Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cell Lines and Transfections
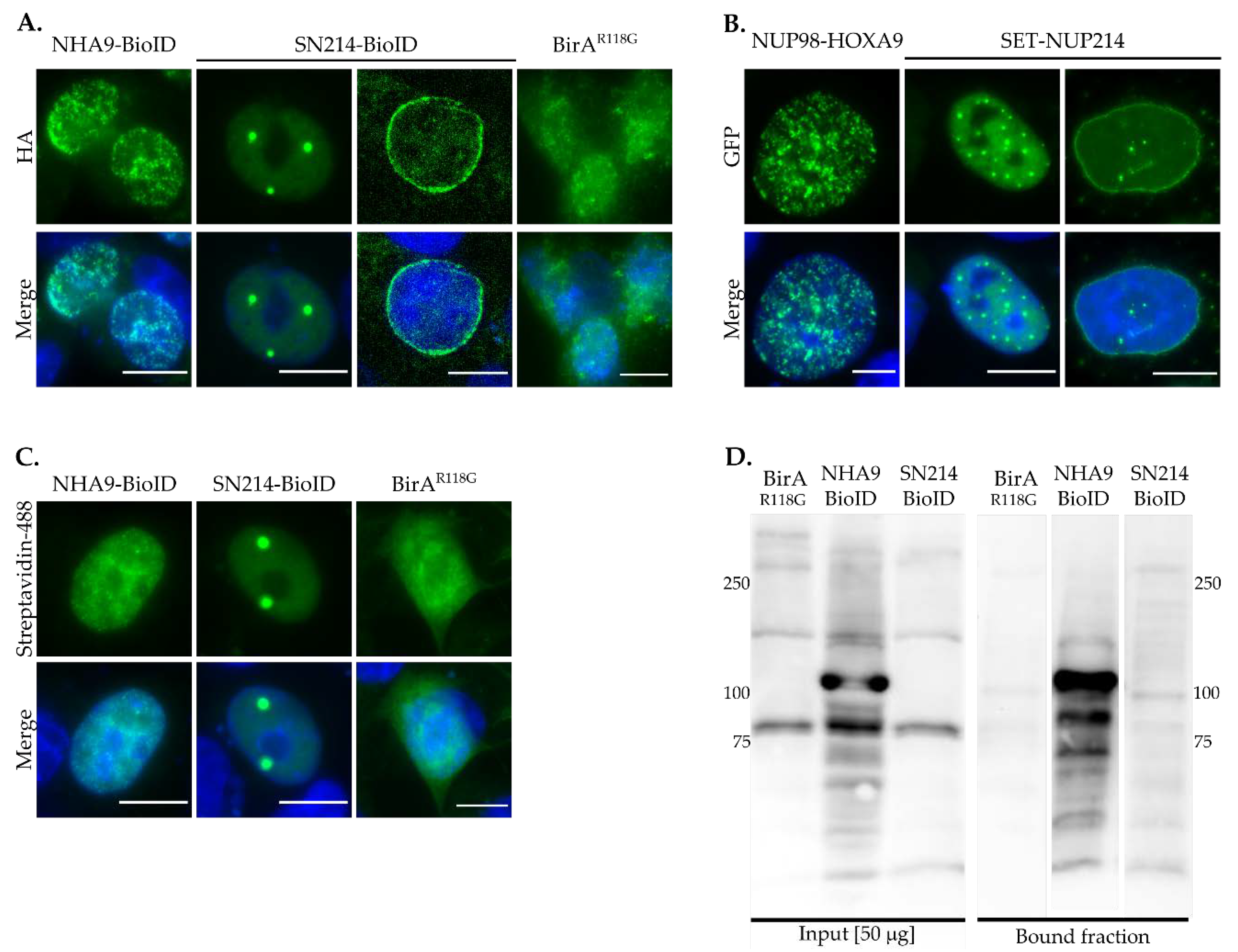
2.3. Immunofluorescence
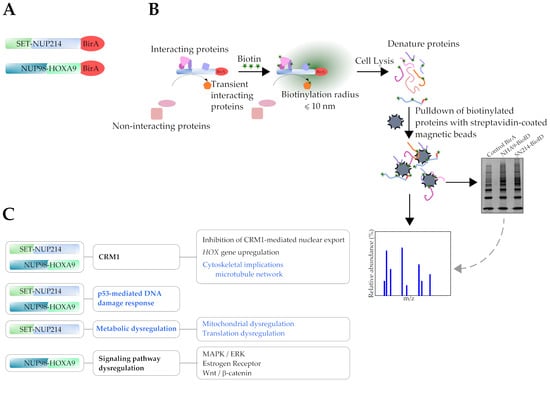
2.4. Pulldown of Biotinylated Proteins
2.5. Mass Spectrometry
2.6. Data Analysis
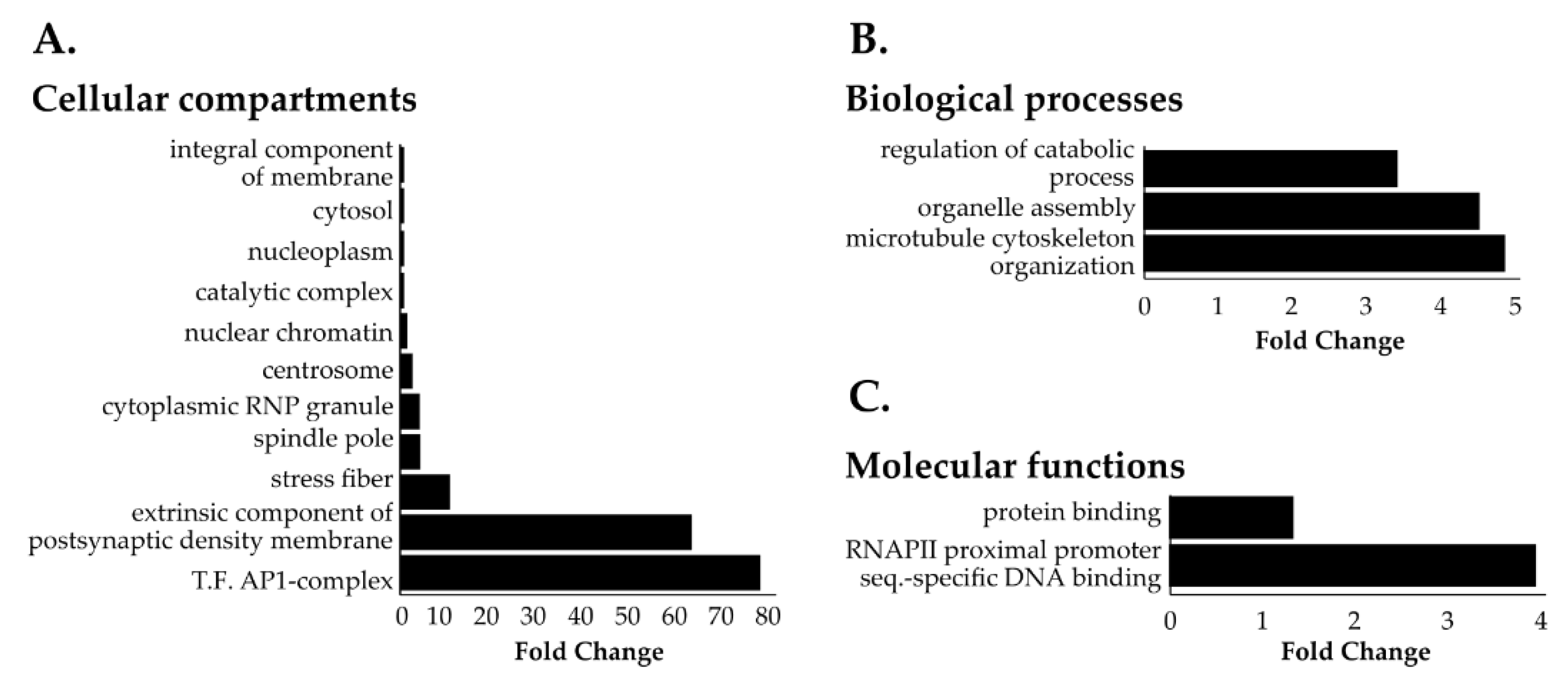
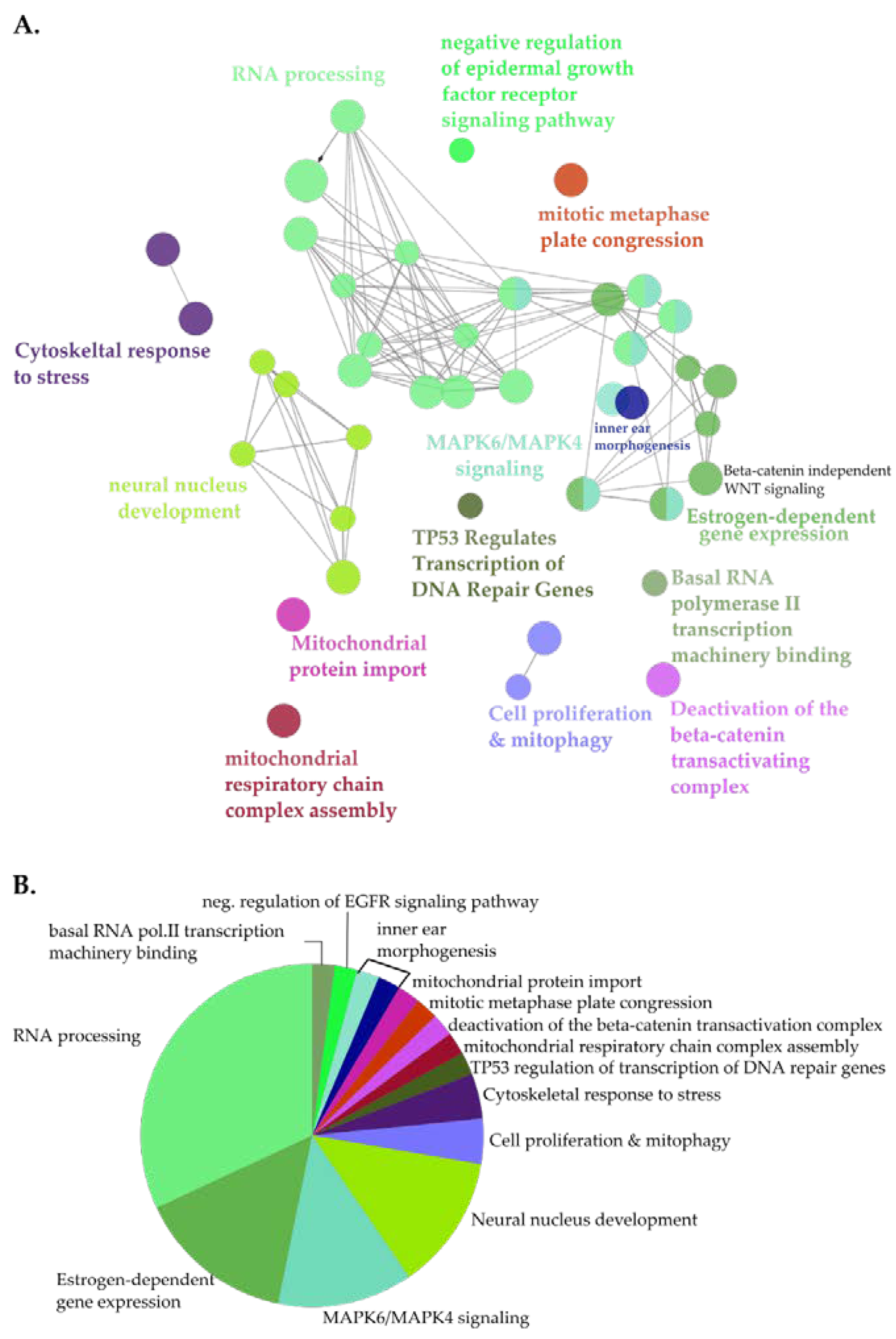
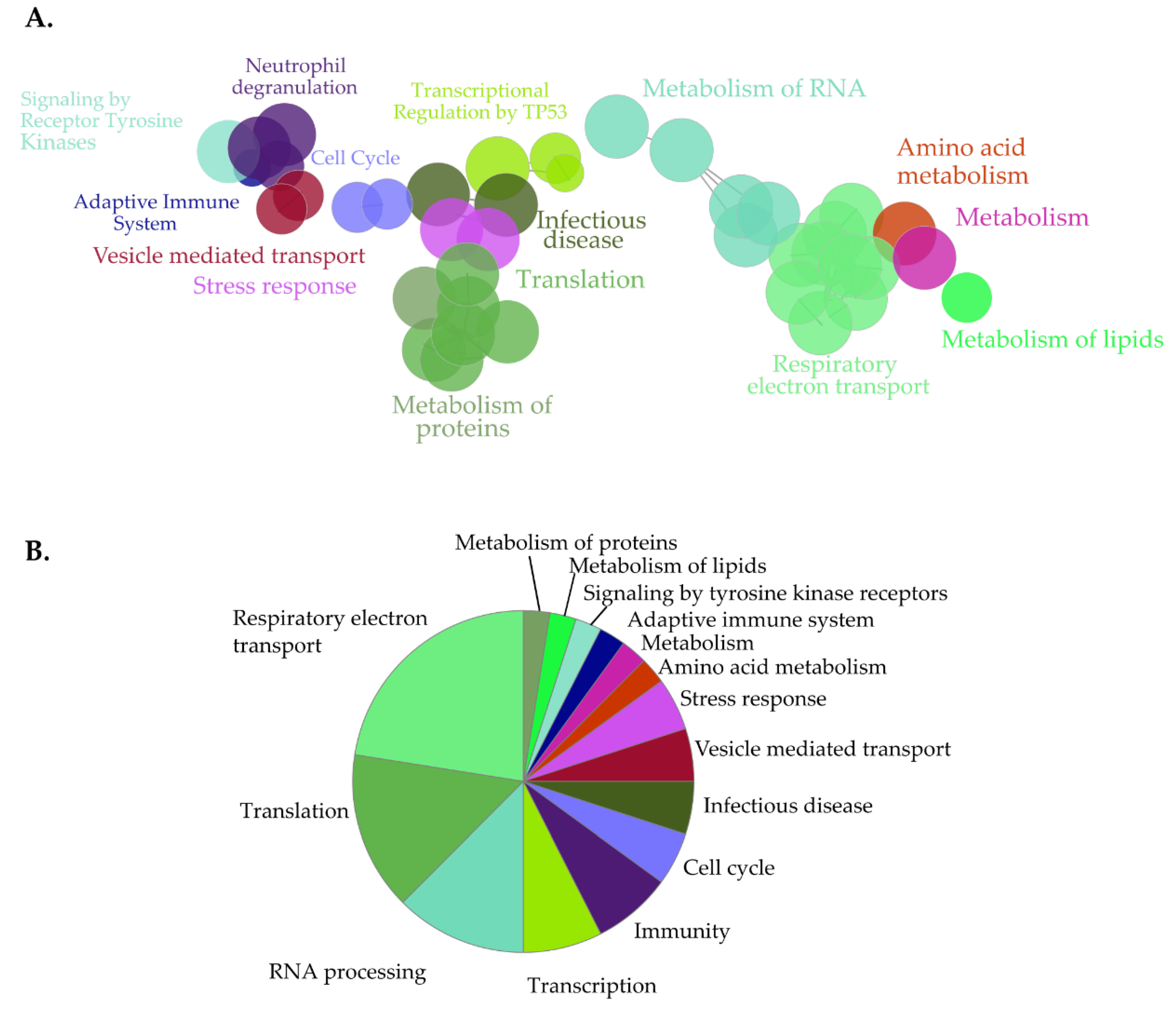
2.7. Gene Ontology and Pathway Analysis
2.8. Screening for Nuclear Export Signals
3. Results
3.1. Subcellular Distribution of BioID Fusion Proteins and Biotinylation Induction
3.2. Identification of Known Proximal Interactors of NUP98-HOXA9 and SET-NUP214
3.3. Identification of Novel Proximal Interactors of NUP98-HOXA9
3.4. NUP98-HOXA9 Proximal Interactors are Enriched in Nuclear Export Signal (NES) Containing Proteins
3.5. Identification of Novel Proximal Interactors of SET-NUP214
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Cloning of SET-NUP214 into the pcDNA3.1 MCS-BirA(R118G)-HA Vector
Appendix A.2. Incubation and Elution of Biotinylated Proteins from Streptavidin-Coated Magnetic Beads
Appendix A.3. Sample Injection and Mass Spectrometer Operation
Appendix A.4. Raw Data Analysis Using MaxQuant
References
- Terry, L.J.; Wente, S. Flexible Gates: Dynamic Topologies and Functions for FG Nucleoporins in Nucleocytoplasmic Transport. Eukaryot. Cell 2009, 8, 1814–1827. [Google Scholar] [CrossRef]
- Paulillo, S.M.; Powers, M.A.; Ullman, K.S.; Fahrenkrog, B. Changes in Nucleoporin Domain Topology in Response to Chemical Effectors. J. Mol. Biol. 2006, 363, 39–50. [Google Scholar] [CrossRef]
- Chatel, G.; Desai, S.H.; Mattheyses, A.L.; Powers, M.A.; Fahrenkrog, B. Domain topology of nucleoporin Nup98 within the nuclear pore complex. J. Struct. Biol. 2012, 177, 81–89. [Google Scholar] [CrossRef]
- Yang, J.; Lyu, X.; Zhu, X.; Meng, X.-G.; Zuo, W.; Ai, H.; Deng, M. Chromosome t(7;11)(p15;p15) translocation in acute myeloid leukemia coexisting with multilineage dyspoiesis and mutations in NRAS and WT1: A case report and literature review. Oncol. Lett. 2017, 13, 3066–3070. [Google Scholar] [CrossRef][Green Version]
- Gough, S.M.; Slape, C.; Aplan, P.D. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef]
- Fahrenkrog, B. Nucleoporin Gene Fusions and Hematopoietic Malignancies. New J. Sci. 2014, 2014, 1–18. [Google Scholar] [CrossRef]
- Martins, N.; Mendes, A.; Fahrenkrog, B. On the Effects of Leukemogenic Nucleoporin Fusion Proteins on Nucleocytoplasmic Transport and Gene Expression; Springer Science and Business Media LLC: New York, NY, USA, 2018; pp. 223–248. [Google Scholar]
