Human Cardiac Organoids for Modeling Genetic Cardiomyopathy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.1.1. Human Cardiac Microvascular Endothelial Cells
2.1.2. Human Cardiac Fibroblast
2.1.3. Cor.4U Cardiomyocytes
2.1.4. Human Induced Pluripotent Stem Cell Lines
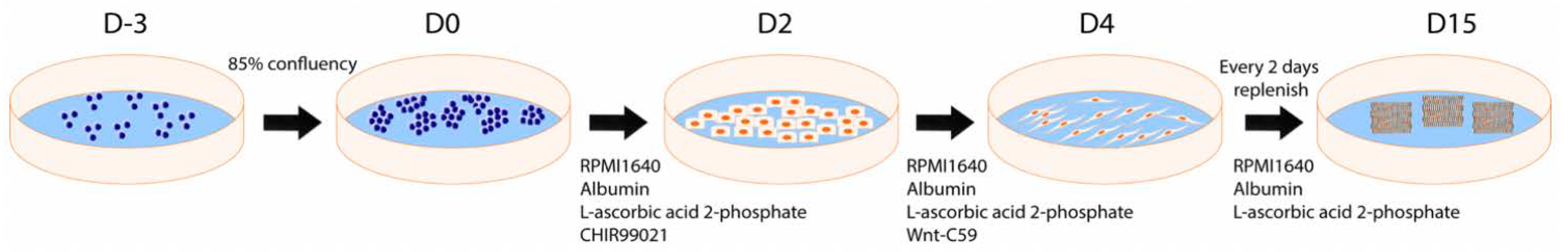
2.2. Cardiomyocyte Differentiation
2.2.1. STEMdiff Differentiation Protocol
2.2.2. CDM3 Differentiation Protocol
2.3. Flow Cytometry
2.4. Real-Time Quantitative PCR Analysis
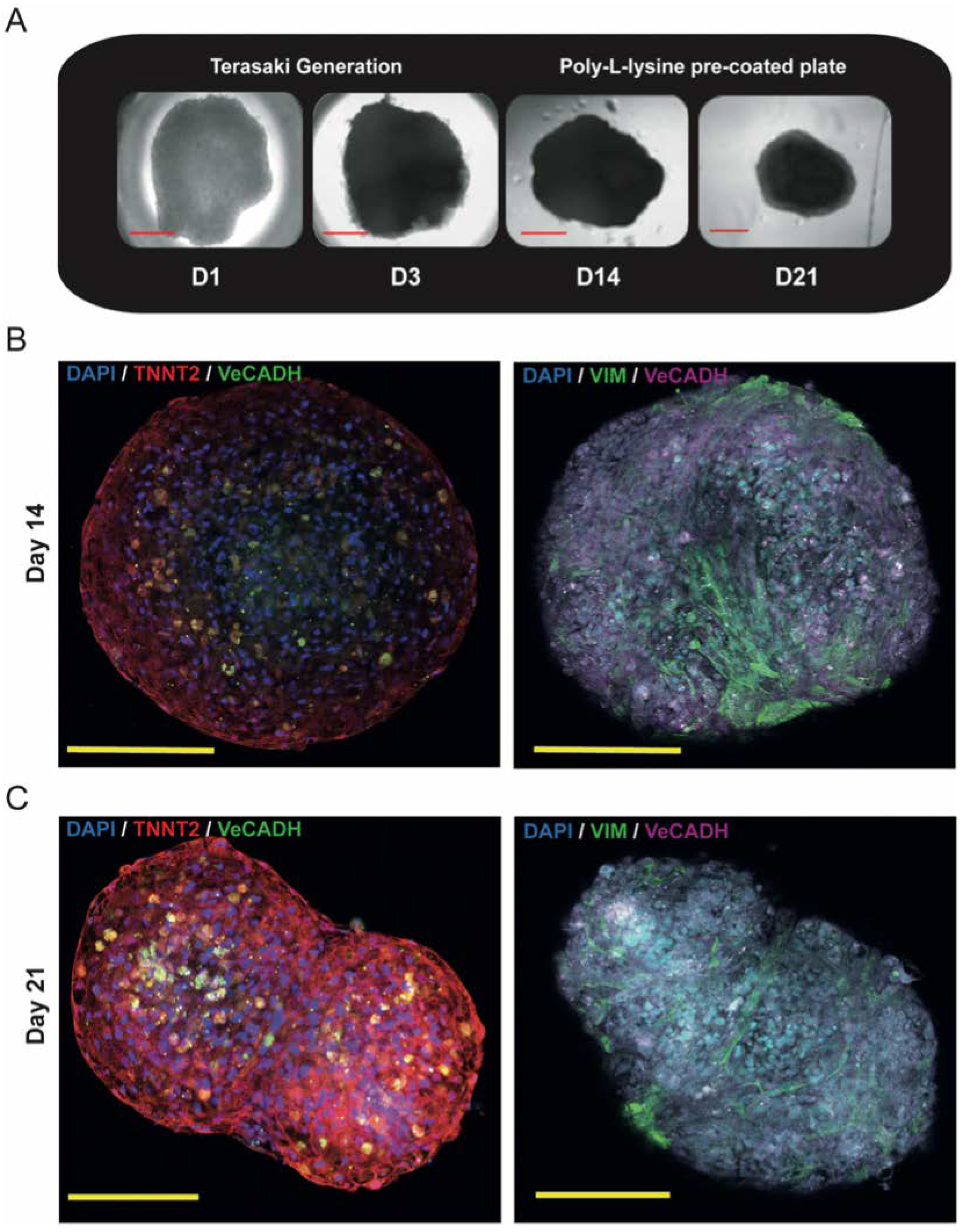
2.5. Organoid Formation Technique
2.6. Cell and Organoid Immunofluorescence Staining
2.7. Calcium Activity Monitoring Assay
2.8. Statistical Analysis
3. Results
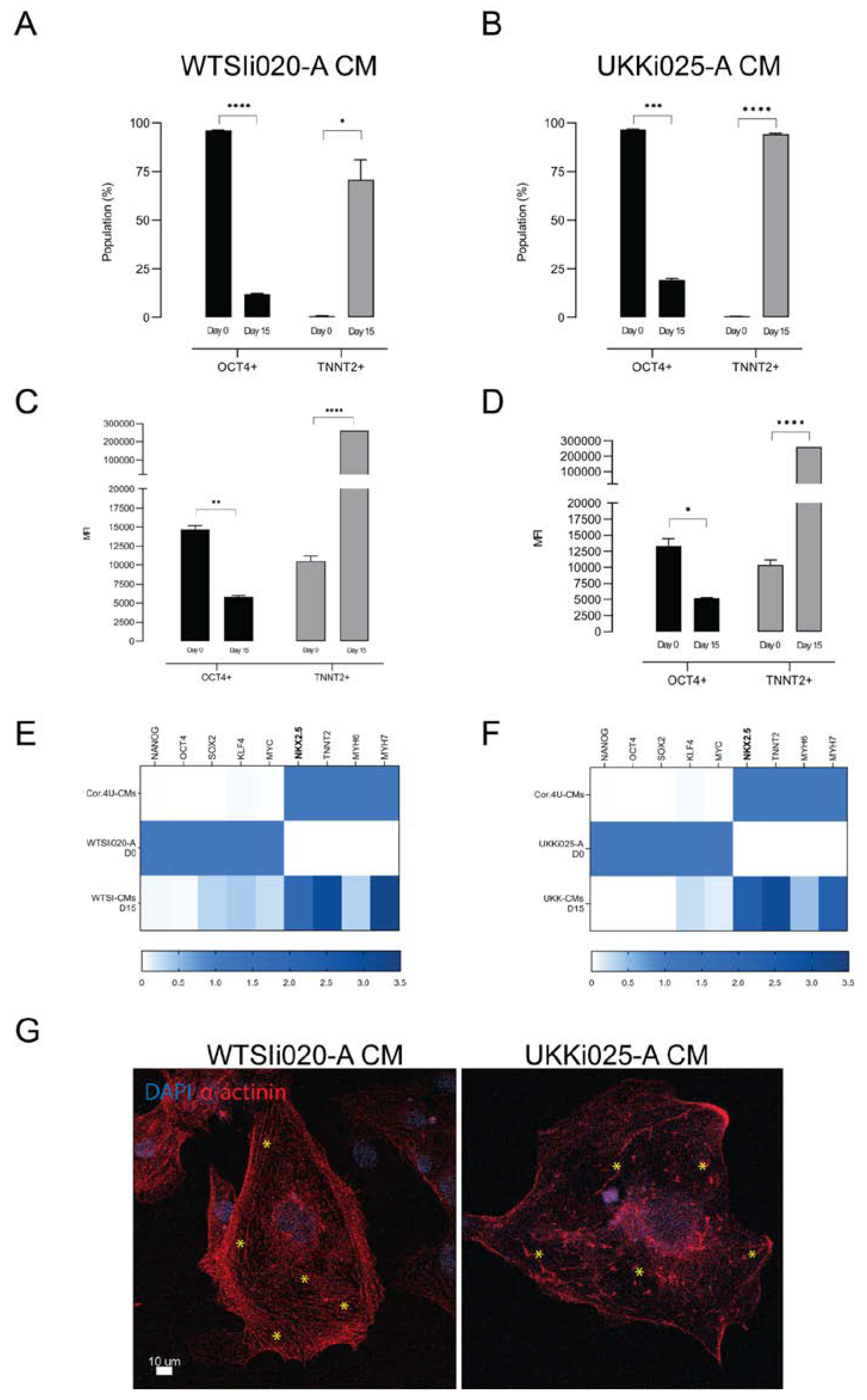
3.1. Differentiation of hiPSCs into Cardiomyocytes
3.2. Characterization of hiPSC-CMs
3.3. Generation of Triculture Cardiac Organoids
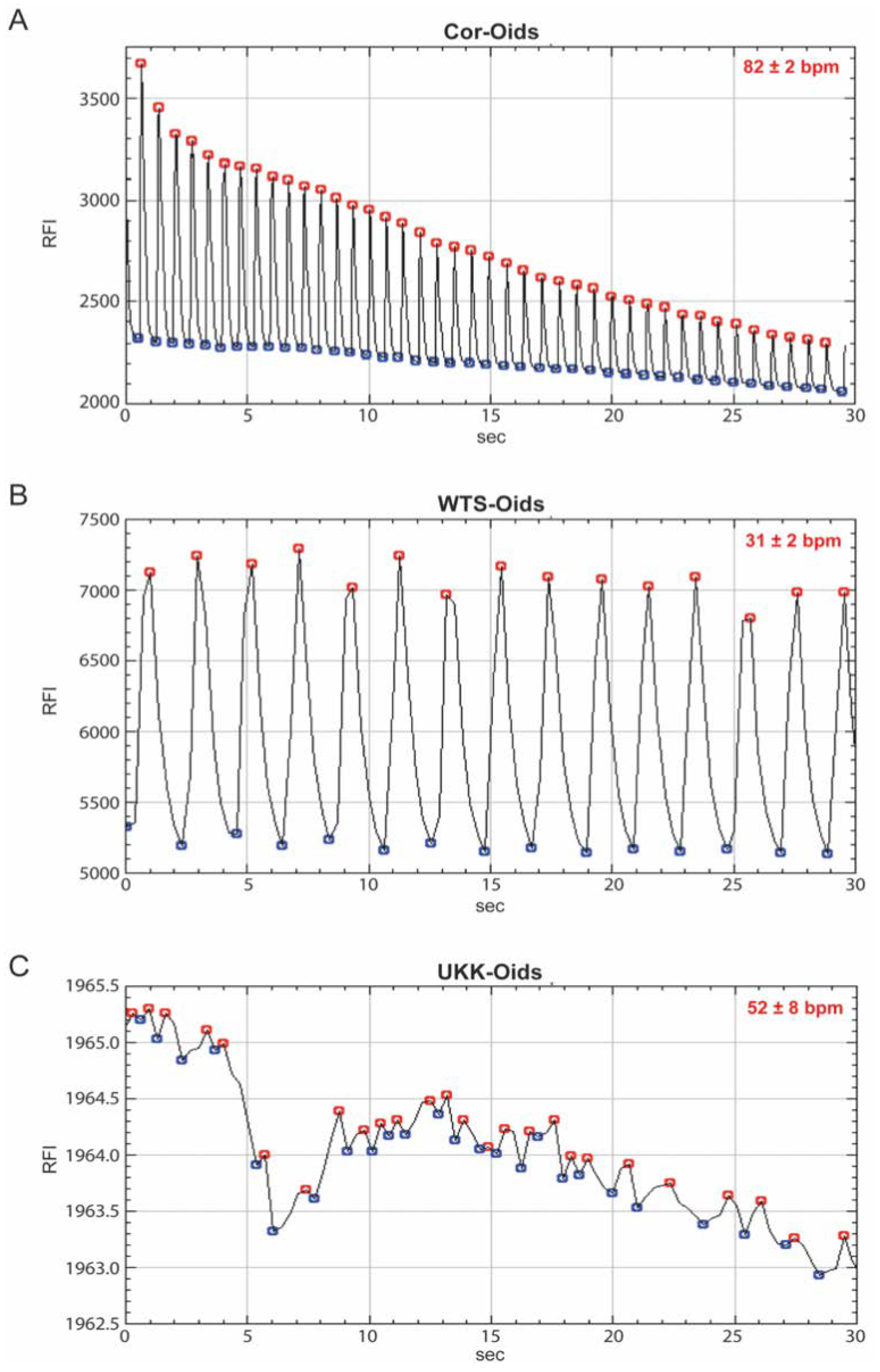
3.3.1. Proof of Concept Experiment: Gold Standard Cor-Oids
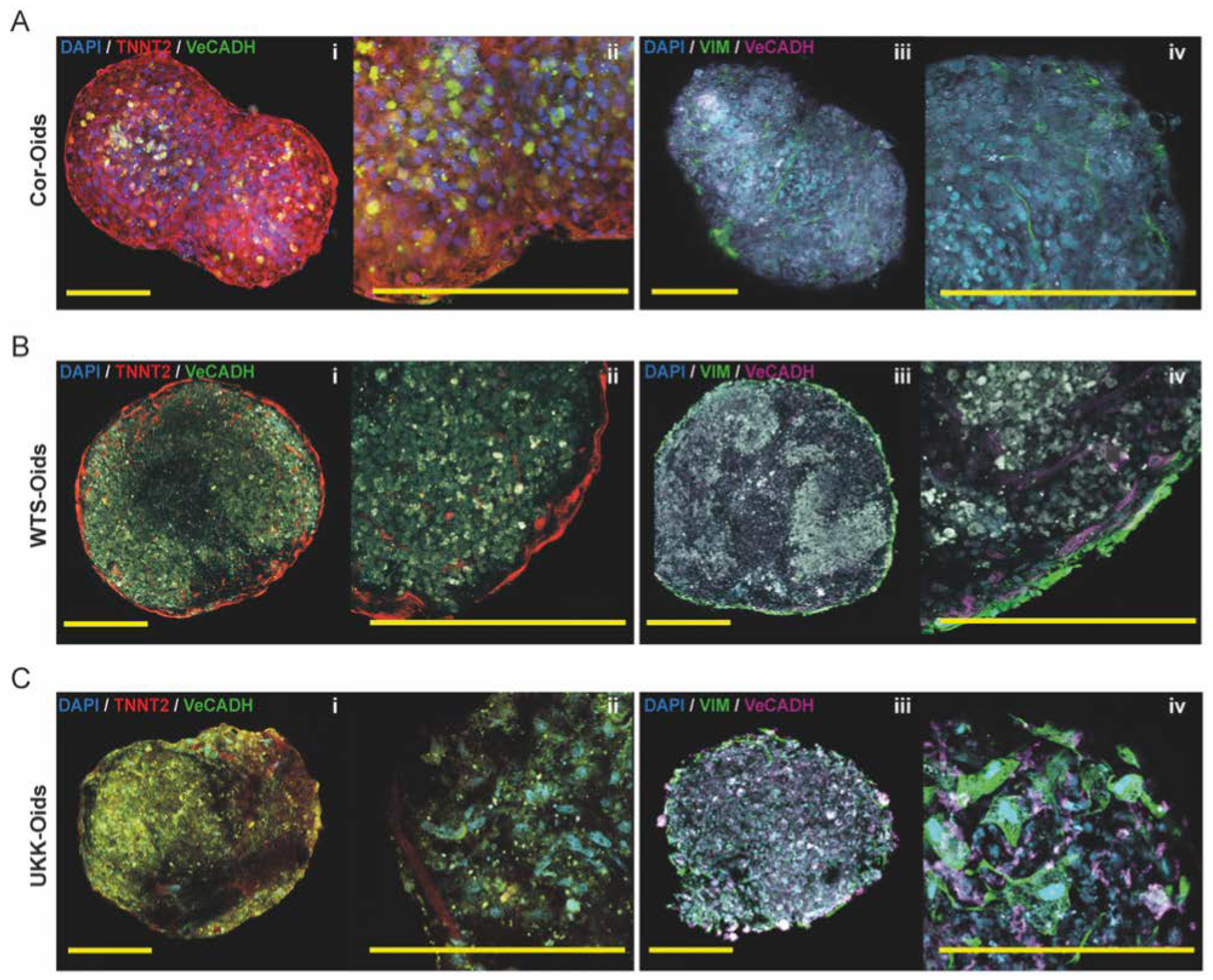
3.3.2. hiPSC-CM-Based Cardiac Organoids
3.3.3. Cardiac Organoids Display Patient-Specific Phenotypic Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CMs | cardiomyocytes |
| Cor-Oids | Cor.4U-based organoids |
| ECM | extracellular Matrix |
| ECs | endothelial cells |
| HCFs | human cardiac fibroblasts |
| HCM | hypertrophic cardiomyopathy |
| HCMECs | human cardiac microvascular ECs |
| hiPSCs | human induced pluripotent stem cells |
| hiPSC-CMs | hiPSC-derived CMs |
| KLF4 | kruppel like factor 4 |
| MFI | median florescence intensity |
| MYC | V-Myc avian myelocytomatosis viral oncogene homolog |
| MYH6 | myosin heavy chain 6 |
| MYH7 | myosin heavy chain 7 |
| NANOG | nanog homeobox protein |
| NKX2.5 | NK2 homeobox 5 |
| OCT4 | octamer-binding transcription factor 4 |
| RT-qPCR | real time quantitative PCR |
| SOX2 | SRY (sex determining region Y)-box |
| TNNT2 | cardiac-specific troponin |
| UKK-CMs | UKKi025-A derived CMs |
| UKK-Oids | UKK-CMs based organoids |
| VeCADH | vascular cadherin |
| VIM | vimentin |
