Abstract
The interaction of oncogenes with cellular proteins is a major determinant of cellular transformation. The NUP98-HOXA9 and SET-NUP214 chimeras result from recurrent chromosomal translocations in acute leukemia. Functionally, the two fusion proteins inhibit nuclear export and interact with epigenetic regulators. The full interactome of NUP98-HOXA9 and SET-NUP214 is currently unknown. We used proximity-dependent biotin identification (BioID) to study the landscape of the NUP98-HOXA9 and SET-NUP214 environments. Our results suggest that both fusion proteins interact with major regulators of RNA processing, with translation-associated proteins, and that both chimeras perturb the transcriptional program of the tumor suppressor p53. Other cellular processes appear to be distinctively affected by the particular fusion protein. NUP98-HOXA9 likely perturbs Wnt, MAPK, and estrogen receptor (ER) signaling pathways, as well as the cytoskeleton, the latter likely due to its interaction with the nuclear export receptor CRM1. Conversely, mitochondrial proteins and metabolic regulators are significantly overrepresented in the SET-NUP214 proximal interactome. Our study provides new clues on the mechanistic actions of nucleoporin fusion proteins and might be of particular relevance in the search for new druggable targets for the treatment of nucleoporin-related leukemia.
1. Introduction
The nucleoporins NUP98 and NUP214 are components of the nuclear pore complex (NPC) and belong to the group of so-called phenylalanine-glycine (FG) rich nucleoporins, which are essential for nucleocytoplasmic transport. Via their FG domains, NUP98 and NUP214 contribute to the selective and semi-permeable NPC barrier, and interact with nuclear transport receptors (NTRs) of the β-karyopherin family, thereby promoting the fast exchange of cargoes between the nucleus and the cytoplasm [1,2,3].
A number of chromosomal translocations involving the NUP98 and NUP214 loci are reported in acute myeloid and lymphoblastic leukemias (AML and ALL, respectively). NUP98- and NUP214-related leukemia are associated with poor overall survival [4,5,6,7], and no specific or targeted therapies are as yet available to improve prognosis. The chromosomal rearrangements of NUP98 and NUP214 result in their fusion with a large range of gene partners, all of which retain the FG domain of the respective nucleoporin [7,8]. The fusion of NUP98 with the homeobox protein Hox-A9 (HOXA9), NUP98-HOXA9, that results from t(7;11)(p15;p15), has been studied as the prototype for the oncogenic mechanisms governing the actions of NUP98 fusions with homeodomain (HD) proteins in AML [9]. HOXA9 is a transcription factor that regulates hematopoietic stem cell expansion and is abundantly expressed in hematopoietic precursor cells, while being progressively silenced during differentiation [10,11]. NUP214 is frequently found in conjunction with the oncogene SET, resulting from either t(9;9)(q34;q34) or an interstitial deletion at 9q34 [12,13,14]. SET-NUP214 is typically linked to ALL, and less frequently to AML [15,16]. SET is a chromatin-binding protein and an epigenetic regulator as part of the inhibitor of acetyltransferases (INHAT) complex [17,18]. Due to its role as an epigenetic modifier, SET is involved in a multitude of cellular functions, including regulation of the cell cycle, gene expression, and apoptosis [19,20,21]. NUP98-HOXA9 and SET-NUP214 share several characteristics: both form nuclear foci that accumulate endogenous proteins [22,23]; both interact with the NTR chromosome region maintenance 1 (CRM1), or exportin 1 (XPO1), and sequester cargo-loaded CRM1-nuclear export complexes to inhibit their translocation to the cytoplasm [23,24,25]. Moreover, NUP98-HOXA9 and SET-NUP214 interact with chromatin-binding proteins, such as the histone methyltransferases mixed lineage leukemia 1 (MLL1) and the disruptor of telomeric silencing 1-like (DOT1L) [26,27,28]. Association of NUP98-HOXA9 and SET-NUP214 with chromatin-bound CRM1 induces over-expression of HOX genes, a hallmark of unfavorable prognosis in leukemia [10,29,30]. The full landscape of NUP98-HOXA9 and SET-NUP214 interactors, however, has not yet been determined.
In recent years, advances in enzyme-mediated protein labelling became a powerful approach to study specific protein–protein interactions (PPIs) [31,32,33]. Proximity-dependent protein biotinylation (BioID) is an enzyme-mediated protein labelling approach that uses a modified version of the Escherichia coli (E. coli) biotin ligase BirA (BirAR118G), which has a lower affinity than wild type BirA for biotinoyl-adenylate (bio-AMP), the active form of biotin that can bind lysine residues [31,34]. Thus, when expressed in-frame with a protein of interest, BirAR118G biotinylates proximal proteins, which can then be purified by streptavidin pulldown and further identified by mass spectrometry [32]. In contrast to other interaction assays, which may be regarded as a snapshot of PPIs at the moment of cell lysis, BioID interrogates both stable and transient PPIs in living cells, thus providing a broader picture of protein interactors. Here, we used a modified BioID approach to study the proximal interactome of NUP98-HOXA9 and SET-NUP214. We identified further common associated partner as well as discrete fusion protein-specific interactors in the environment of NUP98-HOXA9 and SET-NUP214.