- Lam, D.H.; Aplan, P.D. NUP98 gene fusions in hematologic malignancies. Leukemia 2001, 15, 1689–1695. [Google Scholar] [CrossRef] [PubMed]
- Ghannam, G.; Takeda, A.; Camarata, T.; Moore, M.A.; Viale, A.; Yaseen, N.R. The OncogeneNup98-HOXA9Induces Gene Transcription in Myeloid Cells. J. Biol. Chem. 2003, 279, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.T.; Hess, J.L. Role of HOXA9 in leukemia: Dysregulation, cofactors and essential targets. Oncogene 2015, 35, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, G.; Lansdorp, P.M.; Eaves, C.J.; Hogge, D.E.; Dragowska, W.H.; Reid, D.S.; Largman, C.; Lawrence, H.J.; Humphries, R.K. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. USA 1994, 91, 12223–12227. [Google Scholar] [CrossRef] [PubMed]
- von Lindern, M.; van Baal, S.; Wiegant, J.; Raap, A.; Hagemeijer, A.; Grosveld, G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3’ half to different genes: Characterization of the set gene. Mol. Cell. Biol. 1992, 12, 3346–3355. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.; Fahrenkrog, B. NUP214 in Leukemia: It’s More than Transport. Cells 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.-B.; McNamara, P.; Heo, S.; Turner, A.; Lane, W.S.; Chakravarti, D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 2001, 104, 119–130. [Google Scholar] [CrossRef]
- Kutney, S.N.; Hong, R.; Macfarlan, T.S.; Chakravarti, D. A Signaling Role of Histone-binding Proteins and INHAT Subunits pp32 and Set/TAF-Iβ in Integrating Chromatin Hypoacetylation and Transcriptional Repression. J. Biol. Chem. 2004, 279, 30850–30855. [Google Scholar] [CrossRef] [PubMed]
- Switzer, C.H.; Cheng, R.Y.; Vitek, T.M.; Christensen, D.J.; Wink, D.A.; Vitek, M.P. Targeting SET/I2PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene 2011, 30, 2504–2513. [Google Scholar] [CrossRef]
- Ichijo, T.; Chrousos, G.P.; Kino, T. Activated glucocorticoid receptor interacts with the INHAT component Set/TAF-Iβ and releases it from a glucocorticoid-responsive gene promoter, relieving repression: Implications for the pathogenesis of glucocorticoid resistance in acute undifferentiated leukemia with Set-Can translocation. Mol. Cell. Endocrinol. 2008, 283, 19–31. [Google Scholar] [CrossRef]
- Enjoji, S.; Yabe, R.; Tsuji, S.; Yoshimura, K.; Kawasaki, H.; Sakurai, M.; Sakai, Y.; Takenouchi, H.; Yoshino, S.; Hazama, S.; et al. Stemness Is Enhanced in Gastric Cancer by a SET/PP2A/E2F1 Axis. Mol. Cancer Res. 2018, 16, 554–563. [Google Scholar] [CrossRef]
