The Sub-Nuclear Localization of RNA-Binding Proteins in KSHV-Infected Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Viruses
2.2. Immunofluorescence
2.3. Western Blotting
2.4. RNA Fluorescence In Situ Hybridization (FISH)
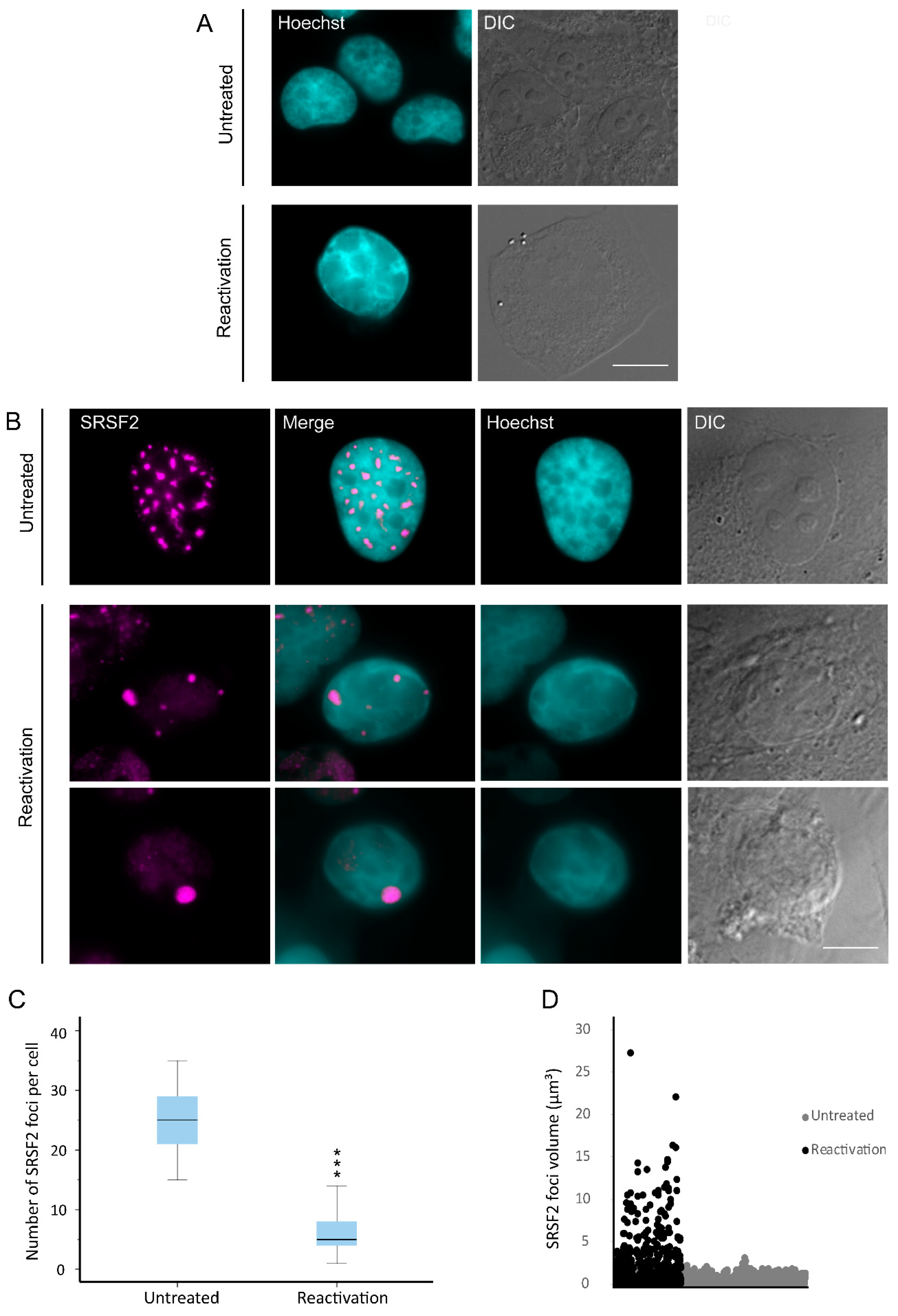
2.5. Image Analysis and Statistical Analysis
3. Results
3.1. Nuclear Speckles Aggregate at Condensed Chromatin Upon Lytic Reactivation of KSHV
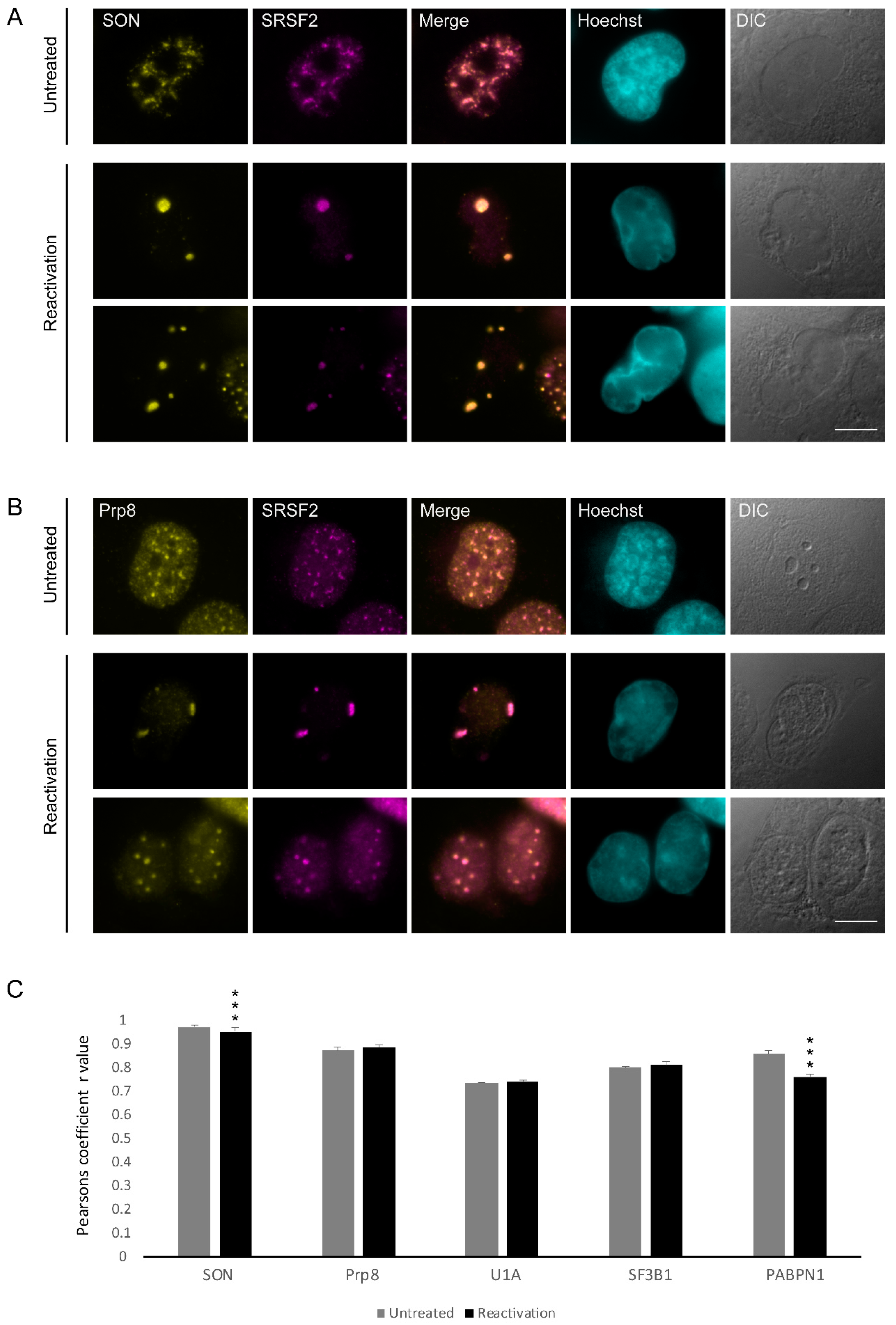
3.2. Changes in the Distribution of Splicing Factors during Lytic Reactivation of KSHV Infection
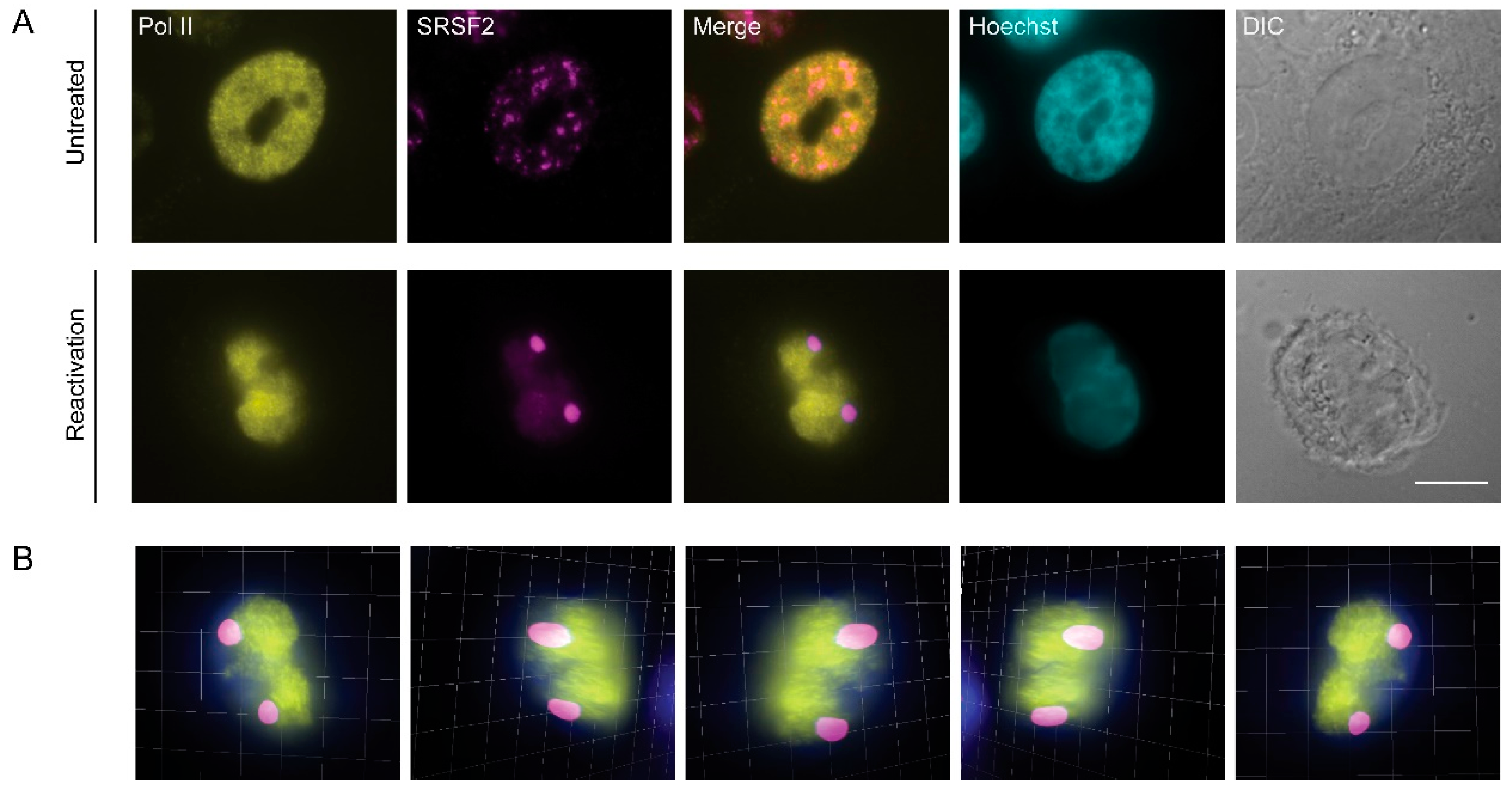
3.3. RNA Pol II Is Not Found in Nuclear Speckles During Lytic Reactivation of KSHV Infection
3.4. Changes in the Distribution of mRNA Export Factors During Lytic Reactivation of KSHV Infection
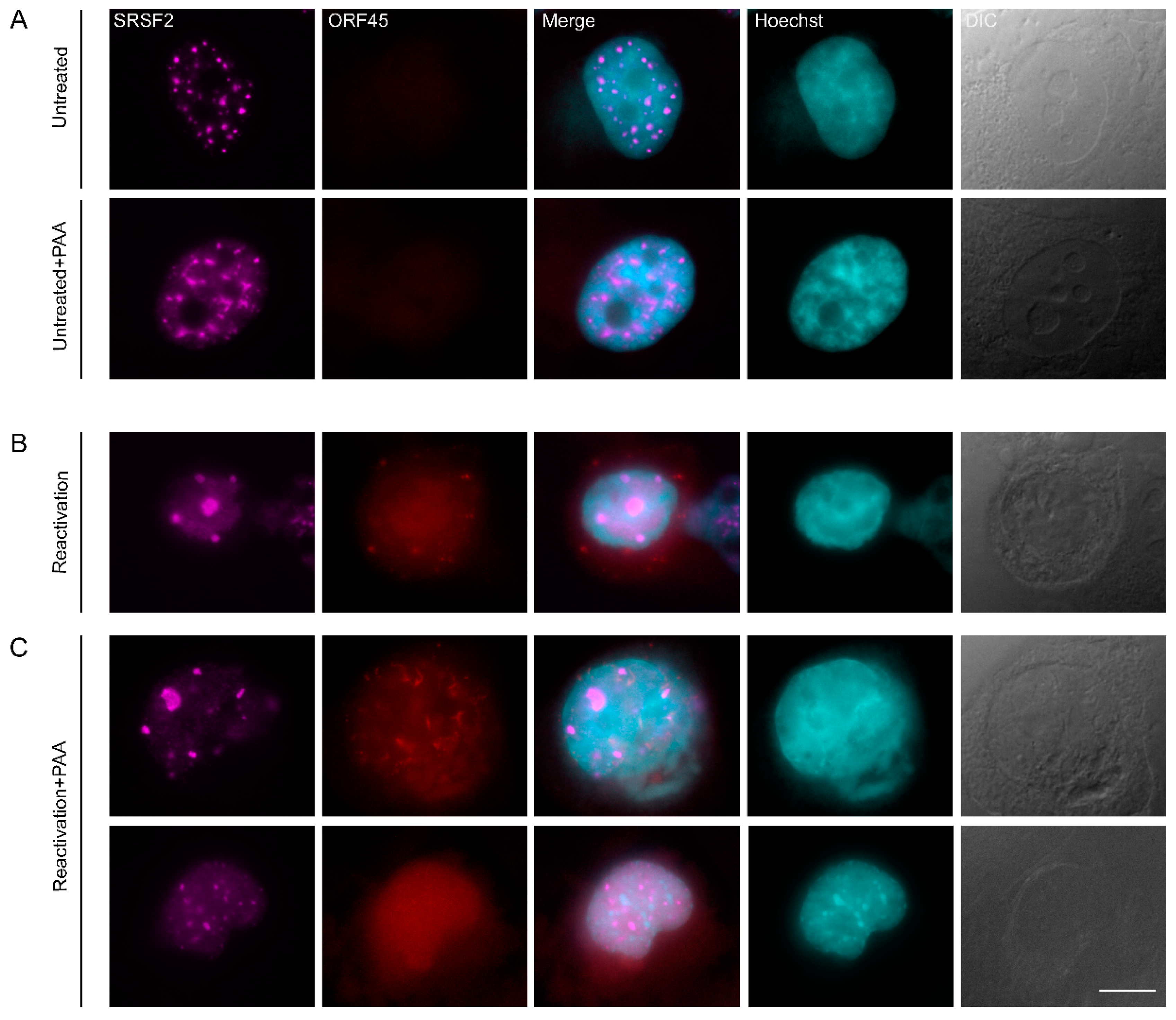
3.5. Inhibition of KSHV DNA Replication Did Not Impair Nuclear Speckle Reorganization Upon Lytic Induction
3.6. Poly(A)+ RNA Nuclear Distribution Changes during Lytic Reactivation of KSHV Infection
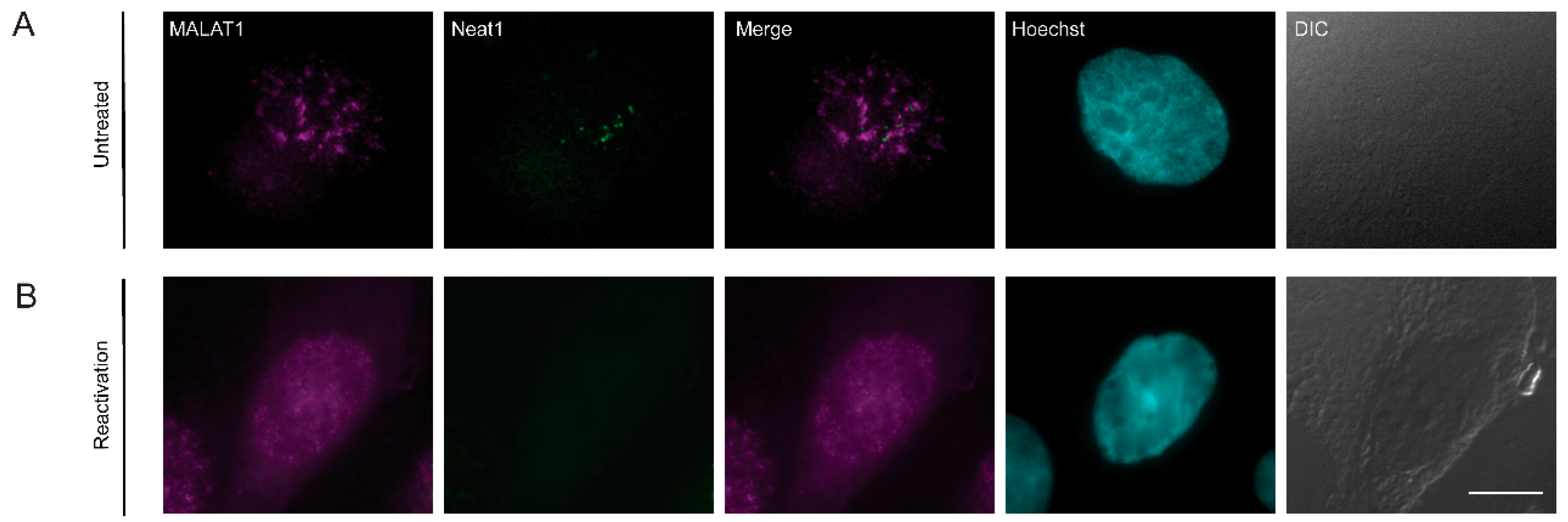
3.7. The Localization of Viral mRNAs during Lytic Reactivation of KSHV Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi’s sarcoma and its associated herpesvirus. Nat. Rev. Cancer 2010, 10, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karass, M.; Grossniklaus, E.; Seoud, T.; Jain, S.; Goldstein, D.A. Kaposi Sarcoma Inflammatory Cytokine Syndrome (KICS): A Rare but Potentially Treatable Condition. Oncologist 2017, 22, 623–625. [Google Scholar] [CrossRef] [Green Version]
- Mariggio, G.; Koch, S.; Schulz, T.F. Kaposi sarcoma herpesvirus pathogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goncalves, P.H.; Ziegelbauer, J.; Uldrick, T.S.; Yarchoan, R. Kaposi sarcoma herpesvirus-associated cancers and related diseases. Curr. Opin. HIV AIDS 2017, 12, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Calabro, M.L.; Sarid, R. Human Herpesvirus 8 and Lymphoproliferative Disorders. Mediterr. J. Hematol. Infect. Dis. 2018, 10, e2018061. [Google Scholar] [CrossRef]
- Dollery, S.J. Towards Understanding KSHV Fusion and Entry. Viruses 2019, 11, 1073. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Majerciak, V.; Zheng, Z.M.; Lan, K. Towards Better Understanding of KSHV Life Cycle: From Transcription and Posttranscriptional Regulations to Pathogenesis. Virol. Sin. 2019, 34, 135–161. [Google Scholar] [CrossRef] [Green Version]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef]