2. Materials and Methods
All experiments were carried out at room temperature (RT) unless otherwise specified.
2.1. Plasmids
pcDNA3.1 MCS-BirA(R118G)-HA was a gift from Dr. Kyle Roux (Addgene plasmid # 36047; [31]) and BirA(R118G)-HA destination vector from Dr. Karl Kramer (Addgene plasmid # 53581). For the cloning of SET-NUP214, total RNA was extracted from LOUCY cells, which carry del(9)(q34.11q34.13) resulting in the SET-NUP214 fusion transcript [35]. The coding sequence of SET-NUP214 was cloned into the pcDNA3.1 MCS-BirA(R118G)-HA vector, as described in Appendix A. The NUP98-HOXA9-BirAR118G construct was generated by Gateway® cloning. The coding sequence of NUP98-HOXA9 was first subcloned from pEGFP-NUP98-HOXA9 [22] into the pENTR/TOPO vector using the TOPO® TA Cloning Kit (Invitrogen, Merelbeke, Belgium) to generate the pENTR/NUP98-HOXA9 Gateway® entry vector. The NUP98-HOXA9 sequence was then subcloned into the BirA(R118G)-HA destination vector using the Gateway ™ LR Clonase™ enzyme mix (Invitrogen).
2.2. Cell Lines and Transfections
HCT-116 cells were a gift from Dr. Denis Lafontaine (Institute of Molecular Biology and Medicine, Université Libre de Bruxelles, Charleroi, Belgium). HCT-116 cells were cultured in McCoy’s 5A medium (LONZATM BioWhittakerTM, Verviers, Belgium), supplemented with 10% FBS and 1% penicillin/streptomycin (P/S, GIBCO, Invitrogen), and cultivated in a humidified incubator at 37 °C with 5% CO2 atmosphere. HCT-116 cells were transfected using the jetPRIME® transfection reagent (Polyplus transfection®, Illkirch, France). Briefly, plasmids were mixed with transfection reagent at a 1:2 (w/v) ratio and incubated for 40 min. Transfection mixes were added to the cell culture and incubated for 24 h. Next, the culture medium was replaced by fresh medium containing 50 µM biotin (Sigma–Aldrich, Overijse, Belgium) and biotinylation was induced for an additional 24 h. Cells were tested for mycoplasma contamination on a regular basis.
2.3. Immunofluorescence
Cells were grown on polylysine-coated glass coverslips and fixed in 2% formaldehyde for 15 min, washed three times for 10 min with PBS, and permeabilized with PBS/2% BSA/0.1% Triton X-100 for 10 min. Cells were washed twice for 10 min in PBS/2% BSA, incubated with Streptavidin-Alexa Fluor ™ 488 conjugate (dilution 1:1000; Invitrogen) or anti-HA antibody (clone 12CA5, dilution 1/50, supernatant of a mouse hybridoma cell line) for 1 h and washed twice in PBS/2% BSA/0.1% Triton X-100. Cells incubated with anti-HA were then incubated with goat anti-mouse IgG Alexa Fluor™ 488 (dilution 1:1000; Invitrogen) for 1 h and washed twice for 10 min with PBS. All cells were mounted with Mowiol-4088 containing DAPI (1 μg/ml) and stored at 4 °C until viewed. Cells were imaged using a Zeiss LSM-710 confocal laser-scanning microscope (Zeiss, Oberkochen, Germany). Images were recorded using the microscope system software and processed using ImageJ v.1.52t (http://imagej.nih.gov) and Inkscape 0.92 Software (http://www.inkscape.org).
