 2008, 26, 61–73. [Google Scholar] [CrossRef]
- Prisk, V.; Huard, J. Muscle injuries and repair: The role of prostaglandins and inflammation. Histol. Histopathol. 2003, 18, 1243–1256. [Google Scholar] [CrossRef]
- Mendias, C.L.; Tatsumi, R.; Allen, R.E. Role of cyclooxygenase-1 and -2 in satellite cell proliferation, differentiation, and fusion. Muscle Nerve 2004, 30, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Jiang, H. Effects of arachidonic acid and its major prostaglandin derivatives on bovine myoblast proliferation, differentiation, and fusion. Domest. Anim. Endocrinol. 2019, 67, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Duchesne, E.; Tremblay, M.H.; Cote, C.H. Mast cell tryptase stimulates myoblast proliferation; a mechanism relying on protease-activated receptor-2 and cyclooxygenase-2. BMC Musculoskelet. Disord. 2011, 12, 235. [Google Scholar] [CrossRef] [Green Version]
- Nagata, K.; Nakamura, T.; Fujihara, S.; Tanaka, E. Ultrasound modulates the inflammatory response and promotes muscle regeneration in injured muscles. Ann. Biomed. Eng. 2013, 41, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Velica, P.; Khanim, F.L.; Bunce, C.M. Prostaglandin D2 inhibits C2C12 myogenesis. Mol. Cell Endocrinol. 2010, 319, 71–78. [Google Scholar] [CrossRef]
- Mo, C.; Romero-Suarez, S.; Bonewald, L.; Johnson, M.; Brotto, M. Prostaglandin E2: From clinical applications to its potential role in bone- muscle crosstalk and myogenic differentiation. Recent Pat. Biotechnol. 2012, 6, 223–229. [Google Scholar] [CrossRef]
- Kramer, G.; Marberger, M. Could inflammation be a key component in the progression of benign prostatic hyperplasia? Curr. Opin. Urol. 2006, 16, 25–29. [Google Scholar]
- Oshima, H.; Oshima, M. The inflammatory network in the gastrointestinal tumor microenvironment: Lessons from mouse models. J. Gastroenterol. 2012, 47, 97–106. [Google Scholar] [CrossRef]
- Fosslien, E. Molecular pathology of cyclooxygenase-2 in neoplasia. Ann. Clin. Lab. Sci 2000, 30, 3–21. [Google Scholar]
- Ferri, P.; Barbieri, E.; Burattini, S.; Guescini, M.; D’Emilio, A.; Biagiotti, L.; Del Grande, P.; De Luca, A.; Stocchi, V.; Falcieri, E. Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle C2C12 myoblasts. J. Cell Biochem. 2009, 108, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.E.; Newcomb, P.V.; Holly, J.M. Multifaceted roles of TNF-alpha in myoblast destruction: A multitude of signal transduction pathways. J. Cell. Physiol. 2004, 198, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Langen, R.C.; Van Der Velden, J.L.; Schols, A.M.; Kelders, M.C.; Wouters, E.F.; Janssen-Heininger, Y.M. Tumor necrosis factor-alpha inhibits myogenic differentiation through MyoD protein destabilization. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2004, 18, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Welc, S.S.; Wehling-Henricks, M.; Tidball, J.G. Myeloid cell-derived tumor necrosis factor-alpha promotes sarcopenia and regulates muscle cell fusion with aging muscle fibers. Aging Cell 2018, 17, e12828. [Google Scholar] [CrossRef]
- Kataoka, Y.; Matsumura, I.; Ezoe, S.; Nakata, S.; Takigawa, E.; Sato, Y.; Kawasaki, A.; Yokota, T.; Nakajima, K.; Felsani, A.; et al. Reciprocal inhibition between MyoD and STAT3 in the regulation of growth and differentiation of myoblasts. J. Biol. Chem. 2003, 278, 44178–44187. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Ma, K.; Wang, H.; Xiao, F.; Gao, Y.; Zhang, W.; Wang, K.; Gao, X.; Ip, N.; Wu, Z. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J. Cell Biol. 2007, 179, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, M.; De Rossi, M.; Barberi, L.; Musaro, A. IL-6 impairs myogenic differentiation by downmodulation of p90RSK/eEF2 and mTOR/p70S6K axes, without affecting AKT activity. Biomed. Res. Int. 2014, 2014, 206026. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; McFarlane, C.; Kambadur, R.; Kukreti, H.; Bonala, S.; Srinivasan, S. Myostatin: Expanding horizons. IUBMB Life 2015, 67, 589–600. [Google Scholar] [CrossRef]