- Purushothaman, P.; Dabral, P.; Gupta, N.; Sarkar, R.; Verma, S.C. KSHV Genome Replication and Maintenance. Front. Microbiol. 2016, 7, 54. [Google Scholar] [CrossRef]
- Hu, J.; Garber, A.C.; Renne, R. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 2002, 76, 11677–11687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, H.; Verma, S.C.; Lampson, M.A.; Cai, Q.; Robertson, E.S. Kaposi’s sarcoma-associated herpesvirus-encoded LANA can interact with the nuclear mitotic apparatus protein to regulate genome maintenance and segregation. J. Virol. 2008, 82, 6734–6746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson-Holley, M.; Colgrove, R.C.; Nalepa, G.; Harper, J.W.; Knipe, D.M. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 2005, 79, 12840–12851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Cha, S.; Jang, J.H.; Kim, Y.; Seo, T. Kaposis sarcoma-associated herpesvirus ORF36 protein induces chromosome condensation and phosphorylation of histone H3. Acta Virol. 2013, 57, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.P.; Lyu, Y.; Chuang, F.; Nakano, K.; Izumiya, C.; Jin, D.; Campbell, M.; Izumiya, Y. Kaposi’s Sarcoma-Associated Herpesvirus Hijacks RNA Polymerase II to Create a Viral Transcriptional Factory. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Stanek, D.; Fox, A.H. Nuclear bodies: News insights into structure and function. Curr. Opin. Cell Biol. 2017, 46, 94–101. [Google Scholar] [CrossRef]
- Lamond, A.I.; Spector, D.L. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003, 4, 605–612. [Google Scholar] [CrossRef]
- Hall, L.L.; Smith, K.P.; Byron, M.; Lawrence, J.B. Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 2006, 288, 664–675. [Google Scholar] [CrossRef] [Green Version]
- Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Xiong, J.; Wang, D.; Fu, X.D. Pre-mRNA splicing: Where and when in the nucleus. Trends Cell Biol. 2011, 21, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Belmont, A.S. Genome organization around nuclear speckles. Curr. Opin. Genet. Dev. 2019, 55, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Hall, L.L.; Lawrence, J.B. Nuclear