Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived microRNA Signatures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort of Healthy Volunteers
2.2. Plasma Preparation
2.3. RNA Extraction and qPCR Analysis
2.4. Determination of Plasma Concentrations
2.5. Statistical Analysis
3. Results
3.1. Anticoagulation Affects Blood-Derived miRNA Signatures
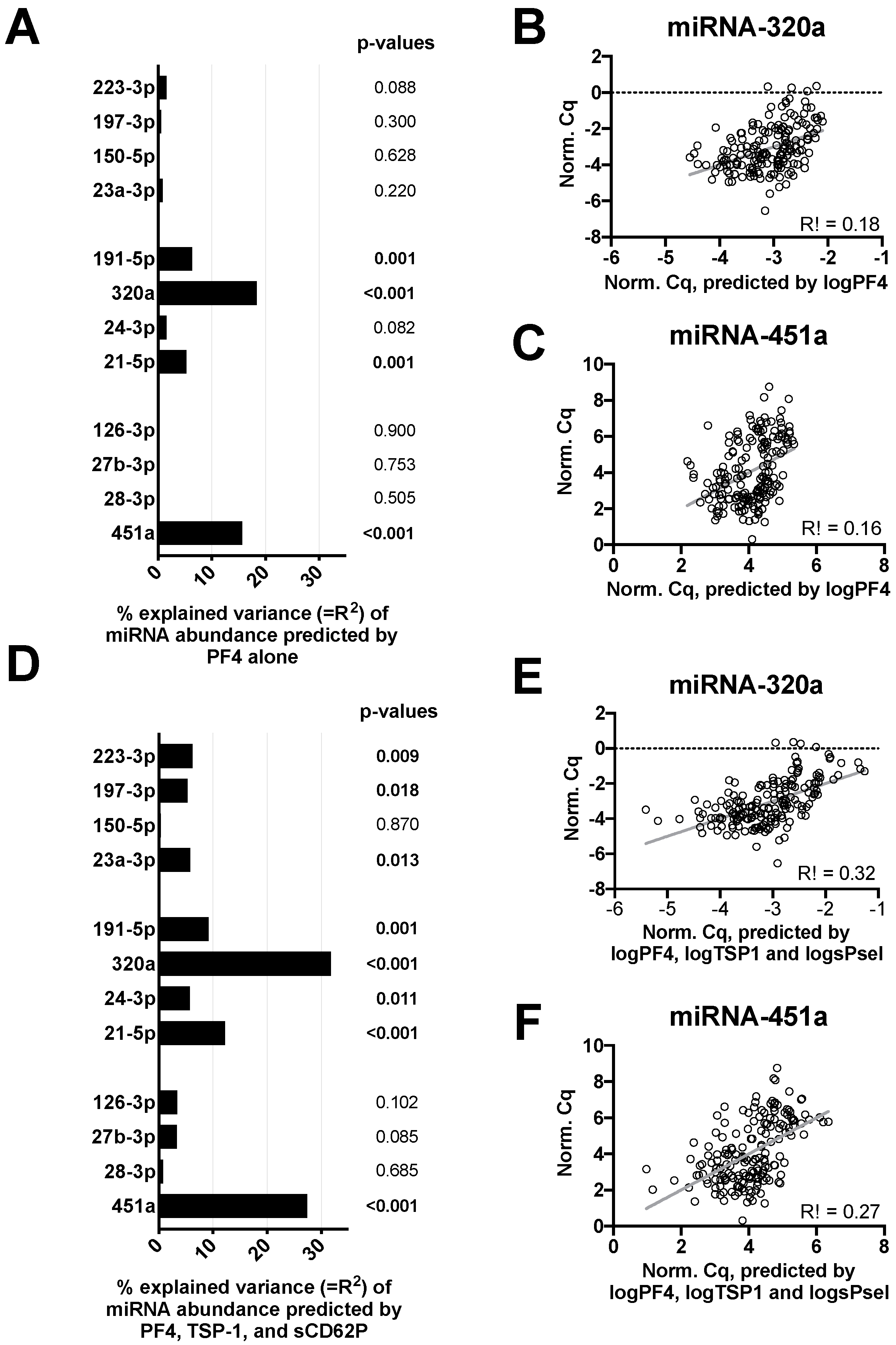
3.2. In Vitro Platelet Activation Increases Variability of microRNA Signatures
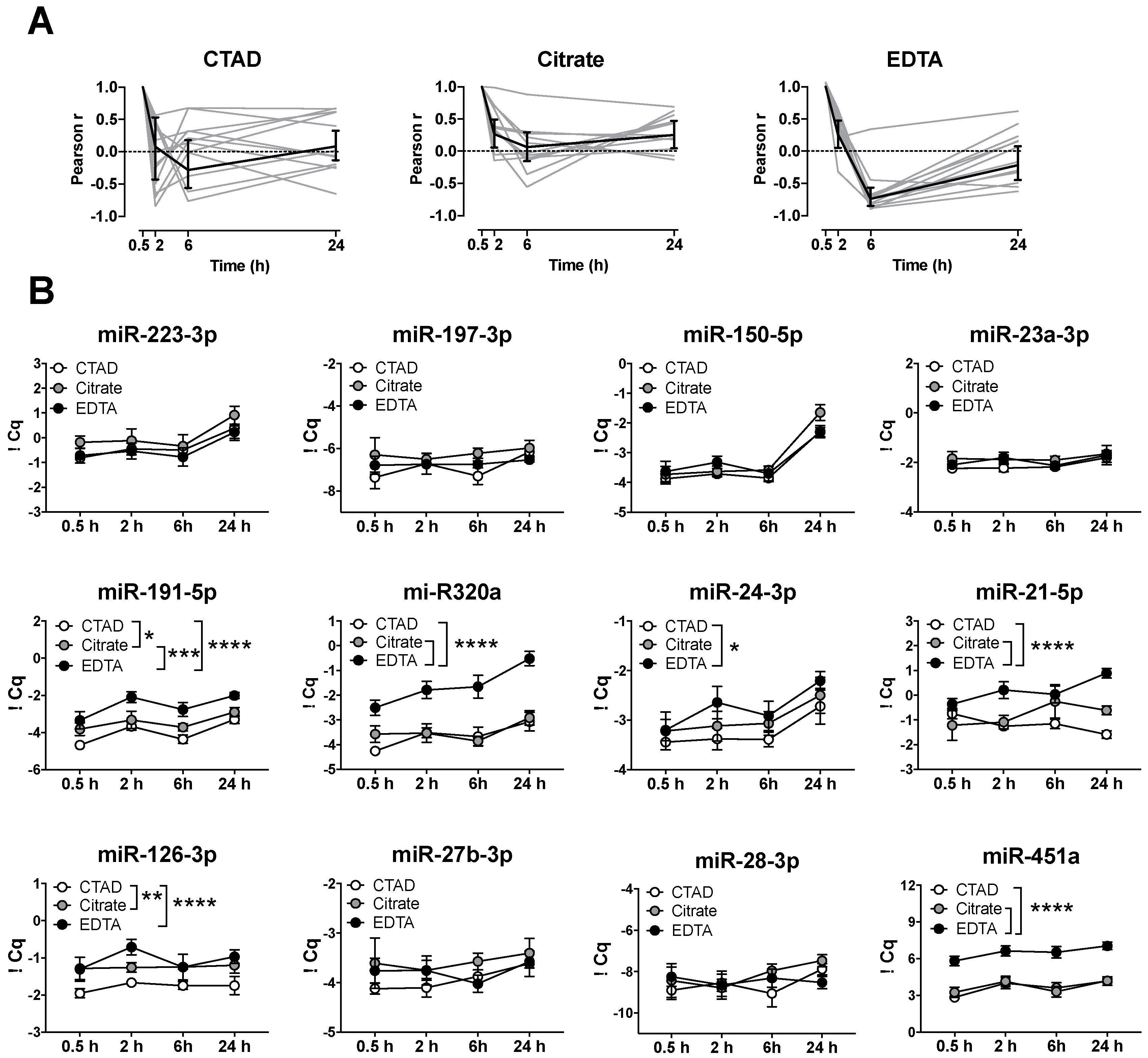
3.3. Storage Time Affects Blood-Derived miRNA Levels
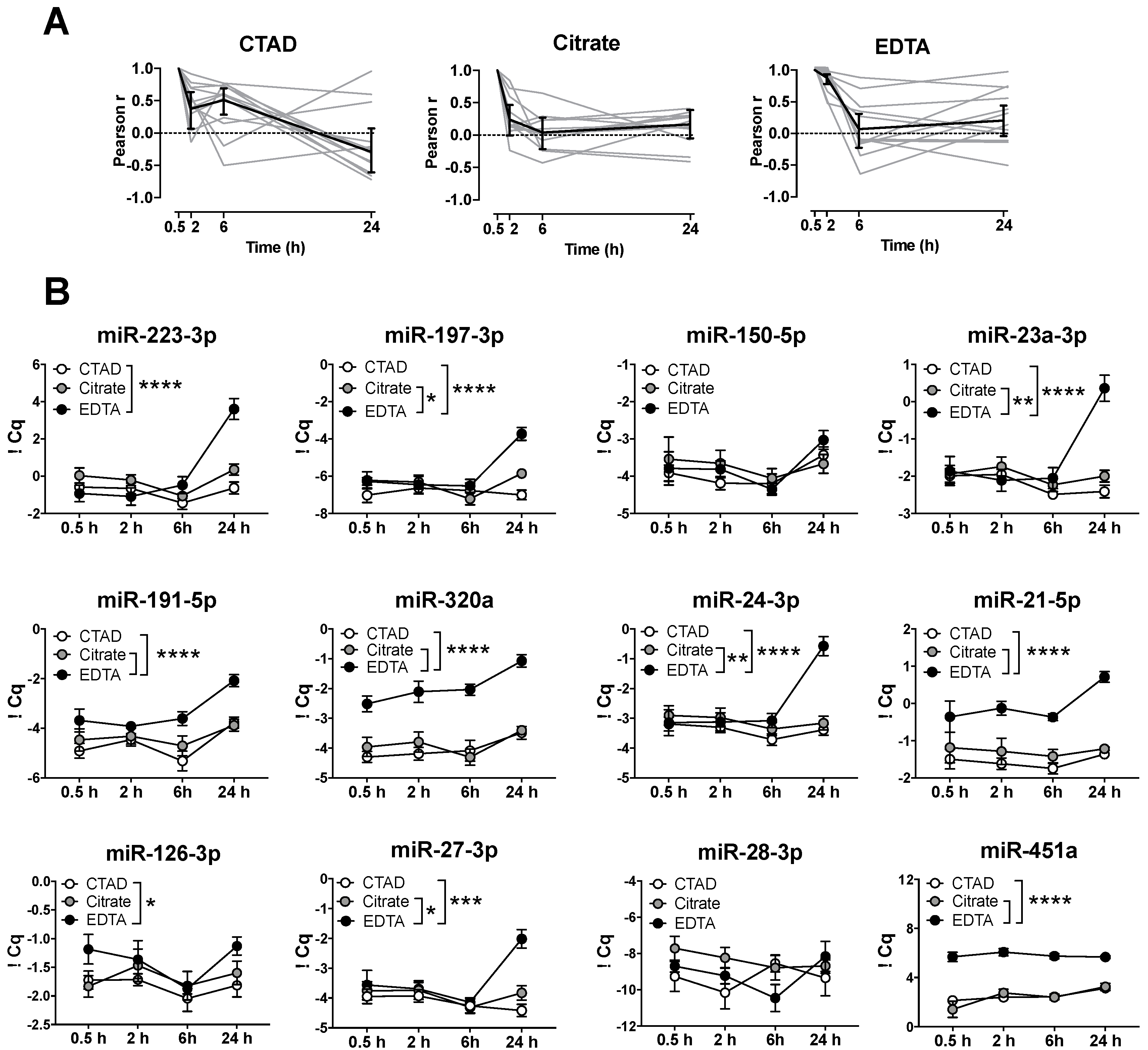
3.4. Storage of Blood at RT Causes A Significant Increase in miRNA Levels after 24 h
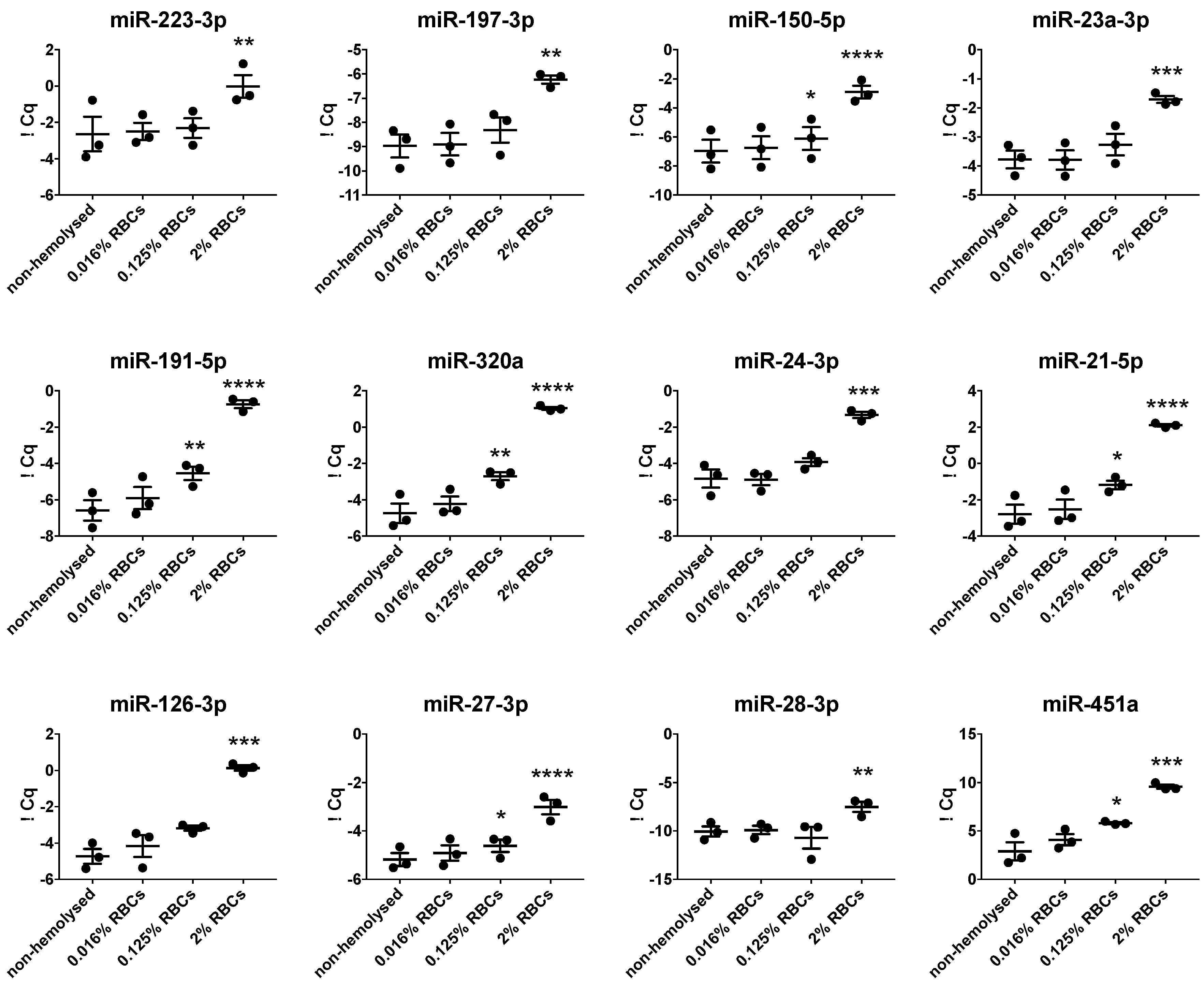
3.5. Hemolysis Affects Blood-Derived miRNA Signatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential biomarkers, therapeutic targets and challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, C.R.; Edelstein, L.C. MicroRNAs in Platelet Physiology and Function. Semin. Thromb. Hemost. 2016, 42, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Sunderland, N.; Skroblin, P.; Barwari, T.; Huntley, R.P.; Lu, R.; Joshi, A.; Lovering, R.C.; Mayr, M. MicroRNA Biomarkers and Platelet Reactivity: The Clot Thickens. Circ. Res. 2017, 120, 418–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholer, N.; Langer, C.; Dohner, H.; Buske, C.; Kuchenbauer, F. Serum microRNAs as a novel class of biomarkers: A comprehensive review of the literature. Exp. Hematol. 2010, 38, 1126–1130. [Google Scholar] [CrossRef]
