Arabidopsis thaliana cbp80, c2h2, and flk Knockout Mutants Accumulate Increased Amounts of Circular RNAs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and RNA Extraction
2.2. Library Preparation and Sequencing
2.3. CircRNA Identification and Quantification
2.4. Transcript-Level Expression and Alternative Splicing Analysis
3. Results
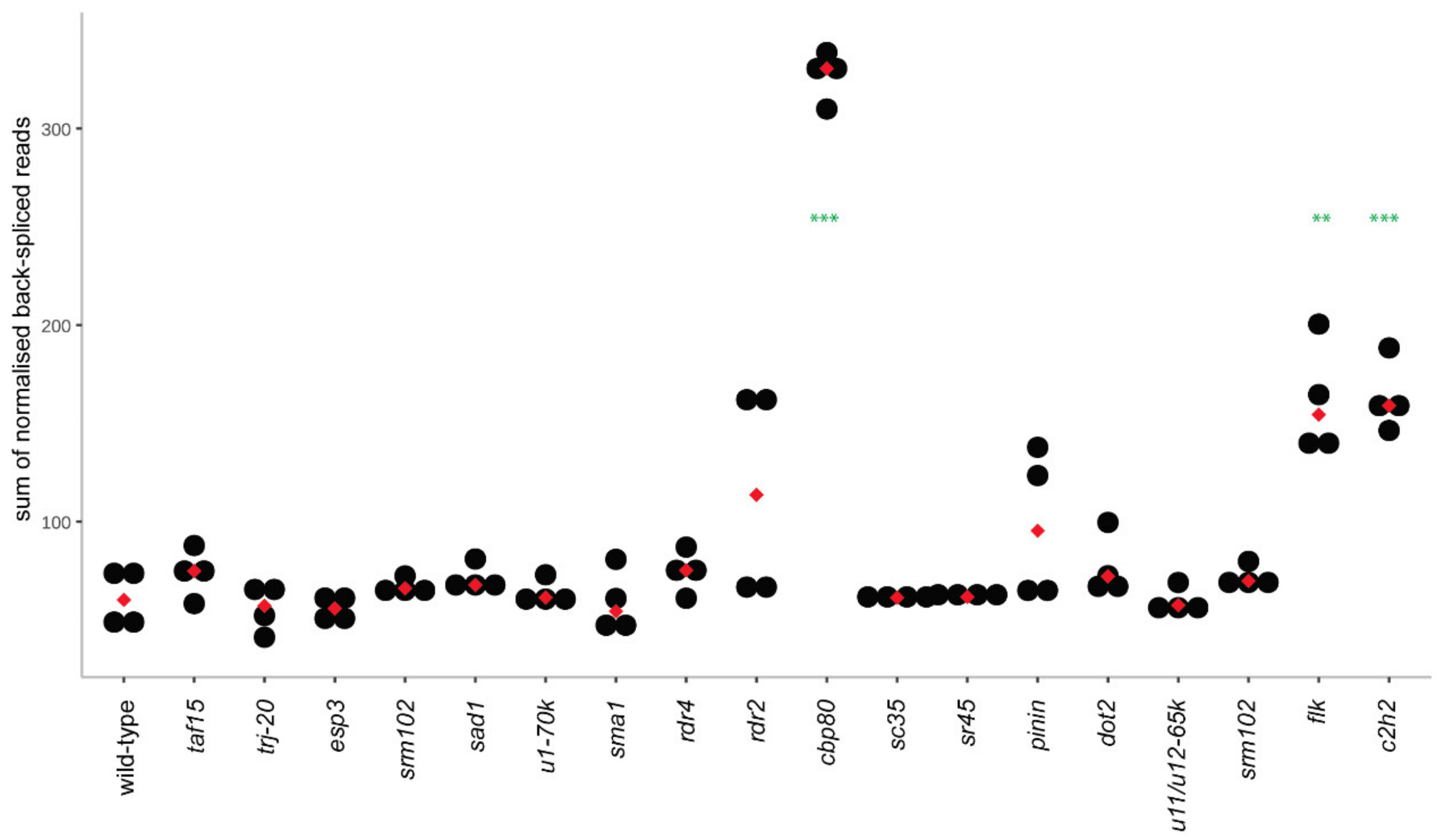
3.1. Selection of the A. thaliana Knock-Out Mutants
3.2. CircRNA Identification in Wild-Type and Mutant Plants
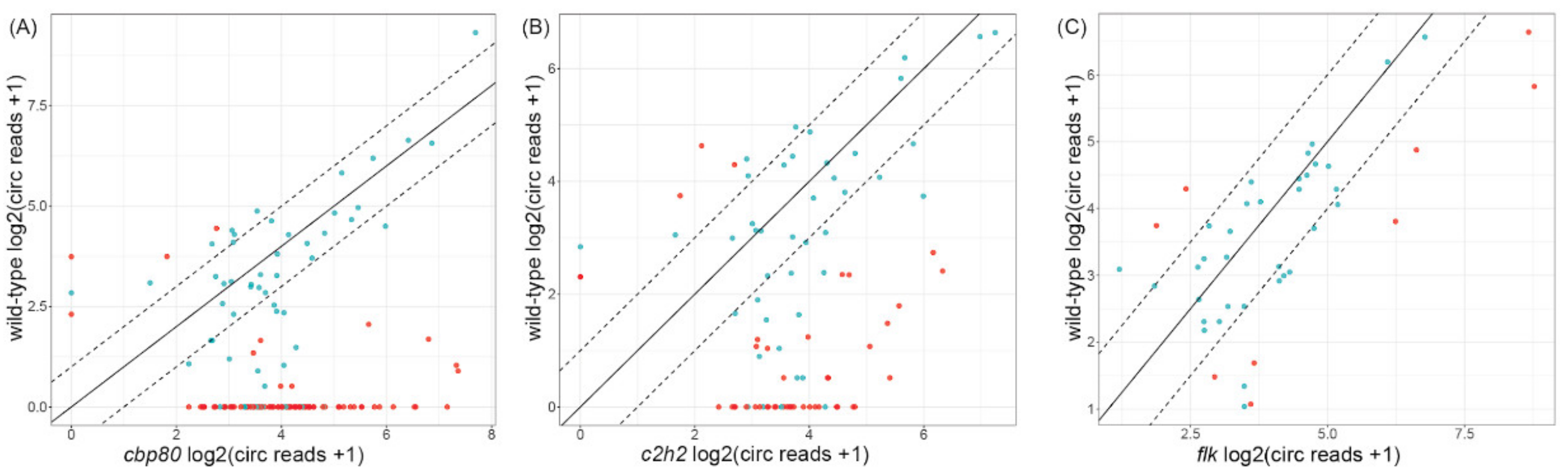
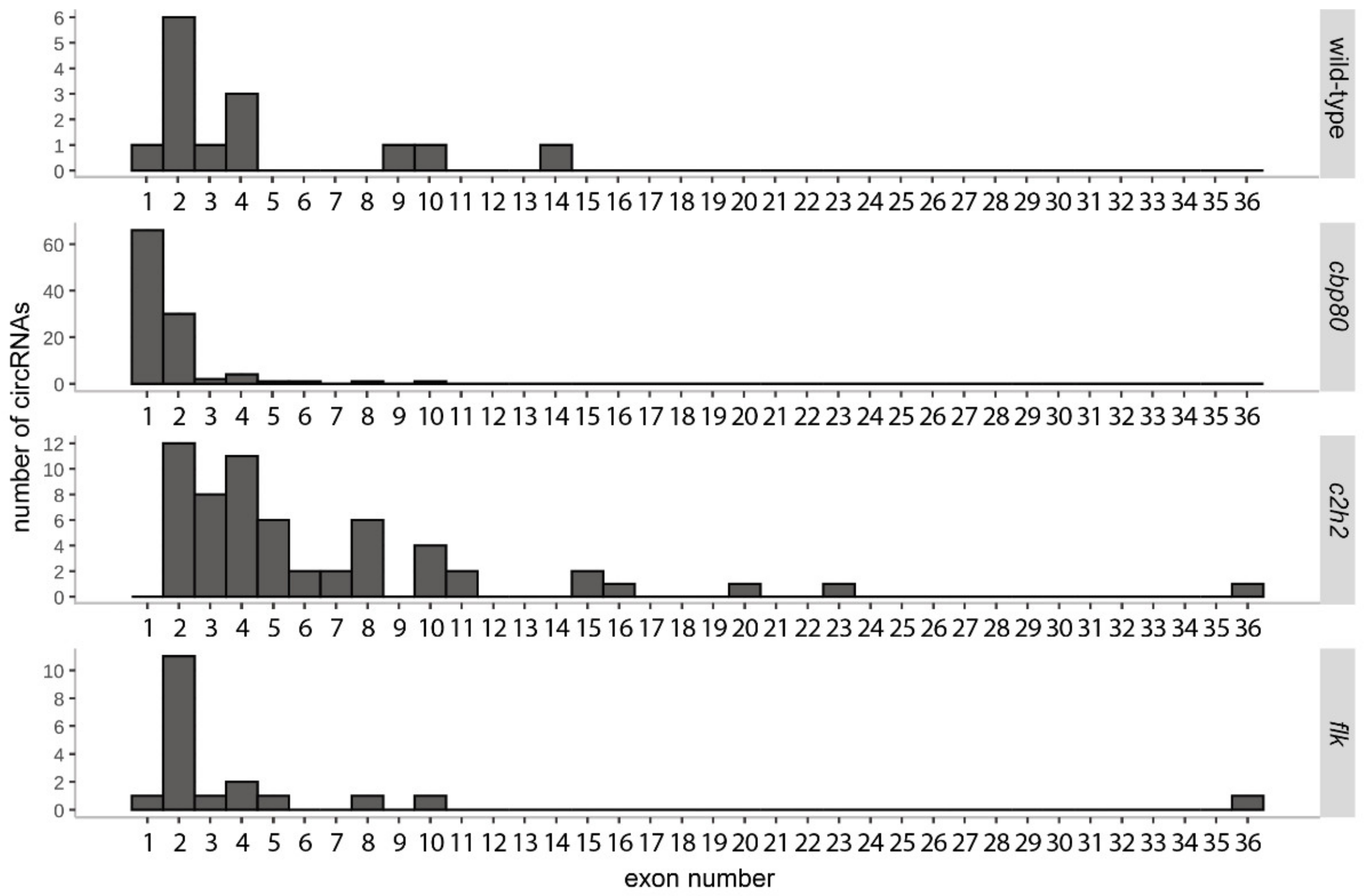
3.3. CircRNAs in the cbp80 Mutant
3.4. CircRNAs in the c2h2 Mutant
3.5. CircRNAs in the flk Mutant
3.6. CircRNA Production as a Form of Alternative Splicing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, P.L.; Bao, Y.; Yee, M.