hubs built on RNAs and clustered organization of the genome. Curr. Opin. Cell Biol. 2020, 64, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Hasenson, S.E.; Shav-Tal, Y. Speculating on the roles of nuclear speckles: How RNA-Protein nuclear assemblies affect gene expression. Bioessays 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Y.; Wang, Y.; Zhang, L.; Brinkman, E.K.; Adam, S.A.; Goldman, R.; van Steensel, B.; Ma, J.; Belmont, A.S. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Venkata, N.C.; Hernandez Gonzalez, G.A.; Khanna, N.; Belmont, A.S. Gene expression amplification by nuclear speckle association. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Huang, S.; Deerinck, T.J.; Ellisman, M.H.; Spector, D.L. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J. Cell Biol. 1994, 126, 877–899. [Google Scholar] [CrossRef]
- Carter, K.C.; Taneja, K.L.; Lawrence, J.B. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 1991, 115, 1191–1202. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misteli, T.; Caceres, J.F.; Spector, D.L. The dynamics of a pre-mRNA splicing factor in living cells. Nature 1997, 387, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Hochberg-Laufer, H.; Neufeld, N.; Brody, Y.; Nadav-Eliyahu, S.; Ben-Yishay, R.; Shav-Tal, Y. Availability of splicing factors in the nucleoplasm can regulate the release of mRNA from the gene after transcription. PLoS Genet. 2019, 15, e1008459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Ann. Rev. Biochem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Shepard, P.J.; Hertel, K.J. The SR protein family. Genome Biol. 2009, 10, 242. [Google Scholar] [CrossRef] [Green Version]
- Valcarcel, J.; Green, M.R. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 1996, 21, 296–301. [Google Scholar] [CrossRef]
- Misteli, T.; Caceres, J.F.; Clement, J.Q.; Krainer, A.R.; Wilkinson, M.F.; Spector, D.L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998, 143, 297–307. [Google Scholar] [CrossRef] [Green Version]
- Kohtz, J.D.; Jamison, S.F.; Will, C.L.; Zuo, P.; Luhrmann, R.; Garcia-Blanco, M.A.; Manley, J.L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 1994, 368, 119–124. [Google Scholar] [CrossRef]