- Mussbacher, M.; Schrottmaier, W.C.; Salzmann, M.; Brostjan, C.; Schmid, J.A.; Starlinger, P.; Assinger, A. Optimized plasma preparation is essential to monitor platelet-stored molecules in humans. PLoS ONE 2017, 12, e0188921. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, A.J.; Gray, W.D.; Hayek, S.S.; Ko, Y.A.; Thomas, S.; Rooney, K.; Awad, M.; Roback, J.D.; Quyyumi, A.; Searles, C.D. Platelets confound the measurement of extracellular miRNA in archived plasma. Sci. Rep. 2016, 6, 32651. [Google Scholar] [CrossRef] [Green Version]
- Willeit, P.; Zampetaki, A.; Dudek, K.; Kaudewitz, D.; King, A.; Kirkby, N.S.; Crosby-Nwaobi, R.; Prokopi, M.; Drozdov, I.; Langley, S.R.; et al. Circulating microRNAs as novel biomarkers for platelet activation. Circ. Res. 2013, 112, 595–600. [Google Scholar] [CrossRef] [Green Version]
- Carino, A.; De Rosa, S.; Sorrentino, S.; Polimeni, A.; Sabatino, J.; Caiazzo, G.; Torella, D.; Spaccarotella, C.; Mongiardo, A.; Strangio, A.; et al. Modulation of Circulating MicroRNAs Levels during the Switch from Clopidogrel to Ticagrelor. Biomed. Res. Int. 2016. [Google Scholar] [CrossRef] [Green Version]
- Tran, J.Q.D.; Pedersen, O.H.; Larsen, M.L.; Grove, E.L.; Kristensen, S.D.; Hvas, A.M.; Nissen, P.H. Platelet microRNA expression and association with platelet maturity and function in patients with essential thrombocythemia. Platelets 2020, 31, 365–372. [Google Scholar] [CrossRef]
- Bao, H.; Chen, Y.X.; Huang, K.; Zhuang, F.; Bao, M.; Han, Y.; Chen, X.H.; Shi, Q.; Yao, Q.P.; Qi, Y.X. Platelet-derived microparticles promote endothelial cell proliferation in hypertension via miR-142-3p. FASEB J. 2018, 32, 3912–3923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Qi, X.; Chen, H.; Li, M.; Gu, J.; Liu, C.; Xue, H.; Wang, L.; Geng, Y.; Qi, P.; et al. Expression of miRNA-26a in platelets is associated with clopidogrel resistance following coronary stenting. Exp. Med. 2016, 12, 518–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dangwal, S.; Stratmann, B.; Bang, C.; Lorenzen, J.M.; Kumarswamy, R.; Fiedler, J.; Falk, C.S.; Scholz, C.J.; Thum, T.; Tschoepe, D. Impairment