| WTSI-CMs | WTSIi020-A derived CMs |
| WTS-Oids | WTSI-CMs based organoids |
References
- Roth, G.A.; Huffman, M.D.; Moran, A.E.; Feigin, V.; Mensah, G.A.; Naghavi, M.; Murray, C.J.L. Global and Regional Patterns in Cardiovascular Mortality From 1990 to 2013. Circulation 2015, 132, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.P.; Brantner, V.V. Spending on New Drug Development. Health Econ. 2010, 19, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Engle, S.J.; Puppala, D. Integrating Human Pluripotent Stem Cells into Drug Development. Cell Stem Cell 2013, 12, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Mathur, A.; Loskill, P.; Hong, S.; Lee, J.; Marcus, S.G.; Dumont, L.; Conklin, B.R.; Willenbring, H.; Lee, L.P.; Healy, K.E. Human Induced Pluripotent Stem Cell-Based Microphysiological Tissue Models of Myocardium and Liver for Drug Development. Stem Cell Res. Ther. 2013, 4, S14. [Google Scholar] [CrossRef]
- Emmert, M.Y.; Wolint, P.; Wickboldt, N.; Gemayel, G.; Weber, B.; Brokopp, C.E.; Boni, A.; Falk, V.; Bosman, A.; Jaconi, M.E.; et al. Human Stem Cell-Based Three-Dimensional Microtissues for Advanced Cardiac Cell Therapies. Biomaterials 2013, 34, 6339–6354. [Google Scholar] [CrossRef]
- Günter, J.; Wolint, P.; Bopp, A.; Steiger, J.; Cambria, E.; Hoerstrup, S.P.; Emmert, M.Y. Microtissues in Cardiovascular Medicine: Regenerative Potential Based on a 3D Microenvironment. Stem Cells Int. 2016, 2016, 1–20. [Google Scholar] [CrossRef]
- Emmert, M.Y.; Hitchcock, R.W.; Hoerstrup, S.P. Cell Therapy, 3D Culture Systems and Tissue Engineering for Cardiac Regeneration. Adv. Drug Deliv. Rev. 2014, 69–70, 254–269. [Google Scholar] [CrossRef]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The Third Dimension Bridges the Gap between Cell Culture and Live Tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef]