- Fu, X.D.; Mayeda, A.; Maniatis, T.; Krainer, A.R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc. Natl. Acad. Sci. USA 1992, 89, 11224–11228. [Google Scholar] [CrossRef] [Green Version]
- Roscigno, R.F.; Garcia-Blanco, M.A. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA 1995, 1, 692–706. [Google Scholar]
- Hertel, K.J.; Graveley, B.R. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 2005, 30, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.; Zhou, Y.; Shiue, L.; Coutinho-Mansfield, G.; Li, H.; Qiu, J.; Huang, J.; Yeo, G.W.; Ares, M., Jr.; Fu, X.D. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell 2013, 50, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Myoung, J.; Ganem, D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 2011, 174, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brulois, K.F.; Chang, H.; Lee, A.S.; Ensser, A.; Wong, L.Y.; Toth, Z.; Lee, S.H.; Lee, H.R.; Myoung, J.; Ganem, D.; et al. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef] [Green Version]
- Bergson, S.; Kalt, I.; Itzhak, I.; Brulois, K.F.; Jung, J.U.; Sarid, R. Fluorescent tagging and cellular distribution of the Kaposi’s sarcoma-associated herpesvirus ORF45 tegument protein. J. Virol. 2014, 88, 12839–12852. [Google Scholar] [CrossRef] [Green Version]
- Bolte, S.; Cordelieres, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Vallery, T.K.; Withers, J.B.; Andoh, J.A.; Steitz, J.A. Kaposi’s Sarcoma-Associated Herpesvirus mRNA Accumulation in Nuclear Foci Is Influenced by Viral DNA Replication and Viral Noncoding Polyadenylated Nuclear RNA. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, N.; Spahr, C.S.; Patterson, S.D.; Bubulya, P.; Neuwald, A.F.; Spector, D.L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 2004, 15, 3876–3890. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Takata, H.; Shibahara, K.; Bubulya, A.; Bubulya, P.A. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 2010, 21, 650–663. [Google Scholar] [CrossRef] [Green Version]