- Fahrenkrog, B.; Martinelli, V.; Nilles, N.; Fruhmann, G.; Chatel, G.; Juge, S.; Sauder, U.; Di Giacomo, D.; Mecucci, C.; Schwaller, J. Expression of Leukemia-Associated Nup98 Fusion Proteins Generates an Aberrant Nuclear Envelope Phenotype. PLoS One 2016, 11, e0152321. [Google Scholar] [CrossRef][Green Version]
- Port, S.A.; Mendes, A.; Valkova, C.; Spillner, C.; Fahrenkrog, B.; Kaether, C.; Kehlenbach, R.H. The Oncogenic Fusion Proteins SET-Nup214 and Sequestosome-1 (SQSTM1)-Nup214 Form Dynamic Nuclear Bodies and Differentially Affect Nuclear Protein and Poly(A)+ RNA Export*. J. Biol. Chem. 2016, 291, 23068–23083. [Google Scholar] [CrossRef]
- Saito, S.; Cigdem, S.; Okuwaki, M.; Nagata, K. Leukemia-Associated Nup214 Fusion Proteins Disturb the XPO1-Mediated Nuclear-Cytoplasmic Transport Pathway and Thereby the NF-κB Signaling Pathway. Mol. Cell. Biol. 2016, 36, 1820–1835. [Google Scholar] [CrossRef]
- Mendes, A.; Juehlen, R.; Martinelli, V.; Fahrenkrog, B. Targeted CRM1-inhibition perturbs leukemogenic NUP214 fusion proteins and exerts anti-cancer effects in leukemia cell lines with NUP214 rearrangements. bioRxiv 2020. [Google Scholar] [CrossRef]
- Takeda, A.; Sarma, N.J.; Abdul-Nabi, A.M.; Yaseen, N.R. Inhibition of CRM1-mediated Nuclear Export of Transcription Factors by Leukemogenic NUP98 Fusion Proteins. J. Biol. Chem. 2010, 285, 16248–16257. [Google Scholar] [CrossRef] [PubMed]
- Van Vlierberghe, P.; Van Grotel, M.; Tchinda, J.; Lee, C.; Beverloo, H.B.; Van Der Spek, P.J.; Stubbs, A.; Cools, J.; Nagata, K.; Fornerod, M.; et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 2008, 111, 4668–4680. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Valerio, D.G.; Eisold, M.E.; Sinha, A.; Koche, R.P.; Hu, W.; Chen, C.-W.; Chu, S.H.; Brien, G.L.; Park, C.Y.; et al. NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell 2016, 30, 863–878. [Google Scholar] [CrossRef] [PubMed]
- Shima, Y.; Yumoto, M.; Katsumoto, T.; Kitabayashi, I. MLL is essential for NUP98-HOXA9-induced leukemia. Leukemia 2017, 31, 2200–2210. [Google Scholar] [CrossRef]
- Oka, M.; Mura, S.; Yamada, K.; Sangel, P.; Hirata, S.; Maehara, K.; Kawakami, K.; Tachibana, T.; Ohkawa, Y.; Kimura, H.; et al. Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes. eLife 2016, 5, 1195. [Google Scholar] [CrossRef]