- Ewart, L.; Dehne, E.-M.; Fabre, K.; Gibbs, S.; Hickman, J.; Hornberg, E.; Ingelman-Sundberg, M.; Jang, K.-J.; Jones, D.R.; Lauschke, V.M.; et al. Application of Microphysiological Systems to Enhance Safety Assessment in Drug Discovery. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 65–82. [Google Scholar] [CrossRef]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. Adv. life Sci. R D 2017, 22, 456–472. [Google Scholar]
- Gomes, M.E.; Rodrigues, M.T.; Domingues, R.M.A.; Reis, R.L. Tissue Engineering and Regenerative Medicine: New Trends and Directions—A Year in Review. Tissue Eng. Part B Rev. 2017, 23, 211–224. [Google Scholar] [CrossRef]
- Nugraha, B.; Buono, M.F.; von Boehmer, L.; Hoerstrup, S.P.; Emmert, M.Y. Human Cardiac Organoids for Disease Modeling. Clin. Pharmacol. Ther. 2019, 105, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Nugraha, B.; Buono, M.F.; Emmert, M.Y. Modelling Human Cardiac Diseases with 3D Organoid. Eur. Heart J. 2018, 39, 4234–4237. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Tzatzalos, E.; Abilez, O.J.; Shukla, P.; Wu, J.C. Engineered Heart Tissues and Induced Pluripotent Stem Cells: Macro- and Microstructures for Disease Modeling, Drug Screening, and Translational Studies. Adv. Drug Deliv. Rev. 2016, 96, 234–244. [Google Scholar] [CrossRef]
- Lan, F.; Lee, A.S.; Liang, P.; Sanchez-Freire, V.; Nguyen, P.K.; Wang, L.; Han, L.; Yen, M.; Wang, Y.; Sun, N.; et al. Abnormal Calcium Handling Properties Underlie Familial Hypertrophic Cardiomyopathy Pathology in Patient-Specific Induced Pluripotent Stem Cells. Cell Stem Cell 2013, 12, 101–113. [Google Scholar] [CrossRef]
- Van Driest, S.L.; Ackerman, M.J.; Ommen, S.R.; Shakur, R.; Will, M.L.; Nishimura, R.A.; Tajik, A.J.; Gersh, B.J. Prevalence and Severity of “Benign” Mutations in the β-Myosin Heavy Chain, Cardiac Troponin T, and α-Tropomyosin Genes in Hypertrophic Cardiomyopathy. Circulation 2002, 106, 3085–3090. [Google Scholar] [CrossRef]
