Thioredoxin-Related Transmembrane Proteins: TMX1 and Little Brothers TMX2, TMX3, TMX4 and TMX5
Abstract
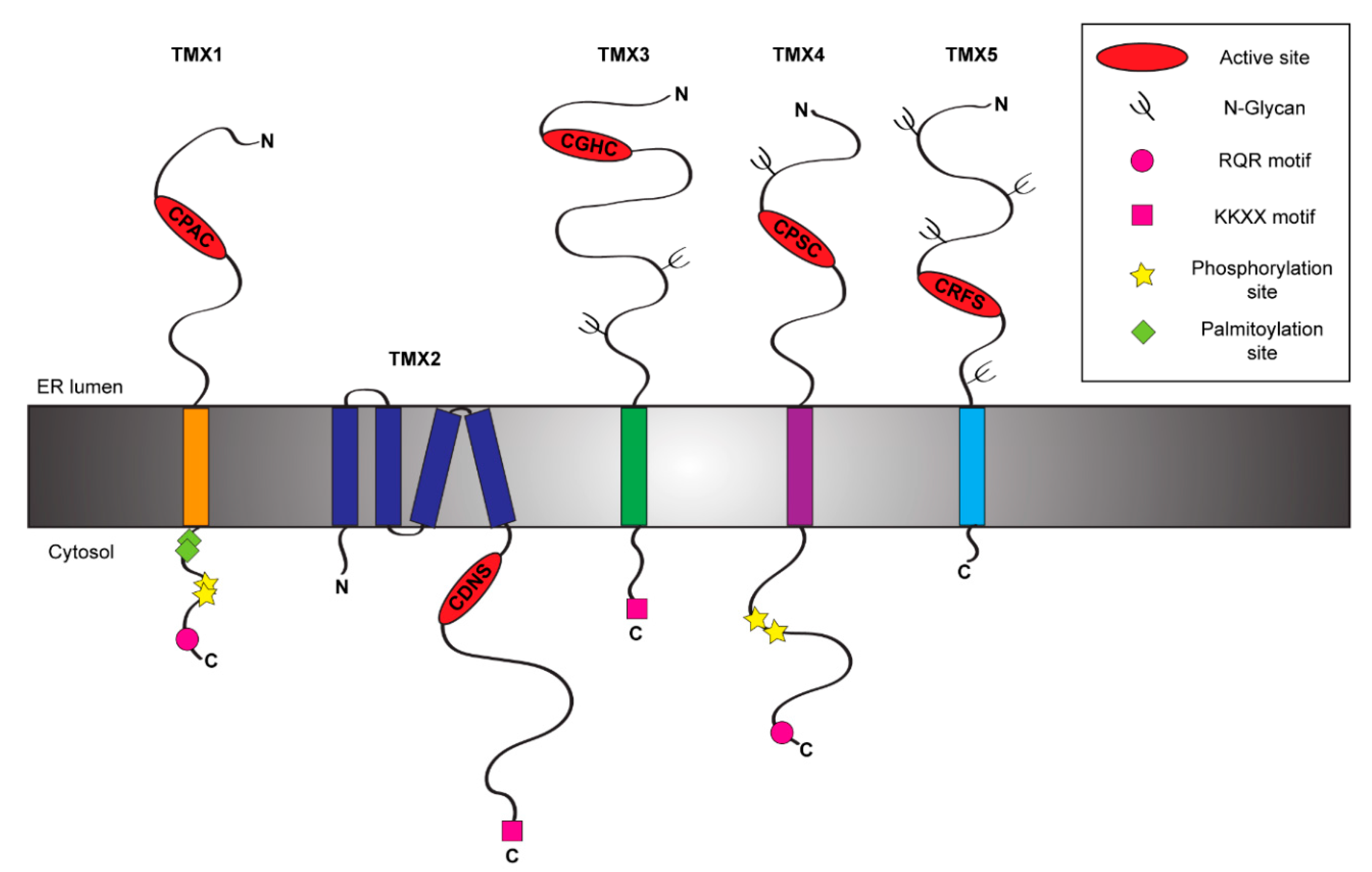
1. Introduction
2. TMX1: A Topology-Specific ER-Resident Reductase
2.1. TMX1 Assists Folding of Membrane-Tethered Polypeptides
2.2. TMX1 Selectively Intervenes in ERAD of Membrane-Tethered Folding-Defective Polypeptides
2.3. MAM-Localized TMX1 Acts as a Regulator of Ca2+ Flux
3. TMX2 and its Cytosolic Active Site
4. TMX3, a Classic PDI
5. TMX4, the Paralogue of TMX1
6. TMX5, a Natural Trapping Mutant Member of the TMX Family
7. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Ellgaard, L.; McCaul, N.; Chatsisvili, A.; Braakman, I. Co- and Post-Translational Protein Folding in the ER. Traffic 2016, 17, 615–638. [Google Scholar] [CrossRef] [PubMed]
- Kosuri, P.; Alegre-Cebollada, J.; Feng, J.; Kaplan, A.; Ingles-Prieto, A.; Badilla, C.L.; Stockwell, B.R.; Sanchez-Ruiz, J.M.; Holmgren, A.; Fernandez, J.M. Protein folding drives disulfide formation. Cell 2012, 151, 794–806. [Google Scholar] [CrossRef] [PubMed]
- Hatahet, F.; Ruddock, L.W. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 2009, 11, 2807–2850. [Google Scholar] [CrossRef]
- Pisoni, G.B.; Molinari, M. Five Questions (with their Answers) on ER-Associated Degradation. Traffic 2016, 17, 341–350. [Google Scholar] [CrossRef]
- Suzuki, Y.; Schmitt, M.J. Redox diversity in ERAD-mediated protein retrotranslocation from the endoplasmic reticulum: A complex puzzle. Biol. Chem. 2015, 396, 539–554. [Google Scholar] [CrossRef]
- Appenzeller-Herzog, C.; Ellgaard, L. The human PDI family: Versatility packed into a single fold. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2008, 1783, 535–548. [Google Scholar] [CrossRef]
- Myllyharju, J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003, 22, 15–24. [Google Scholar] [CrossRef]
- Shrimal, S.; Gilmore, R. Oligosaccharyltransferase structures provide novel insight into the mechanism of asparagine-linked glycosylation in prokaryotic and eukaryotic cells. Glycobiology 2019, 29, 288–297. [Google Scholar] [CrossRef]
- Okumura, M.; Kadokura, H.; Inaba, K. Structures and functions of protein disulfide isomerase family members involved in proteostasis in the endoplasmic reticulum. Free Radic. Biol. Med. 2015, 83, 314–322. [Google Scholar] [CrossRef]
- Hatahet, F.; Ruddock, L.W. Substrate recognition by the protein disulfide isomerases. FEBS J. 2007, 274, 5223–5234. [Google Scholar] [CrossRef] [PubMed]
- Oguro, A.; Imaoka, S. Thioredoxin-related transmembrane protein 2 (TMX2) regulates the Ran protein gradient and importin-beta-dependent nuclear cargo transport. Sci. Rep. 2019, 9, 15296. [Google Scholar] [CrossRef] [PubMed]