- Oka, M.; Mura, S.; Otani, M.; Miyamoto, Y.; Nogami, J.; Maehara, K.; Harada, A.; Tachibana, T.; Yoneda, Y.; Ohkawa, Y. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. eLife 2019, 8. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Galon, J.; Bindea, G. Automated exploration of gene ontology term and pathway networks with ClueGO-REST. Bioinformatics 2019, 35, 3864–3866. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- La Cour, T.; Kiemer, L.; Gupta, R.; Skriver, K.; Brunak, S.; Mølgaard, A. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Marquis, K.; Pei, J.; Fu, S.-C.; Cağatay, T.; Grishin, N.V.; Chook, Y.M. LocNES: A computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 2014, 31, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Powers, M.A. Nup98-Homeodomain Fusions Interact with Endogenous Nup98 during Interphase and Localize to Kinetochores and Chromosome Arms during Mitosis. Mol. Biol. Cell 2010, 21, 1585–1596. [Google Scholar] [CrossRef]
- Rio-Machin, A.; Gómez-López, G.; Muñoz, J.; Garcia, F.; Maiques-Diaz, A.; Alvarez, S.; Salgado, R.N.; Shrestha, M.; Torres-Ruiz, R.; Haferlach, C.; et al. The molecular pathogenesis of the NUP98-HOXA9 fusion protein in acute myeloid leukemia. Leukemia 2017, 31, 2000–2005. [Google Scholar] [CrossRef][Green Version]
- Kasper, L.H.; Brindle, P.; Schnabel, C.A.; Pritchard, C.E.J.; Cleary, M.L.; Van Deursen, J.M.A. CREB Binding Protein Interacts with Nucleoporin-Specific FG Repeats That Activate Transcription and Mediate NUP98-HOXA9 Oncogenicity. Mol. Cell. Biol. 1999, 19, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2016, 45, D777–D783. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2018, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Aguilera, A.; Arranz, L.; Pérez, D.M.; García-García, A.; Stavropoulou, V.; Kubovcakova, L.; Isern, J.; Martín-Salamanca, S.; Langa, X.; Skoda, R.C.; et al. Estrogen Signaling Selectively Induces Apoptosis of Hematopoietic Progenitors and Myeloid Neoplasms without Harming Steady-State Hematopoiesis. Cell Stem Cell 2014, 15, 791–804. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2016, 36, 1461–1473. [Google Scholar] [CrossRef]
- Zolotukhin, A.S.; Felber, B.K. Nucleoporins Nup98 and Nup214 Participate in Nuclear Export of Human Immunodeficiency Virus Type 1 Rev. J. Virol. 1999, 73, 120–127. [Google Scholar] [CrossRef]
- Lee, Y.; Pei, J.; Baumhardt, J.M.; Chook, Y.M.; Grishin, N.V. Structural prerequisites for CRM1-dependent nuclear export signaling peptides: Accessibility, adapting conformation, and the stability at the binding site. Sci. Rep. 2019, 9, 6627. [Google Scholar] [CrossRef]