- Landstrom, A.P.; Ackerman, M.J. Mutation Type Is Not Clinically Useful in Predicting Prognosis in Hypertrophic Cardiomyopathy. Circulation 2010, 122, 2441–2450. [Google Scholar] [CrossRef]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Cozzarelli Prize Winner: Robust Cardiomyocyte Differentiation from Human Pluripotent Stem Cells via Temporal Modulation of Canonical Wnt Signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M.; et al. Chemically Defined Generation of Human Cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef]
- Richards, D.J.; Coyle, R.C.; Tan, Y.; Jia, J.; Wong, K.; Toomer, K.; Menick, D.R.; Mei, Y. Inspiration from Heart Development: Biomimetic Development of Functional Human Cardiac Organoids. Biomaterials 2017, 142, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Archer, C.R.; Sargeant, R.; Basak, J.; Pilling, J.; Barnes, J.R.; Pointon, A. Characterization and Validation of a Human 3D Cardiac Microtissue for the Assessment of Changes in Cardiac Pathology. Sci. Rep. 2018, 8, 10160. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Nguyen, H.V.; Tsang, S.H. Skin Biopsy and Patient-Specific Stem Cell Lines. Methods Mol. Biol. 2016, 1353, 77–88. [Google Scholar] [PubMed]
- Saini, H.; Navaei, A.; Van Putten, A.; Nikkhah, M. 3D Cardiac Microtissues Encapsulated with the Co-Culture of Cardiomyocytes and Cardiac Fibroblasts. Adv. Healthc. Mater. 2015, 4, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Kaneko, T.; Nomura, F. On-Chip Pre-Clinical Cardiac Toxicity: Testing Compounds beyond Herg and Qt Using HES/HiPS Cardiomyocyte Re-Entry Cell Network Model on a Chip. In Proceedings of the 14th International Conference on Miniaturized Systems for Chemistry and Life Sciences 2010, MicroTAS 2010, Groningen, The Netherlands, 3–7 October 2010. [Google Scholar]
- Burridge, P.W.; Metzler, S.A.; Nakayama, K.H.; Abilez, O.J.; Simmons, C.S.; Bruce, M.A.; Matsuura, Y.; Kim, P.; Wu, J.C.; Butte, M.; et al. Multi-Cellular Interactions Sustain Long-Term Contractility of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Am. J. Transl. Res. 2014, 6, 724. [Google Scholar]
- Devalla, H.D.; Passier, R. Cardiac Differentiation of Pluripotent Stem Cells and Implications for Modeling the Heart in Health and Disease. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef]