- Haugstetter, J.; Blicher, T.; Ellgaard, L. Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J. Biol. Chem. 2005, 280, 8371–8380. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, Y.; Araki, K.; Iemura, S.; Natsume, T.; Hoseki, J.; Nagata, K. Novel thioredoxin-related transmembrane protein TMX4 has reductase activity. J. Biol. Chem. 2010, 285, 7135–7142. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Maattanen, P.; Thomas, D.Y.; Gehring, K. A structural overview of the PDI family of proteins. FEBS J. 2010, 277, 3924–3936. [Google Scholar] [CrossRef]
- Matsuo, Y.; Akiyama, N.; Nakamura, H.; Yodoi, J.; Noda, M.; Kizaka-Kondoh, S. Identification of a novel thioredoxin-related transmembrane protein. J. Biol. Chem. 2001, 276, 10032–10038. [Google Scholar] [CrossRef]
- Pisoni, G.B.; Ruddock, L.W.; Bulleid, N.; Molinari, M. Division of labor among oxidoreductases: TMX1 preferentially acts on transmembrane polypeptides. Mol. Biol. Cell 2015, 26, 3390–3400. [Google Scholar] [CrossRef]
- Guerra, C.; Brambilla Pisoni, G.; Solda, T.; Molinari, M. The reductase TMX1 contributes to ERAD by preferentially acting on membrane-associated folding-defective polypeptides. Biochem. Biophys. Res. Commun. 2018, 503, 938–943. [Google Scholar] [CrossRef]
- Akiyama, N.; Matsuo, Y.; Sai, H.; Noda, M.; Kizaka-Kondoh, S. Identification of a series of transforming growth factor beta-responsive genes by retrovirus-mediated gene trap screening. Mol. Cell Biol. 2000, 20, 3266–3273. [Google Scholar] [CrossRef]
- Matsuo, Y.; Nishinaka, Y.; Suzuki, S.; Kojima, M.; Kizaka-Kondoh, S.; Kondo, N.; Son, A.; Sakakura-Nishiyama, J.; Yamaguchi, Y.; Masutani, H.; et al. A human transmembrane oxidoreductase of the thioredoxin family: The possible role in disulfide-linked protein folding in the endoplasmic reticulum. Arch. Biochem. Biophys. 2004, 423, 81–87. [Google Scholar] [CrossRef]
- Roth, D.; Lynes, E.; Riemer, J.; Hansen, H.G.; Althaus, N.; Simmen, T.; Ellgaard, L. A di-arginine motif contributes to the ER localization of the type I transmembrane ER oxidoreductase TMX4. Biochem. J. 2009, 425, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Lynes, E.M.; Bui, M.; Yap, M.C.; Benson, M.D.; Schneider, B.; Ellgaard, L.; Berthiaume, L.G.; Simmen, T. Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J. 2012, 31, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, In Vivo, and Site-Specific Phosphorylation Dynamics in Signaling Networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Galligan, J.J.; Petersen, D.R. The human protein disulfide isomerase gene family. Hum. Genom. 