- Grainger, R.J.; Beggs, J.D. Prp8 protein: At the heart of the spliceosome. RNA 2005, 11, 533–557. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Spector, D.L. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc. Natl. Acad. Sci. USA 1992, 89, 305–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmo-Fonseca, M.; Pepperkok, R.; Sproat, B.S.; Ansorge, W.; Swanson, M.S.; Lamond, A.I. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 1991, 10, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Gama-Carvalho, M.; Krauss, R.D.; Chiang, L.; Valcarcel, J.; Green, M.R.; Carmo-Fonseca, M. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 1997, 137, 975–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majerciak, V.; Yamanegi, K.; Allemand, E.; Kruhlak, M.; Krainer, A.R.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 2008, 82, 2792–2801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bregman, D.B.; Du, L.; van der Zee, S.; Warren, S.L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 1995, 129, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Mortillaro, M.J.; Blencowe, B.J.; Wei, X.; Nakayasu, H.; Du, L.; Warren, S.L.; Sharp, P.A.; Berezney, R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 1996, 93, 8253–8257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mintz, P.J.; Patterson, S.D.; Neuwald, A.F.; Spahr, C.S.; Spector, D.L. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999, 18, 4308–4320. [Google Scholar] [CrossRef] [Green Version]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000, 19, 6860–6869. [Google Scholar] [CrossRef] [Green Version]
- Kohler, A.; Hurt, E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007, 8, 761–773. [Google Scholar] [CrossRef]
- Viphakone, N.; Sudbery, I.; Griffith, L.; Heath, C.G.; Sims, D.; Wilson, S.A. Co-transcriptional Loading of RNA Export Factors Shapes the Human Transcriptome. Mol. Cell 2019, 75, 310–323 e318. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazy-Titelman, A.; Shav-Tal, Y.; Kehlenbach, R.H. Into the basket and beyond: The journey of mRNA through the nuclear pore complex. Biochem. J. 2020, 477, 23–44. [Google Scholar] [CrossRef]
- Hoyt, C.C.; Bouchard, R.J.; Tyler, K.L. Novel nuclear herniations induced by nuclear localization of a viral protein. J. Virol. 2004, 78, 6360–6369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, P.; Engels, M.; Senn, C.; Tobler, K.; Ziegler, U.; Schraner, E.M.; Loepfe, E.; Ackermann, M.; Mueller, M.; Walther, P. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 2005, 79, 1071–1083. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.C.; Robishaw, E.E.; Overby, L.R. Inhibition of DNA polymerase from herpes simplex virus-infected wi-38 cells by phosphonoacetic Acid. J. Virol. 1975, 15, 1281–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overby, L.R.; Robishaw, E.E.; Schleicher, J.B.; Rueter, A.; Shipkowitz, N.L.; Mao, J.C. Inhibition of herpes simplex virus replication by phosphonoacetic acid. Antimicrob. Agents Chemother. 1974, 6, 360–365. [Google Scholar] [CrossRef] [Green Version]
- Krause, S.; Fakan, S.; Weis, K.; Wahle, E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 1994, 214, 75–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [Green Version]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef] [Green Version]
- Kirshner, J.R.; Lukac, D.M.; Chang, J.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 2000, 74, 3586–3597. [Google Scholar] [CrossRef] [Green Version]
- Majerciak, V.; Yamanegi, K.; Nie, S.H.; Zheng, Z.M. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 2006, 281, 28365–28378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majerciak, V.; Zheng, Z.M. Kaposi’s sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 2009, 14, 1516–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brody, Y.; Neufeld, N.; Bieberstein, N.; Causse, S.Z.; Bohnlein, E.M.; Neugebauer, K.M.; Darzacq, X.; Shav-Tal, Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011, 9, e1000573. [Google Scholar] [CrossRef] [PubMed]
- Darzacq, X.; Shav-Tal, Y.; de Turris, V.; Brody, Y.; Shenoy, S.M.; Phair, R.D.; Singer, R.H. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007, 14, 796–806. [Google Scholar] [CrossRef]
- Majerciak, V.; Lu, M.; Li, X.; Zheng, Z.M. Attenuation of the suppressive activity of cellular splicing factor SRSF3 by Kaposi sarcoma-associated herpesvirus ORF57 protein is required for RNA splicing. RNA 2014, 20, 1747–1758. [Google Scholar] [CrossRef] [Green Version]
- Boyne, J.R.; Colgan, K.J.; Whitehouse, A. Recruitment of the complete hTREX complex is required for Kaposi’s sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 2008, 4, e1000194. [Google Scholar] [CrossRef] [Green Version]