- Wolint, P.; Bopp, A.; Woloszyk, A.; Tian, Y.; Evrova, O.; Hilbe, M.; Giovanoli, P.; Calcagni, M.; Hoerstrup, S.P.; Buschmann, J.; et al. Cellular Self-Assembly into 3D Microtissues Enhances the Angiogenic Activity and Functional Neovascularization Capacity of Human Cardiopoietic Stem Cells. Angiogenesis 2018, 22, 37–52. [Google Scholar] [CrossRef]
- Sun, N.; Yazawa, M.; Liu, J.; Han, L.; Sanchez-Freire, V.; Abilez, O.J.; Navarrete, E.G.; Hu, S.; Wang, L.; Lee, A.; et al. Patient-Specific Induced Pluripotent Stem Cells as a Model for Familial Dilated Cardiomyopathy. Sci. Transl. Med. 2012, 4, 130ra47. [Google Scholar] [CrossRef]
- Wang, Z.; Oron, E.; Nelson, B.; Razis, S.; Ivanova, N. Distinct Lineage Specification Roles for NANOG, OCT4, and SOX2 in Human Embryonic Stem Cells. Cell Stem Cell 2012, 10, 440–454. [Google Scholar] [CrossRef]
- den Hartogh, S.C.; Wolstencroft, K.; Mummery, C.L.; Passier, R. A Comprehensive Gene Expression Analysis at Sequential Stages of in Vitro Cardiac Differentiation from Isolated MESP1-Expressing-Mesoderm Progenitors. Sci. Rep. 2016, 6, 19386. [Google Scholar] [CrossRef] [PubMed]
- Vega-Crespo, A.; Truong, B.; Hermann, K.J.; Awe, J.P.; Chang, K.M.; Lee, P.C.; Schoenberg, B.E.; Wu, L.; Byrne, J.A.; Lipshutz, G.S. Investigating the Functionality of an OCT4-Short Response Element in Human Induced Pluripotent Stem Cells. Mol. Ther. Methods Clin. Dev. 2016, 3, 16050. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liao, Y.-J.; Chen, Y.-S.; Lee, J.-X.; Chen, L.-R.; Yang, J.-R. Effects of Klf4 and C-Myc Knockdown on Pluripotency Maintenance in Porcine Induced Pluripotent Stem Cell. Cell J. 2018, 19, 640–646. [Google Scholar] [PubMed]
- Burridge, P.W.; Thompson, S.; Millrod, M.A.; Weinberg, S.; Yuan, X.; Peters, A.; Mahairaki, V.; Koliatsos, V.E.; Tung, L.; Zambidis, E.T. A Universal System for Highly Efficient Cardiac Differentiation of Human Induced Pluripotent Stem Cells That Eliminates Interline Variability. PLoS ONE 2011, 6, e18293. [Google Scholar] [CrossRef]