2012, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Masutani, H.; Son, A.; Kizaka-Kondoh, S.; Yodoi, J. Physical and functional interaction of transmembrane thioredoxin-related protein with major histocompatibility complex class I heavy chain: Redox-based protein quality control and its potential relevance to immune responses. Mol. Biol. Cell 2009, 20, 4552–4562. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Matsuo, Y.; Irie, K.; Kiyonari, H.; Okuyama, H.; Nakamura, H.; Son, A.; Lopez-Ramos, D.A.; Tian, H.; Oka, S.; Okawa, K.; et al. The protective role of the transmembrane thioredoxin-related protein TMX in inflammatory liver injury. Antioxid. Redox Signal. 2013, 18, 1263–1272. [Google Scholar] [CrossRef]
- Hatahet, F.; Ruddock, L.W. Modulating proteostasis: Peptidomimetic inhibitors and activators of protein folding. Curr. Pharm. Des. 2009, 15, 2488–2507. [Google Scholar] [CrossRef]
- Roos, G.; Garcia-Pino, A.; Van belle, K.; Brosens, E.; Wahni, K.; Vandenbussche, G.; Wyns, L.; Loris, R.; Messens, J. The Conserved Active Site Proline Determines the Reducing Power of Staphylococcus aureus Thioredoxin. J. Mol. Biol. 2007, 368, 800–811. [Google Scholar] [CrossRef]
- Pasetto, M.; Barison, E.; Castagna, M.; Della Cristina, P.; Anselmi, C.; Colombatti, M. Reductive activation of type 2 ribosome-inactivating proteins is promoted by transmembrane thioredoxin-related protein. J. Biol. Chem. 2012, 287, 7367–7373. [Google Scholar] [CrossRef]
- Schulman, S.; Wang, B.; Li, W.; Rapoport, T.A. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl. Acad. Sci. USA 2010, 107, 15027–15032. [Google Scholar] [CrossRef]
- Jessop, C.E.; Chakravarthi, S.; Garbi, N.; Hammerling, G.J.; Lovell, S.; Bulleid, N.J. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007, 26, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Lamriben, L.; Graham, J.B.; Adams, B.M.; Hebert, D.N. N-Glycan-based ER Molecular Chaperone and Protein Quality Control System: The Calnexin Binding Cycle. Traffic 2016, 17, 308–326. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.; Galli, C.; Piccaluga, V.; Pieren, M.; Paganetti, P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J. Cell Biol. 2002, 158, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, T.; Simmen, T. Endoplasmic reticulum chaperones tweak the mitochondrial calcium rheostat to control metabolism and cell death. Cell Calcium 2018, 70, 64–75. [Google Scholar] [CrossRef]
- Janssens, S.; Bultynck, G.; Krols, M. ER–Mitochondria contact sites: A new regulator of cellular calcium flux comes into play. J. Cell Biol. 2016, 214, 367–370. [Google Scholar]
- Raturi, A.; Gutierrez, T.; Ortiz-Sandoval, C.; Ruangkittisakul, A.; Herrera-Cruz, M.S.; Rockley, J.P.; Gesson, K.; Ourdev, D.; Lou, P.H.; Lucchinetti, E.; et al. TMX1 determines cancer cell metabolism as a thiol-based modulator of ER-mitochondria Ca2+ flux. J. Cell Biol. 2016, 214, 433–444. [Google Scholar] [CrossRef]