of Wound Healing in Patients With Type 2 Diabetes Mellitus Influences Circulating MicroRNA Patterns via Inflammatory Cytokines. Arter. Thromb. Vasc. Biol. 2015, 35, 1480–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Ma, J.; Wang, Q.; Wu, F.; Ping, J.; Ming, L. Combination of Circulating miRNA-320a/b and D-Dimer Improves Diagnostic Accuracy in Deep Vein Thrombosis Patients. Med. Sci. Monit. 2018, 24, 2031–2037. [Google Scholar] [CrossRef]
- Maegdefessel, L.; Spin, J.M.; Raaz, U.; Eken, S.M.; Toh, R.; Azuma, J.; Adam, M.; Nakagami, F.; Heymann, H.M.; Chernogubova, E.; et al. miR-24 limits aortic vascular inflammation and murine abdominal aneurysm development. Nat. Commun. 2014, 5, 5214. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Sundquist, K.; Svensson, P.J.; Rastkhani, H.; Palmer, K.; Memon, A.A.; Sundquist, J.; Zoller, B. Association of recurrent venous thromboembolism and circulating microRNAs. Clin. Epigenetics 2019, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Xie, N.; Cui, H.; Banerjee, S.; Tan, Z.; Salomao, R.; Fu, M.; Abraham, E.; Thannickal, V.J.; Liu, G. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J. Immunol. 2014, 193, 327–334. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; He, S.; Liu, B.; Liu, C. MiR-27-3p regulates TLR2/4-dependent mouse alveolar macrophage activation by targetting PPARgamma. Clin. Sci. 2018, 132, 943–958. [Google Scholar] [CrossRef]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [Green Version]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef]