- Zhang, D.; Pu, W.T. Exercising Engineered Heart Muscle to Maturity. Nat. Rev. Cardiol. 2018, 15, 383–384. [Google Scholar] [CrossRef]
- Yoshida, S.; Miyagawa, S.; Fukushima, S.; Kawamura, T.; Kashiyama, N.; Ohashi, F.; Toyofuku, T.; Toda, K.; Sawa, Y. Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes by Soluble Factors from Human Mesenchymal Stem Cells. Mol. Ther. 2018, 26, 2681–2695. [Google Scholar] [CrossRef]
- Davidovic, G.; Iric-Cupic, V.; Milanov, S.; Dimitijevic, A.; Petrovic-Janicijevic, M. When Heart Goes “BOOM” to Fast. Heart Rate Greater than 80 as Mortality Predictor in Acute Myocardial Infarction. Am. J. Cardiovasc. Dis. 2013, 3, 120. [Google Scholar]
- Giacomelli, E.; Bellin, M.; Sala, L.; van Meer, B.J.; Tertoolen, L.G.J.; Orlova, V.V.; Mummery, C.L. Three-Dimensional Cardiac Microtissues Composed of Cardiomyocytes and Endothelial Cells Co-Differentiated from Human Pluripotent Stem Cells. Development 2017, 144, 1008–1017. [Google Scholar] [CrossRef]
- Savoji, H.; Mohammadi, M.H.; Rafatian, N.; Toroghi, M.K.; Wang, E.Y.; Zhao, Y.; Korolj, A.; Ahadian, S.; Radisic, M. Cardiovascular Disease Models: A Game Changing Paradigm in Drug Discovery and Screening. Biomaterials 2019, 198, 3–26. [Google Scholar] [CrossRef]
- Yin, X.; Mead, B.E.; Safaee, H.; Langer, R.; Karp, J.M.; Levy, O. Engineering Stem Cell Organoids. Cell Stem Cell 2016, 18, 25–38. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Thavandiran, N.; Dubois, N.; Mikryukov, A.; Masse, S.; Beca, B.; Simmons, C.A.; Deshpande, V.S.; McGarry, J.P.; Chen, C.S.; Nanthakumar, K.; et al. Design and Formulation of Functional Pluripotent Stem Cell-Derived Cardiac Microtissues. Proc. Natl. Acad. Sci. USA 2013, 110, E4698–E4707. [Google Scholar] [CrossRef] [PubMed]
- Stevens, K.R.; Kreutziger, K.L.; Dupras, S.K.; Korte, F.S.; Regnier, M.; Muskheli, V.; Nourse, M.B.; Bendixen, K.; Reinecke, H.; Murry, C.E. Physiological Function and Transplantation of Scaffold-Free and Vascularized Human Cardiac Muscle Tissue. Proc. Natl. Acad. Sci. USA 2009, 106, 16568–16573. [Google Scholar] [CrossRef]