- Appenzeller-Herzog, C.; Simmen, T. ER-luminal thiol/selenol-mediated regulation of Ca2+ signalling. Biochem. Soc. Trans. 2016, 44, 452–459. [Google Scholar] [CrossRef]
- Don, A.S.; Hogg, P.J. Mitochondria as cancer drug targets. Trends Mol. Med. 2004, 10, 372–378. [Google Scholar] [CrossRef]
- Phan, V.; Schmidt, J.; Matyash, V.; Malchow, S.; Thanisch, M.; Lorenz, C.; Diepolder, I.; Schulz, J.B.; Stenzel, W.; Roos, A.; et al. Characterization of Naïve and Vitamin C-Treated Mouse Schwann Cell Line MSC80: Induction of the Antioxidative Thioredoxin Related Transmembrane Protein 1. J. Proteome Res. 2018, 17, 2925–2936. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, C.; Chen, J.; Peng, S.; Cao, Y.; Ying, K.; Xie, Y.; Mao, Y. Cloning and Identification of a Novel cDNA Coding Thioredoxin-Related Transmembrane Protein 2. Biochem. Genet. 2003, 41, 99–106. [Google Scholar] [CrossRef]
- Vandervore, L.V.; Schot, R.; Milanese, C.; Smits, D.J.; Kasteleijn, E.; Fry, A.E.; Pilz, D.T.; Brock, S.; Borklu-Yucel, E.; Post, M.; et al. TMX2 Is a Crucial Regulator of Cellular Redox State, and Its Dysfunction Causes Severe Brain Developmental Abnormalities. Am. J. Hum. Genet. 2019, 105, 1126–1147. [Google Scholar] [CrossRef]
- Quimby, B.B.; Corbett, A.H. Nuclear transport mechanisms. Cell. Mol. Life Sci. 2001, 58, 1766–1773. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.G.; Wang, L.; Breuss, M.W.; Green, J.D.; Stanley, V.; Yang, X.; Ross, D.; Traynor, B.J.; Alhashem, A.M.; Azam, M.; et al. Recurrent homozygous damaging mutation in TMX2, encoding a protein disulfide isomerase, in four families with microlissencephaly. J. Med. Genet. 2020, 57, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.E.; Cushion, T.D.; Pilz, D.T. The genetics of lissencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.J.; Haney, M.S.; Morgens, D.W.; Jovičić, A.; Couthouis, J.; Li, A.; Ousey, J.; Ma, R.; Bieri, G.; Tsui, C.K.; et al. CRISPR–Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet. 2018, 50, 603–612. [Google Scholar] [CrossRef]
- Poulton, C.J.; Schot, R.; Kia, S.K.; Jones, M.; Verheijen, F.W.; Venselaar, H.; de Wit, M.-C.Y.; de Graaff, E.; Bertoli-Avella, A.M.; Mancini, G.M.S. Microcephaly with Simplified Gyration, Epilepsy, and Infantile Diabetes Linked to Inappropriate Apoptosis of Neural Progenitors. Am. J. Hum. Genet. 2011, 89, 265–276. [Google Scholar] [CrossRef]
- Haugstetter, J.; Maurer, M.A.; Blicher, T.; Pagac, M.; Wider, G.; Ellgaard, L. Structure-Function Analysis of the Endoplasmic Reticulum Oxidoreductase TMX3 Reveals Interdomain Stabilization of the N-terminal Redox-active Domain. J. Biol. Chem. 2007, 282, 33859–33867. [Google Scholar] [CrossRef]