- Schmitt, C.; Von Kobbe, C.; Bachi, A.; Panté, N.; Rodrigues, J.P.; Boscheron, C.; Rigaut, G.; Wilm, M.; Seraphin, B.; Carmo-Fonseca, M.; et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999, 18, 4332–4347. [Google Scholar] [CrossRef]
- Trotman, L.C.; Mosberger, N.; Fornerod, M.; Stidwill, R.P.; Greber, U.F. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001, 3, 1092–1100. [Google Scholar] [CrossRef]
- Von Moeller, H.; Basquin, C.; Conti, E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009, 16, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Bernad, R.; van der Velde, H.; Fornerod, M.; Pickersgill, H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 2004, 24, 2373–2384. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, B.; Barral, Y. The cell biology of open and closed mitosis. Nucleus 2013, 4, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Kırlı, K.; Karaca, S.; Dehne, H.J.; Samwer, M.; Pan, K.-T.; Lenz, C.; Urlaub, H.; Görlich, D. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. eLife 2015, 4. [Google Scholar] [CrossRef]
- Griffis, E.; Altan, N.; Lippincott-Schwartz, J.; Powers, M.A. Nup98 Is a Mobile Nucleoporin with Transcription-dependent Dynamics. Mol. Biol. Cell 2002, 13, 1282–1297. [Google Scholar] [CrossRef]
- Singer, S.; Zhao, R.; Barsotti, A.M.; Ouwehand, A.; Fazollahi, M.; Coutavas, E.; Breuhahn, K.; Neumann, O.; Longerich, T.; Pusterla, T.; et al. Nuclear Pore Component Nup98 Is a Potential Tumor Suppressor and Regulates Posttranscriptional Expression of Select p53 Target Genes. Mol. Cell 2012, 48, 799–810. [Google Scholar] [CrossRef]
- Cazzalini, O.; Scovassi, A.I.; Savio, M.; Stivala, L.A.; Prosperi, E. Multiple roles of the cell cycle inhibitor p21CDKN1A in the DNA damage response. Mutat. Res. Mutat. Res. 2010, 704, 12–20. [Google Scholar] [CrossRef]
- Wang, N.; Kon, N.; Lasso, G.; Jiang, L.; Leng, W.; Zhu, W.-G.; Qin, J.; Honig, B.; Gu, W. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 2016, 538, 118–122. [Google Scholar] [CrossRef]
- Prokocimer, M.; Molchadsky, A.; Rotter, V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: Projections on diagnostic workup and therapy. Blood 2017, 130, 699–712. [Google Scholar] [CrossRef]