- Voges, H.K.; Mills, R.J.; Elliott, D.A.; Parton, R.G.; Porrello, E.R.; Hudson, J.E. Development of a Human Cardiac Organoid Injury Model Reveals Innate Regenerative Potential. Development 2017, 144, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Varzideh, F.; Pahlavan, S.; Ansari, H.; Halvaei, M.; Kostin, S.; Feiz, M.S.; Latifi, H.; Aghdami, N.; Braun, T.; Baharvand, H. Human Cardiomyocytes Undergo Enhanced Maturation in Embryonic Stem Cell-Derived Organoid Transplants. Biomaterials 2019, 192, 537–550. [Google Scholar] [CrossRef]
- Williams, I.M.; Wu, J.C. Generation of Endothelial Cells From Human Pluripotent Stem Cells. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1317–1329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tian, L.; Shen, M.; Tu, C.; Wu, H.; Gu, M.; Paik, D.T.; Wu, J.C. Generation of Quiescent Cardiac Fibroblasts From Human Induced Pluripotent Stem Cells for In Vitro Modeling of Cardiac Fibrosis. Circ. Res. 2019, 125, 552–566. [Google Scholar] [CrossRef] [PubMed]
| hiPSC Line | Gender | Age | Race | Remarks |
|---|---|---|---|---|
| WTSIi020-A | Female | 40–44 | Caucasian | Derived from healthy donor |
| UKKi025-A | Female | 40–44 | Caucasian | Derived from hypertrophic cardiomyopathy (HCM) donor with MYH7 mutation. The mutation is a heterozygous missense mutation of the β-myosin heavy chain MYH7 gene located in exon 19, at chromosome location 14q11.2. There is a change of the single nucleotide sequence at position 2156 from G to A which causes the exchange of a nonpolar amino acid for a positively charged one (Arginine changed into Glutamine) in codon 719. |
| Gene Category | Symbol | Gene Name | Taqman Primer ID |
|---|---|---|---|
| Housekeeping | 18S | Eukaryotic 18S rRNA | Hs99999901_s1 |
| Pluripotency | NANOG | Nanog Homeobox Protein | Hs02387400_g1 |
| OCT4 (POU5F1) | Octamer-binding Transcription Factor 4 | Hs00999632_g1 | |
| SOX2 | SRY (Sex determining Region Y)-box 2 | Hs01053049_s1 | |