- Huh, J.Y.; Dincer, F.; Mesfum, E.; Mantzoros, C.S. Irisin stimulates muscle growth-related genes and regulates adipocyte differentiation and metabolism in humans. Int. J. Obes. 2014, 38, 1538–1544. [Google Scholar] [CrossRef]
- Chang, J.S.; Kong, I.D. Irisin prevents dexamethasone-induced atrophy in C2C12 myotubes. Pflügers Archiv 2020, 472, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Cho, W.J.; Kim, E.J.; Lee, S.J.; Kim, H.D.; Shin, H.J.; Lim, W.K. Involvement of SPARC in in vitro differentiation of skeletal myoblasts. Biochem. Biophys. Res. Commun. 2000, 271, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Petersson, S.J.; Jorgensen, L.H.; Andersen, D.C.; Norgaard, R.C.; Jensen, C.H.; Schroder, H.D. SPARC is up-regulated during skeletal muscle regeneration and inhibits myoblast differentiation. Histol. Histopathol. 2013, 28, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Shiba, H.; Fujita, T.; Doi, N.; Nakamura, S.; Nakanishi, K.; Takemoto, T.; Hino, T.; Noshiro, M.; Kawamoto, T.; Kurihara, H.; et al. Differential effects of various growth factors and cytokines on the syntheses of DNA, type I collagen, laminin, fibronectin, osteonectin/secreted protein, acidic and rich in cysteine (SPARC), and alkaline phosphatase by human pulp cells in culture. J. Cell. Physiol. 1998, 174, 194–205. [Google Scholar] [CrossRef]
- O’Leary, M.F.; Wallace, G.R.; Bennett, A.J.; Tsintzas, K.; Jones, S.W. IL-15 promotes human myogenesis and mitigates the detrimental effects of TNFalpha on myotube development. Sci. Rep. 2017, 7, 12997. [Google Scholar] [CrossRef]
- Bugera, E.M.; Duhamel, T.A.; Peeler, J.D.; Cornish, S.M. The systemic myokine response of decorin, interleukin-6 (IL-6) and interleukin-15 (IL-15) to an acute bout of blood flow restricted exercise. Eur. J. Appl. Physiol. 2018, 118, 2679–2686. [Google Scholar] [CrossRef] [PubMed]

| Gen Name | Primer Sequence |
|---|---|
| Cox2 | F: TTCCAATCCATGTCAAAACCGT |
| R: AGTCCGGGTACAGTCACACTT | |
| GAPDH | F: AGGTCGGTGTGAACGGATTTG |
| R: TGTAGACCATGTAGTTGAGGTCA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez, A.M.; DeOcesano-Pereira, C.; Teixeira, C.; Moreira, V. IL-1β and TNF-α Modulation of Proliferated and Committed Myoblasts: IL-6 and COX-2-Derived Prostaglandins as Key Actors in the Mechanisms Involved. Cells 2020, 9, 2005. https://doi.org/10.3390/cells9092005
Alvarez AM, DeOcesano-Pereira C, Teixeira C, Moreira V. IL-1β and TNF-α Modulation of Proliferated and Committed Myoblasts: IL-6 and COX-2-Derived Prostaglandins as Key Actors in the Mechanisms Involved. Cells. 2020; 9(9):2005. https://doi.org/10.3390/cells9092005
Chicago/Turabian StyleAlvarez, Angela M., Carlos DeOcesano-Pereira, Catarina Teixeira, and Vanessa Moreira. 2020. "IL-1β and TNF-α Modulation of Proliferated and Committed Myoblasts: IL-6 and COX-2-Derived Prostaglandins as Key Actors in the Mechanisms Involved" Cells 9, no. 9: 2005. https://doi.org/10.3390/cells9092005
APA StyleAlvarez, A. M., DeOcesano-Pereira, C., Teixeira, C., & Moreira, V. (2020). IL-1β and TNF-α Modulation of Proliferated and Committed Myoblasts: IL-6 and COX-2-Derived Prostaglandins as Key Actors in the Mechanisms Involved. Cells, 9(9), 2005. https://doi.org/10.3390/cells9092005