- Wilkinson, B.; Gilbert, H.F. Protein disulfide isomerase. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2004, 1699, 35–44. [Google Scholar] [CrossRef]
- Fox, J.; Lu, Z.; Barrows, L. Thiol-disulfide Oxidoreductases TRX1 and TMX3 Decrease Neuronal Atrophy in a Lentiviral Mouse Model of Huntington’s Disease. PLoS Curr. 2015. [Google Scholar] [CrossRef]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: A clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef]
- Li, M.; Wen, Y.; Wen, H.; Gui, C.; Huang, F.; Zeng, Z. Discovery of PPP2R3A and TMX3 pathogenic variants in a Zhuang family with coronary artery disease using whole-exome sequencing. Int. J. Clin. Exp. Pathol. 2018, 11, 3678–3684. [Google Scholar] [PubMed]
- Abraham, E.; Chao, R.; Nevin, L.; Agarwal, P.; Riemer, J.; Bai, X.; Delaney, A.; Akana, M.; JimenezLopez, N.; Bardakjian, T.; et al. A Male with Unilateral Microphthalmia Reveals a Role for TMX3 in Eye Development. PLoS ONE 2010, 5, e10565. [Google Scholar]
- Verma, A.S.; Fitzpatrick, D.R. Anophthalmia and microphthalmia. Orphanet J. Rare Dis. 2007, 2, 47. [Google Scholar] [CrossRef]
- Molinari, M.; Helenius, A. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature 1999, 402, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.C.; Baboo, S.; Lindsay, C.; Brusman, L.; Martinez-Bartolome, S.; Tapia, O.; Zhang, X.; Yates, J.R., 3rd; Gerace, L. Identification of new transmembrane proteins concentrated at the nuclear envelope using organellar proteomics of mesenchymal cells. Nucleus 2019, 10, 126–143. [Google Scholar] [CrossRef]
- Clark, H.F. The Secreted Protein Discovery Initiative (SPDI), a Large-Scale Effort to Identify Novel Human Secreted and Transmembrane Proteins: A Bioinformatics Assessment. Genome Res. 2003, 13, 2265–2270. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, D.F.; Wang, X.; Liang, J.; Sitia, R.; Wang, C.C.; Wang, X. Crystal Structure of the ERp44-Peroxiredoxin 4 Complex Reveals the Molecular Mechanisms of Thiol-Mediated Protein Retention. Structure 2016, 24, 1755–1765. [Google Scholar] [CrossRef]
- Hartill, V.; Szymanska, K.; Sharif, S.M.; Wheway, G.; Johnson, C.A. Meckel–Gruber Syndrome: An Update on Diagnosis, Clinical Management, and Research Advances. Front. Pediatrics 2017, 5, 244. [Google Scholar] [CrossRef]
- Shaheen, R.; Szymanska, K.; Basu, B.; Patel, N.; Ewida, N.; Faqeih, E.; Al Hashem, A.; Derar, N.; Alsharif, H.; Aldahmesh, M.A.; et al. Characterizing the morbid genome of ciliopathies. Genome Biol. 2016, 17, 1–11. [Google Scholar] [CrossRef]
- Radhakrishnan, P.; Nayak, S.S.; Shukla, A.; Lindstrand, A.; Girisha, K.M. Meckel syndrome: Clinical and mutation profile in six fetuses. Clin. Genet. 2019, 96, 560–565. [Google Scholar] [CrossRef]