- Malik, P.; Blackbourn, D.J.; Clements, J.B. The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 2004, 279, 33001–33011. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.M.; Green, J.; das Neves, R.P.; Wallace, H.A.; Smith, A.J.; Hughes, J.; Gray, N.; Taylor, S.; Wood, W.G.; Higgs, D.R.; et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 2008, 182, 1083–1097. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.E.; Barghusen, S.C.; Leser, G.P.; Spear, P.G. Redistribution of nuclear ribonucleoprotein antigens during herpes simplex virus infection. J. Cell Biol. 1987, 105, 2069–2082. [Google Scholar] [CrossRef] [Green Version]
- Phelan, A.; Carmo-Fonseca, M.; McLaughlan, J.; Lamond, A.I.; Clements, J.B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. USA 1993, 90, 9056–9060. [Google Scholar] [CrossRef] [Green Version]
- Sandri-Goldin, R.M.; Hibbard, M.K.; Hardwicke, M.A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 1995, 69, 6063–6076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, L.; Godinez, W.J.; Kim, I.H.; Tektonidis, M.; de Lanerolle, P.; Eils, R.; Rohr, K.; Knipe, D.M. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. USA 2011, 108, E136–E144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochberg-Laufer, H.; Schwed-Gross, A.; Neugebauer, K.M.; Shav-Tal, Y. Uncoupling of nucleo-cytoplasmic RNA export and localization during stress. Nucleic Acids Res. 2019, 47, 4778–4797. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, S.; Ip, J.Y.; Shioi, G.; Tripathi, V.; Zong, X.; Hirose, T.; Prasanth, K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012, 18, 1487–1499. [Google Scholar] [CrossRef] [Green Version]
- Fei, J.; Jadaliha, M.; Harmon, T.S.; Li, I.T.S.; Hua, B.; Hao, Q.; Holehouse, A.S.; Reyer, M.; Sun, Q.; Freier, S.M.; et al. Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J. Cell Sci. 2017, 130, 4180–4192. [Google Scholar] [CrossRef] [Green Version]
- O’Keefe, R.T.; Mayeda, A.; Sadowski, C.L.; Krainer, A.R.; Spector, D.L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 1994, 124, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, J.A.; Carmo-Fonseca, M.; Lamond, A.I. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J. Cell Biol. 1994, 126, 11–23. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M.; Tollervey, D.; Pepperkok, R.; Barabino, S.M.; Merdes, A.; Brunner, C.; Zamore, P.D.; Green, M.R.; Hurt, E.; Lamond, A.I. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J. 1991, 10, 195–206. [Google Scholar] [CrossRef]