- Chiaretti, S.; Brugnoletti, F.; Tavolaro, S.; Bonina, S.; Paoloni, F.; Marinelli, M.; Patten, N.; Bonifacio, M.; Kropp, M.G.; Sica, S.; et al. TP53 mutations are frequent in adult acute lymphoblastic leukemia cases negative for recurrent fusion genes and correlate with poor response to induction therapy. Haematologica 2013, 98, e59–e61. [Google Scholar] [CrossRef]
- Xu, H.; Menéndez, S.; Schlegelberger, B.; Bae, N.; Aplan, P.D.; Göhring, G.; DeBlasio, T.R.; Nimer, S.D. Loss of p53 accelerates the complications of myelodysplastic syndrome in a NUP98-HOXD13–driven mouse model. Blood 2012, 120, 3089–3097. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Goolsby, C.; Yaseen, N.R. NUP98-HOXA9 Induces Long-term Proliferation and Blocks Differentiation of Primary Human CD34+ Hematopoietic. Cells Cancer Res. 2006, 66, 6628–6637. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Nabi, A.M.; Yassin, E.R.; Varghese, N.; Deshmukh, H.; Yaseen, N.R. In Vitro Transformation of Primary Human CD34+ Cells by AML Fusion Oncogenes: Early Gene Expression Profiling Reveals Possible Drug Target in AML. PLoS One 2010, 5, e12464. [Google Scholar] [CrossRef]
- Yassin, E.R.; Sarma, N.J.; Abdul-Nabi, A.M.; Dombrowski, J.; Han, Y.; Takeda, A.; Yaseen, N.R. Dissection of the Transformation of Primary Human Hematopoietic Cells by the Oncogene NUP98-HOXA9. PLoS One 2009, 4, e6719. [Google Scholar] [CrossRef][Green Version]
- Alberti, S.; Matějů, D.; Mediani, L.; Carra, S. Granulostasis: Protein Quality Control of RNP Granules. Front. Mol. Neurosci. 2017, 10, 45920. [Google Scholar] [CrossRef]
- Pushpalatha, K.V.; Besse, F. Local Translation in Axons: When Membraneless RNP Granules Meet Membrane-Bound Organelles. Front. Mol. Biosci. 2019, 6, 129. [Google Scholar] [CrossRef]
- Ding, X.; Yang, Z.; Zhou, F.; Wang, F.; Li, X.; Chen, C.; Li, X.; Hu, X.; Xiang, S.; Zhang, J. Transcription factor AP-2α regulates acute myeloid leukemia cell proliferation by influencing Hoxa gene expression. Int. J. Biochem. Cell Biol. 2013, 45, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.X.; Spanos, C.; Kojidani, T.; Lynch, E.; Rappsilber, J.; Hiraoka, Y.; Haraguchi, T.; Sawin, K.E. Exportin Crm1 is repurposed as a docking protein to generate microtubule organizing centers at the nuclear pore. eLife 2018, 7, e33465. [Google Scholar] [CrossRef]
- Arduíno, D.M.; Esteves, A.R.; Cardoso, S.M. Mitochondria drive autophagy pathology via microtubule disassembly. Autophagy 2013, 9, 112–114. [Google Scholar] [CrossRef]