- Ridnõi, K.; Šois, M.; Vaidla, E.; Pajusalu, S.; Kelder, L.; Reimand, T.; Õunap, K. A prenatally diagnosed case of Meckel–Gruber syndrome with novel compound heterozygous pathogenic variants in the TXNDC15 gene. Mol. Genet. Genom. Med. 2019, 7, e614. [Google Scholar] [CrossRef] [PubMed]
- Leightner, A.C.; Hommerding, C.J.; Peng, Y.; Salisbury, J.L.; Gainullin, V.G.; Czarnecki, P.G.; Sussman, C.R.; Harris, P.C. The Meckel syndrome protein meckelin (TMEM67) is a key regulator of cilia function but is not required for tissue planar polarity. Hum. Mol. Genet. 2013, 22, 2024–2040. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poet, G.J.; Oka, O.B.; van Lith, M.; Cao, Z.; Robinson, P.J.; Pringle, M.A.; Arner, E.S.; Bulleid, N.J. Cytosolic thioredoxin reductase 1 is required for correct disulfide formation in the ER. EMBO J. 2017, 36, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Ali Khan, H.; Mutus, B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front. Chem. 2014, 2, 70. [Google Scholar] [CrossRef]
- Tsai, B.; Rodighiero, C.; Lencer, W.I.; Rapoport, T.A. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 2001, 104, 937–948. [Google Scholar] [CrossRef]
- Anelli, T. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 2002, 21, 835–844. [Google Scholar] [CrossRef]
- Anelli, T.; Alessio, M.; Bachi, A.; Bergamelli, L.; Bertoli, G.; Camerini, S.; Mezghrani, A.; Ruffato, E.; Simmen, T.; Sitia, R. Thiol-mediated protein retention in the endoplasmic reticulum: The role of ERp44. EMBO J. 2003, 22, 5015–5022. [Google Scholar] [CrossRef]
| Protein | TRX-Like Domains | Active Site | Activities | Biological Functions |
|---|---|---|---|---|
| TMX1 | a | CPAC | R | Protein folding and ERAD Ca2+ flux regulation |
| TMX2 | a | SNDC | ? | Nuclear protein import Ca2+ flux regulation |
| TMX3 | abb’ | CGHC | O | ? |
| TMX4 | a | CPSC | R | Protein folding |
| TMX5 | a | CRFS | ? | ? |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerra, C.; Molinari, M. Thioredoxin-Related Transmembrane Proteins: TMX1 and Little Brothers TMX2, TMX3, TMX4 and TMX5. Cells 2020, 9, 2000. https://doi.org/10.3390/cells9092000
Guerra C, Molinari M. Thioredoxin-Related Transmembrane Proteins: TMX1 and Little Brothers TMX2, TMX3, TMX4 and TMX5. Cells. 2020; 9(9):2000. https://doi.org/10.3390/cells9092000
Chicago/Turabian StyleGuerra, Concetta, and Maurizio Molinari. 2020. "Thioredoxin-Related Transmembrane Proteins: TMX1 and Little Brothers TMX2, TMX3, TMX4 and TMX5" Cells 9, no. 9: 2000. https://doi.org/10.3390/cells9092000
APA StyleGuerra, C., & Molinari, M. (2020). Thioredoxin-Related Transmembrane Proteins: TMX1 and Little Brothers TMX2, TMX3, TMX4 and TMX5. Cells, 9(9), 2000. https://doi.org/10.3390/cells9092000