- Wu, F.; Guo, N.J.; Tian, H.; Marohn, M.; Gearhart, S.; Bayless, T.M.; Brant, S.R.; Kwon, J.H. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm. Bowel Dis. 2011, 17, 241–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hur, W.; Lee, J.H.; Kim, S.W.; Kim, J.H.; Bae, S.H.; Kim, M.; Hwang, D.; Kim, Y.S.; Park, T.; Um, S.J.; et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 2015, 64, 265–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blondal, T.; Jensby Nielsen, S.; Baker, A.; Andreasen, D.; Mouritzen, P.; Wrang Teilum, M.; Dahlsveen, I.K. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Mayr, M. Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb. Haemost. 2012, 108, 592–598. [Google Scholar] [PubMed] [Green Version]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [Green Version]
- Schulte, C.; Molz, S.; Appelbaum, S.; Karakas, M.; Ojeda, F.; Lau, D.M.; Hartmann, T.; Lackner, K.J.; Westermann, D.; Schnabel, R.B.; et al. miRNA-197 and miRNA-223 Predict Cardiovascular Death in a Cohort of Patients with Symptomatic Coronary Artery Disease. PLoS ONE 2015, 10, e0145930. [Google Scholar] [CrossRef] [Green Version]
- Emmrich, S.; Henke, K.; Hegermann, J.; Ochs, M.; Reinhardt, D.; Klusmann, J.H. miRNAs can increase the efficiency of ex vivo platelet generation. Ann. Hematol. 2012, 91, 1673–1684. [Google Scholar] [CrossRef]
- Gu, Y.; Ampofo, E.; Menger, M.D.; Laschke, M.W. miR-191 suppresses angiogenesis by activation of NF-kappaB signaling. FASEB J. 2017, 31, 3321–3333. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.Y.; Chen, Y.L.; Jao, Y.C.; Hsieh, I.S.; Chang, K.C.; Hong, T.M. miR-320 regulates tumor angiogenesis driven by vascular endothelial cells in oral cancer by silencing neuropilin 1. Angiogenesis 2014, 17, 247–260. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.J.; Liu, T.; Zhang, H.; Yang, S.J. Plasma microRNA-21 is a potential diagnostic biomarker of acute myocardial infarction. Eur. Rev. Med. Pharm. Sci. 2016, 20, 323–329. [Google Scholar]