- Mackeh, R.; Perdiz, D.; Lorin, S.; Codogno, P.; Poüs, C. Autophagy and microtubules - new story, old players. J. Cell Sci. 2013, 126, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Schwarzerová, K.; Bellinvia, E.; Martinek, J.; Sikorová, L.; Dostál, V.; Libusová, L.; Bokvaj, P.; Fischer, L.; Schmit, A.C.; Nick, P. Tubulin is actively exported from the nucleus through the Exportin1/CRM1 pathway. Sci. Rep. 2019, 9, 5725. [Google Scholar] [CrossRef]
- Akoumianaki, T.; Kardassis, D.; Polioudaki, H.; Georgatos, S.; Theodoropoulos, P.A. Nucleocytoplasmic shuttling of soluble tubulin in mammalian cells. J. Cell Sci. 2009, 122, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, R.; Whitmarsh, A.J. Mitochondrial Proteins Moonlighting in the Nucleus. Trends Biochem. Sci. 2015, 40, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Kotiadis, V.; Duchen, M.R.; Osellame, L.D. Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1840, 1254–1265. [Google Scholar] [CrossRef]
- Schnapp, B.J.; Reese, T.S. Dynein is the motor for retrograde axonal transport of organelles. Proc. Natl. Acad. Sci. USA 1989, 86, 1548–1552. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.; Morita, M.; Hirao, T.; Yamaguchi, M.; Shiratori, R.; Kikuya, M.; Chibana, H.; Ito, K. Shift in energy metabolism caused by glucocorticoids enhances the effect of cytotoxic anti-cancer drugs against acute lymphoblastic leukemia cells. Oncotarget 2017, 8, 94271–94285. [Google Scholar] [CrossRef][Green Version]
- Sandén, C.; Ageberg, M.; Petersson, J.; Lennartsson, A.; Gullberg, U. Forced expression of the DEK-NUP214 fusion protein promotes proliferation dependent on upregulation of mTOR. BMC Cancer 2013, 13, 440. [Google Scholar] [CrossRef]
- Guillen, N.; Wieske, M.; Otto, A.; Mian, A.A.; Rokicki, M.; Guy, C.; Alvares, C.; Hole, P.; Held, H.; Ottmann, O.G.; et al. Subtractive Interaction Proteomics Reveal a Network of Signaling Pathways Activated by an Oncogenic Transcription Factor in Acute Myeloid Leukemia. SSRN Electron. J. 2018. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Zhang, J. mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int. J. Mol. Sci. 2019, 20, 755. [Google Scholar] [CrossRef] [PubMed]
- Sen, B.; Johnson, F.M. Regulation of Src Family Kinases in Human Cancers. J. Signal Transduct. 2011, 2011, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Greuber, E.K.; Smith-Pearson, P.; Wang, J.; Pendergast, A.M. Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat. Rev. Cancer 2013, 13, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018, 3, 5. [Google Scholar] [CrossRef]