-C.; Barrett, S.P.; Hogan, G.J.; Olsen, M.N.; Dinneny, J.R.; Brown, P.O.; Salzman, J. Circular RNA Is Expressed across the Eukaryotic Tree of Life. PLoS ONE 2014, 9, e90859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2012, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lasda, E.; Parker, R. Circular RNAs: diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, S.; Liu, Z.; Yang, X.; Zhou, J.; Yu, H.; Zhang, R.; Li, H. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018, 414, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; I Jensen, T.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017, 7, 4183–4191. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Peng, Y.; Song, X.; Zheng, Y.; Cheng, H.; Lai, W. circCOL3A1-859267 regulates type I collagen expression by sponging miR-29c in human dermal fibroblasts. Eur. J. Dermatol. EJD 2018, 28, 613–620. [Google Scholar]
- Liu, J.; Li, D.; Luo, H.; Zhu, X. Circular RNAs: The star molecules in cancer. Mol. Asp. Med. 2019, 70, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of circ RNA s. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, D.; E Wilusz, J. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014, 28, 2233–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Wang, L.; Li, S.; Zhang, Q.; Zhang, Q.; Tang, W.; Wang, K.; Song, K.; Sablok, G.; Sun, X.; et al. AtCircDB: a tissue-specific database for Arabidopsis circular RNAs. Briefings Bioinform. 2017, 20, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Wang, B.-B.; Brendel, V. The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 2004, 5, R102. [Google Scholar] [CrossRef] [Green Version]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth Stage–Based Phenotypic Analysis of Arabidopsis. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- I Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 002832. [Google Scholar] [CrossRef] [Green Version]
- ABRC. Available online: http://abrc.osu.edu (accessed on 4 June 2019).