- Xie, W.; Li, L.; Zhang, M.; Cheng, H.P.; Gong, D.; Lv, Y.C.; Yao, F.; He, P.P.; Ouyang, X.P.; Lan, G.; et al. MicroRNA-27 Prevents Atherosclerosis by Suppressing Lipoprotein Lipase-Induced Lipid Accumulation and Inflammatory Response in Apolipoprotein E Knockout Mice. PLoS ONE 2016, 11, e0157085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girardot, M.; Pecquet, C.; Boukour, S.; Knoops, L.; Ferrant, A.; Vainchenker, W.; Giraudier, S.; Constantinescu, S.N. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood 2010, 116, 437–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Tan, Y.S.; Cheng, W.C.; Kingsbury, T.J.; Heimfeld, S.; Civin, C.I. MIR144 and MIR451 regulate human erythropoiesis via RAB14. Br. J. Haematol. 2015, 168, 583–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leecharoenkiat, K.; Tanaka, Y.; Harada, Y.; Chaichompoo, P.; Sarakul, O.; Abe, Y.; Smith, D.R.; Fucharoen, S.; Svasti, S.; Umemura, T. Plasma microRNA-451 as a novel hemolytic marker for beta0-thalassemia/HbE disease. Mol. Med. Rep. 2017, 15, 2495–2502. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [Green Version]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.J.; Linnstaedt, S.; Palma, J.; Park, J.C.; Ntrivalas, E.; Kwak-Kim, J.Y.; Gilman-Sachs, A.; Beaman, K.; Hastings, M.L.; Martin, J.N.; et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J. Mol. Diagn. 2012, 14, 71–80. [Google Scholar] [CrossRef] [Green Version]
- Nagalla, S.; Shaw, C.; Kong, X.; Kondkar, A.A.; Edelstein, L.C.; Ma, L.; Chen, J.; McKnight, G.S.; Lopez, J.A.; Yang, L.; et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood 2011, 117, 5189–5197. [Google Scholar] [CrossRef]