- Von Lindern, M.; Poustka, A.; Lerach, H.; Grosveld, G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol. Cell. Biol. 1990, 10, 4016–4026. [Google Scholar] [CrossRef] [PubMed]
- Von Lindern, M.; Breems, D.; Van Baal, S.; Adriaansen, H.; Grosveld, G. Characterization of the translocation breakpoint sequences of twoDEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and aSET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosom. Cancer 1992, 5, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Cheah, J.S.; Yamada, S. A simple elution strategy for biotinylated proteins bound to streptavidin conjugated beads using excess biotin and heat. Biochem. Biophys. Res. Commun. 2017, 493, 1522–1527. [Google Scholar] [CrossRef] [PubMed]
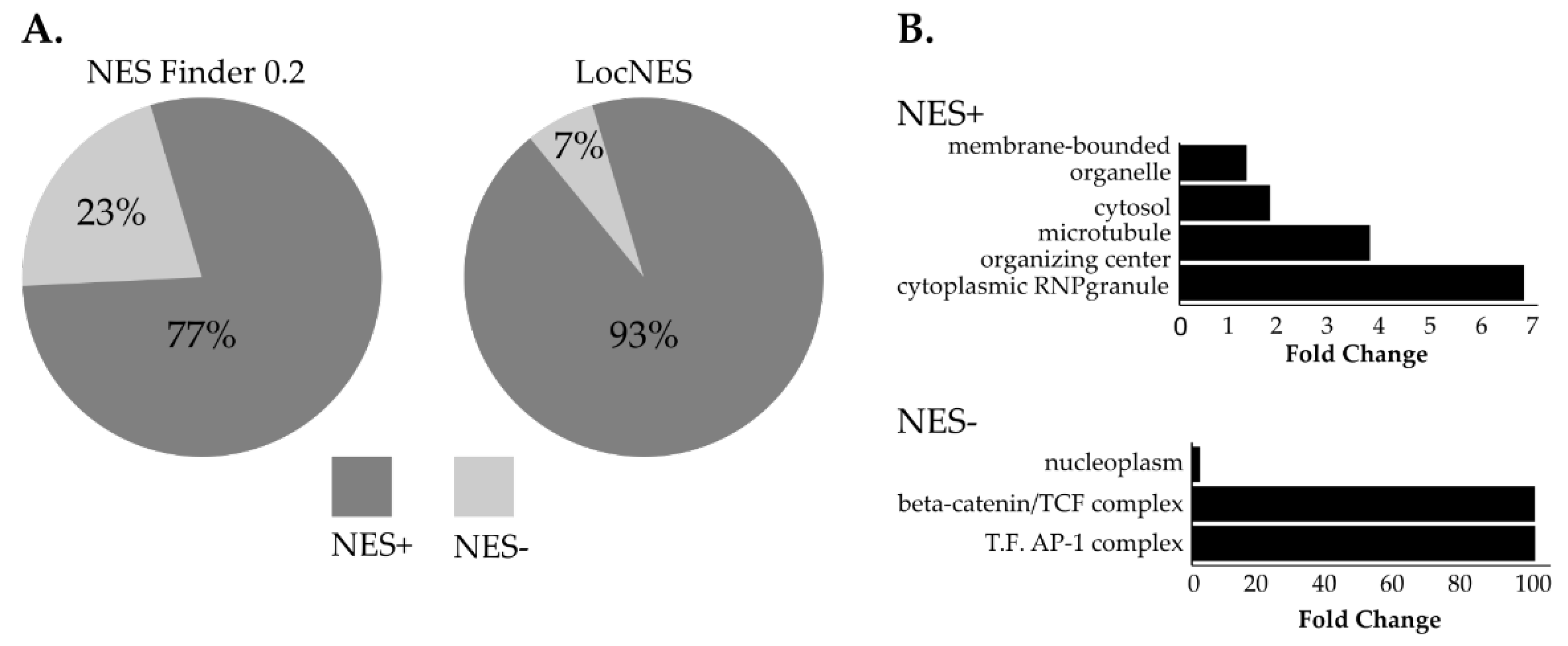

| Protein | Gene | NHA9-BioID | F.C. | Ref. |
|---|---|---|---|---|
| Nucleoporin NUP98 | NUP98 | +/+ | −1.84 | [46] |
| Chromosome region maintenance 1/Exportin 1 | CRM1/XPO1 | +/+ | −2.13 | [24,29] |
| mRNA export factor 1 | RAE1 | +/+ | 6.40 | [46] |
| Nuclear factor of activated T cells | NFAT5 | +/- | 24.81 | [24] |
| Histone-lysine N-methyltransferase 2A/Mixed lineage leukemia 1 | KMT2A/ MLL1 | +/+ | −0.91 | [28] |
| Host cell factor 1 | HCFC1 | +/+ | −1.86 | [27] |
| Histone deacetylase 1 | HDAC1 | +/+ | −2.05 | [47] |
| O-GlcNac transferase subunit p110 | OGT | -/- | [27] | |
| CREB binding protein | CREBBP/p300 | -/- | [47,48] |
| Protein | Gene | SN214-BioID | F.C. | Ref. |
|---|---|---|---|---|
| Nuclear RNA export factor 1 | NXF1/ TAP | +/+ | 1.37 | [25] |
| Chromosome region maintenance 1/Exportin 1 | CRM1/XPO1 | +/+ | 0.16 | [23,25] |
| Nucleoporin 62 | NUP62 | +/+ | 1.79 | [23] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, A.; Jühlen, R.; Bousbata, S.; Fahrenkrog, B. Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach. Cells 2020, 9, 1666. https://doi.org/10.3390/cells9071666
Mendes A, Jühlen R, Bousbata S, Fahrenkrog B. Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach. Cells. 2020; 9(7):1666. https://doi.org/10.3390/cells9071666
Chicago/Turabian StyleMendes, Adélia, Ramona Jühlen, Sabrina Bousbata, and Birthe Fahrenkrog. 2020. "Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach" Cells 9, no. 7: 1666. https://doi.org/10.3390/cells9071666
APA StyleMendes, A., Jühlen, R., Bousbata, S., & Fahrenkrog, B. (2020). Disclosing the Interactome of Leukemogenic NUP98-HOXA9 and SET-NUP214 Fusion Proteins Using a Proteomic Approach. Cells, 9(7), 1666. https://doi.org/10.3390/cells9071666