- Hansen, T.B. Improved circRNA Identification by Combining Prediction Algorithms. Front. Cell Dev. Biol. 2018, 6. [Google Scholar] [CrossRef]
- Szabo, L.; Salzman, J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 2016, 17, 679–692. [Google Scholar] [CrossRef] [Green Version]
- Philips, A.; Nowis, K.; Stelmaszczuk, M.; Jackowiak, P.; Podkowiński, J.; Handschuh, L. Only a small fraction of circular RNAs is reproducibly formed in Arabidopsis thaliana seedlings and organs. BMC Plant Biol. 2020. Conditionally Accepted. [Google Scholar]
- Chu, Q.; Zhang, X.; Zhu, X.; Liu, C.; Mao, L.; Ye, C.; Zhu, Q.-H.; Fan, L. PlantcircBase: A Database for Plant Circular RNAs. Mol. Plant 2017, 10, 1126–1128. [Google Scholar] [CrossRef]
- Zhao, W.; Chu, S.; Jiao, Y. Present Scenario of Circular RNAs (circRNAs) in Plants. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Laubinger, S.; Sachsenberg, T.; Zeller, G.; Busch, W.; Lohmann, J.U.; Rätsch, G.; Weigel, D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 8795–8800. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Yang, J.-Y.; Xu, J.; Jang, I.-C.; Prigge, M.J.; Chua, N.-H.; Kurasawa, K.; Matsui, A.; Yokoyama, R.; Kuriyama, T.; et al. Two Cap-Binding Proteins CBP20 and CBP80 are Involved in Processing Primary MicroRNAs. Plant Cell Physiol. 2008, 49, 1634–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raczynska, K.D.; Simpson, C.G.; Ciesiolka, A.; Szewc, L.; Lewandowska, D.; McNicol, J.; Szweykowska-Kulinska, Z.; Brown, J.W.S.; Jarmolowski, A. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2009, 38, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux, V.; Murata, Y.; Young, J.J.; Kwak, J.M.; Mackesy, D.Z.; I Schroeder, J. Localization, Ion Channel Regulation, and Genetic Interactions during Abscisic Acid Signaling of the Nuclear mRNA Cap-Binding Protein, ABH11. Plant Physiol. 2002, 130, 1276–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, J.M.; Breton, G.; I Schroeder, J. mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J. 2007, 50, 1049–1062. [Google Scholar] [CrossRef]
- Kuhn, J.M.; Hugouvieux, V.; I Schroeder, J. mRNA Cap Binding Proteins: Effects on Abscisic Acid Signal Transduction, mRNA Processing, and Microarray Analyses. In Nuclear pre-mRNA Processing in Plants; Springer: Berlin, Germany, 2008; pp. 139–150. [Google Scholar]
- Bush, M.S.; Hutchins, A.P.; Jones, J.D.; Naldrett, M.J.; Jarmolowski, A.; Lloyd, C.W.; Doonan, J.H. Selective recruitment of proteins to 5′ cap complexes during the growth cycle in Arabidopsis. Plant J. 2009, 59, 400–412. [Google Scholar] [CrossRef]
- Liang, N.; Tatomer, D.C.; Luo, Z.; Wu, H.; Yang, L.; Chen, L.-L.; Cherry, S.; E Wilusz, J. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell 2017, 68, 940–954.e3. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Philips, A.; Nowis, K.; Stelmaszczuk, M.; Podkowiński, J.; Handschuh, L.; Jackowiak, P.; Figlerowicz, M. Arabidopsis thaliana cbp80, c2h2, and flk Knockout Mutants Accumulate Increased Amounts of Circular RNAs. Cells 2020, 9, 1937. https://doi.org/10.3390/cells9091937
Philips A, Nowis K, Stelmaszczuk M, Podkowiński J, Handschuh L, Jackowiak P, Figlerowicz M. Arabidopsis thaliana cbp80, c2h2, and flk Knockout Mutants Accumulate Increased Amounts of Circular RNAs. Cells. 2020; 9(9):1937. https://doi.org/10.3390/cells9091937
Chicago/Turabian StylePhilips, Anna, Katarzyna Nowis, Michal Stelmaszczuk, Jan Podkowiński, Luiza Handschuh, Paulina Jackowiak, and Marek Figlerowicz. 2020. "Arabidopsis thaliana cbp80, c2h2, and flk Knockout Mutants Accumulate Increased Amounts of Circular RNAs" Cells 9, no. 9: 1937. https://doi.org/10.3390/cells9091937
APA StylePhilips, A., Nowis, K., Stelmaszczuk, M., Podkowiński, J., Handschuh, L., Jackowiak, P., & Figlerowicz, M. (2020). Arabidopsis thaliana cbp80, c2h2, and flk Knockout Mutants Accumulate Increased Amounts of Circular RNAs. Cells, 9(9), 1937. https://doi.org/10.3390/cells9091937