- Antoniou, M.; Carmo-Fonseca, M.; Ferreira, J.; Lamond, A.I. Nuclear organization of splicing snRNPs during differentiation of murine erythroleukemia cells in vitro. J. Cell Biol. 1993, 123, 1055–1068. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Cohen, M.; Lapter, S.; Dye, B.; Patton, J.G.; Vandekerckhove, J.; Zipori, D. Nuclear relocalization of the pre-mRNA splicing factor PSF during apoptosis involves hyperphosphorylation, masking of antigenic epitopes, and changes in protein interactions. Mol. Biol. Cell 2001, 12, 2328–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shav-Tal, Y.; Lee, B.C.; Bar-Haim, S.; Schori, H.; Zipori, D. Reorganization of nuclear factors during myeloid differentiation. J. Cell. Biochem. 2001, 81, 379–392. [Google Scholar] [CrossRef]
- Rino, J.; Carvalho, T.; Braga, J.; Desterro, J.M.; Luhrmann, R.; Carmo-Fonseca, M. A stochastic view of spliceosome assembly and recycling in the nucleus. PLoS Comput. Biol. 2007, 3, 2019–2031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Wang, L.; Wang, J.; Chen, S.; Shi, M.; Cheng, H. Intronless mRNAs transit through nuclear speckles to gain export competence. J. Cell Biol. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkalay, E.; Gam Ze Letova Refael, C.; Shoval, I.; Kinor, N.; Sarid, R.; Shav-Tal, Y. The Sub-Nuclear Localization of RNA-Binding Proteins in KSHV-Infected Cells. Cells 2020, 9, 1958. https://doi.org/10.3390/cells9091958
Alkalay E, Gam Ze Letova Refael C, Shoval I, Kinor N, Sarid R, Shav-Tal Y. The Sub-Nuclear Localization of RNA-Binding Proteins in KSHV-Infected Cells. Cells. 2020; 9(9):1958. https://doi.org/10.3390/cells9091958
Chicago/Turabian StyleAlkalay, Ella, Chen Gam Ze Letova Refael, Irit Shoval, Noa Kinor, Ronit Sarid, and Yaron Shav-Tal. 2020. "The Sub-Nuclear Localization of RNA-Binding Proteins in KSHV-Infected Cells" Cells 9, no. 9: 1958. https://doi.org/10.3390/cells9091958
APA StyleAlkalay, E., Gam Ze Letova Refael, C., Shoval, I., Kinor, N., Sarid, R., & Shav-Tal, Y. (2020). The Sub-Nuclear Localization of RNA-Binding Proteins in KSHV-Infected Cells. Cells, 9(9), 1958. https://doi.org/10.3390/cells9091958