- Lieb, V.; Weigelt, K.; Scheinost, L.; Fischer, K.; Greither, T.; Marcou, M.; Theil, G.; Klocker, H.; Holzhausen, H.J.; Lai, X.; et al. Serum levels of miR-320 family members are associated with clinical parameters and diagnosis in prostate cancer patients. Oncotarget 2018, 9, 10402–10416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Best, M.G.; Wesseling, P.; Wurdinger, T. Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018, 78, 3407–3412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foye, C.; Yan, I.K.; David, W.; Shukla, N.; Habboush, Y.; Chase, L.; Ryland, K.; Kesari, V.; Patel, T. Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS ONE 2017, 12, e0189165. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| miRNA | Source | Function | Reference |
|---|---|---|---|
| miR-223-3p | platelets, hematopoietic cells | associated with decreased responsiveness to clopidogrel treatment in patients with coronary artery disease, miRNA-223 knockout in mice are still controversial | [26] |
| miR-197-3p | platelets, hematopoietic cells | predication of cardiovascular death in patients that suffered from symptomatic coronary artery disease | [27] |
| miR-150-5p | platelets, leukocytes | platelet maturation, decreased after switch from clopidogrel to ticagrelor | [9] |
| miR-23a-3p | platelets, hematopoietic cells | repressor of megakaryocyte development and differentiation | [28] |
| miR-191-5p | endothelial cells, platelets | identified as inhibitor of blood vessel development | [29] |
| miR-320a | cardiomyocytes, endothelial cells | cardiomyocyte apoptosis in cardiac ischemia/reperfusion injury, suppression of proliferation and migration of endothelial cells | [30] |
| miR-24-3p | macrophages, smooth muscle cells, platelets | inverse correlation with aortic aneurism | [15] |
| miR-21-5p | endothelial cells, smooth muscle cells, platelets, cardiomyocytes | increased in patients with acute myocardial infarct and angina pectoris | [31] |