| KLF4 | Kruppel Like Factor 4 | Hs00359936_m1 | |
| MYC | V-Myc Avian Myelocytomatosis Viral Oncogene Homolog | Hs00153408_m1 | |
| Cardiac Development | NKX2.5 | NK2 Homeobox 5 | Hs00231763_m1 |
| Cardiac Structure | TNNT2 | Cardiac-specific Troponin | Hs00165960_m1 |
| MYH6 | Myosin Heavy Chain 6 | Hs00411908_m1 | |
| MYH7 | Myosin Heavy Chain 7 | Hs01110632_m1 |
| WTSIi020-A | UKKi025-A | ||||||
|---|---|---|---|---|---|---|---|
| Batch No. | Protocol | Passage | Confluence | CMs Quality | Passage | Confluence | CMs Quality |
| 1st | STEMdiff | 38 | 95% | Low | 42 | 95% | Mid |
| 2nd | STEMdiff | 39 | 95% | Differentiation Failed | 41 | 95% | Mid |
| 3rd | STEMdiff | 39 | 95% | × | 41 | 95% | × |
| CDM3 | 39 | 85% | Mid | 41 | 85% | High | |
| 4th | CDM3 | 40 | 85% | High | 41 | 90% | High |
| 5th | CDM3 | 42 | 90% | High | 42 | 95% | High |
| Organoid Type | Visible Contractions and Relaxations | Sustained Beating Time |
|---|---|---|
| Cor-Oids | Day 2,3 | ~3 Weeks |
| WTS-Oids | Day 7,8 | ~2 Weeks |
| UKK-Oids | Day 12–15 | ~1 Week |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filippo Buono, M.; von Boehmer, L.; Strang, J.; P. Hoerstrup, S.; Y. Emmert, M.; Nugraha, B. Human Cardiac Organoids for Modeling Genetic Cardiomyopathy. Cells 2020, 9, 1733. https://doi.org/10.3390/cells9071733
Filippo Buono M, von Boehmer L, Strang J, P. Hoerstrup S, Y. Emmert M, Nugraha B. Human Cardiac Organoids for Modeling Genetic Cardiomyopathy. Cells. 2020; 9(7):1733. https://doi.org/10.3390/cells9071733
Chicago/Turabian StyleFilippo Buono, Michele, Lisa von Boehmer, Jaan Strang, Simon P. Hoerstrup, Maximilian Y. Emmert, and Bramasta Nugraha. 2020. "Human Cardiac Organoids for Modeling Genetic Cardiomyopathy" Cells 9, no. 7: 1733. https://doi.org/10.3390/cells9071733
APA StyleFilippo Buono, M., von Boehmer, L., Strang, J., P. Hoerstrup, S., Y. Emmert, M., & Nugraha, B. (2020). Human Cardiac Organoids for Modeling Genetic Cardiomyopathy. Cells, 9(7), 1733. https://doi.org/10.3390/cells9071733