| miR-126-3p | endothelial cells, platelets | vessel integrity, angiogenesis, wound repair, apoptosis | [19] |
| miR-27-3p | endothelial cells, adipocytes, platelets | adipogenesis, regulation of endothelial–mesenchymal transition, thrombin-induced synthesis of thrombospondin-1 | [32] |
| miR-28-3p | cardiomyocytes, platelets | regulate the thrombopoietin receptor, marker for the diagnosis of pulmonary embolisms, promotes myocardial ischemia | [33] |
| miR-451a | erythrocytes | associated with hemolysis and erythropoiesis | [34,35] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mussbacher, M.; Krammer, T.L.; Heber, S.; Schrottmaier, W.C.; Zeibig, S.; Holthoff, H.-P.; Pereyra, D.; Starlinger, P.; Hackl, M.; Assinger, A. Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived microRNA Signatures. Cells 2020, 9, 1915. https://doi.org/10.3390/cells9081915
Mussbacher M, Krammer TL, Heber S, Schrottmaier WC, Zeibig S, Holthoff H-P, Pereyra D, Starlinger P, Hackl M, Assinger A. Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived microRNA Signatures. Cells. 2020; 9(8):1915. https://doi.org/10.3390/cells9081915
Chicago/Turabian StyleMussbacher, Marion, Teresa L. Krammer, Stefan Heber, Waltraud C. Schrottmaier, Stephan Zeibig, Hans-Peter Holthoff, David Pereyra, Patrick Starlinger, Matthias Hackl, and Alice Assinger. 2020. "Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived microRNA Signatures" Cells 9, no. 8: 1915. https://doi.org/10.3390/cells9081915
APA StyleMussbacher, M., Krammer, T. L., Heber, S., Schrottmaier, W. C., Zeibig, S., Holthoff, H.-P., Pereyra, D., Starlinger, P., Hackl, M., & Assinger, A. (2020). Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived microRNA Signatures. Cells, 9(8), 1915. https://doi.org/10.3390/cells9081915