2.4. Pulldown of Biotinylated Proteins
2 × 106 cells were plated in a 10 cm2 cell-culture dish, grown for 24 h and subsequently processed for transfection and biotinylation induction as described above. Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1mM EGTA, 1.5 mM MgCl2, 0.4% sodium dodecyl sulfate (SDS), 1 µl/ml benzonaze [25 U/ml], 1% Nonidet-P40, and protease inhibitor cocktail tablets (Roche, Basel, Switzerland). Bradford assay was used to determine protein concentration and 500 μg of protein were incubated with 50 µl of SeraMag™ magnetic Streptavidin-coated beads ([10 mg/ml]; GE Healthcare, Chicago, Illinois, USA). Protein-beads incubation and recovery of biotinylated proteins were carried out as detailed in Appendix A. The entire eluate containing biotinylated proteins was subjected to sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. Proteins were detected using horseradish-conjugated streptavidin (HRP-Strep; Thermo Fischer Scientific, Merelbeke, Belgium). For protein identification by mass spectrometry, the same amount of protein was used. After incubation of whole protein extract with streptavidin-coated magnetic beads, samples were resuspended in on-bead tryptic digestion buffer (20 mM Tris-HCl, pH 8.0, 2 mM CaCl2) and processed for large-scale analysis by tandem mass spectrometry (LC-MS/MS).
2.5. Mass Spectrometry
Proteins were digested on the beads with 1 µg of trypsin (Promega, Leiden, Netherlands) at 37 °C for 4 h while spinning at 161× g. Beads were removed, an additional 1 µg of trypsin was added, and proteins were further digested at 37 °C, overnight. The resulting peptide mixture was purified using OMIX C18 pipette tips (Agilent, Santa Clara, California, USA). The purified peptides were dried completely and re-suspended in 20 µl loading solvent (0.1% TFA in water/ acetonitrile, 2/98 (v/v)) of which 5 µl were injected for LC-MS/MS analysis on an Ultimate 3000 RSLCnano ProFLow system connected online to a Q Exactive HF mass spectrometer (Thermo, Waltham, MA, USA). Peptide trapping and elution and mass spectrometer operation details are described in Appendix A (Sample injection and mass spectrometer operation).
2.6. Data Analysis
Data analysis was performed with MaxQuant (version 1.6.3.4; Max Planck Institute of Biochemistry, Germany [36]) using the Andromeda search engine with default search settings including a false discovery rate (FDR) set at 1% on both the peptide and protein level. Spectra were searched against the human UniProt Tax ID: 9606 proteins in the UniProt/Swiss-Prot reference database (database release version of January 2019, www.uniprot.org) supplemented with the BirA fusion proteins, as detailed in Appendix A (Data Analysis). Further data analysis was performed using the Perseus software (version 1.6.2.1, Max Plank Institute of Biochemistry, Germany) after loading the protein groups file from MaxQuant [37]. First, proteins identified by site and reverse database hits and potential contaminants were removed. Label-free quantification (LFQ) values were then used to normalize protein abundance among NUP98-HOXA9-BirAR118G (NHA9-BioID) and SET-NUP214-BirAR118G (SN214-BioID) proximal interactors, relative to BirAR118G alone (control). Given the potential differences in the expression of the BioID proteins (due to transient transfection), we performed an internal normalization to calculate the LFQ value of each individual protein that results from differences in NHA9-BioID and SN214-BioID protein expression. Subsequently, an external normalization to calculate the LFQ value of each individual protein relative to the control BirAR118G (NHA9-BioID/BirAR118G and SN214-BioID/BirAR118G) was carried out. The results were then converted to log2 and expressed as fold change (F.C.) relative to the control (Figure S1).
2.7. Gene Ontology and Pathway Analysis
Gene ontology (GO) of NHA9-BioID and SN214-BioID proximal interactors was performed by the GO consortium-associated Protein Analysis Through Evolutionary Relationships (PANTHER) Classification System (version 14.1.), available online (www.pantherdb.org) using the default parameters: Fisher’s exact test, with the Benjamini–Hochberg FDR correction for multiple testing and the background reference list: Homo sapiens whole genome [38,39,40]. Clustered enrichment analysis was performed with the Cytoscape software (v3.7.1) plugin ClueGO (v2.5.5), to calculate enrichment of terms as right-sided tests based on the hypergeometric distribution [41]. ClueGO uses Cohen’s kappa statistics to link the terms in the network and to determine the association strength between the terms, which is an indication of term overlapping to define functional clusters [41,42]. For the analysis of proximal interactors of NUP98-HOXA9 and SET-NUP214, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and REACTOME Pathways together with GO vocabularies (GO Biological Processes (GOBP), GO Molecular Function (GOMF), and GO Cellular Compartments (GOCC)) were used, and functional representations of non-redundant and over-represented terms within the input protein sets were generated. Human Ensemble Gene ID identifiers (ENSG IDs) were mapped to the selected annotations (GO, updated on 27th February, 2019, KEGG, updated on 19th December, 2019, and REACTOME pathways, updated on 19th December 2019). The following settings were used during enrichment analysis: p-value cut off 0.01, and Bonferroni step-down correction for multiple comparisons.
2.8. Screening for Nuclear Export Signals
The presence of classical NESs was assessed using the online available software NES finder 0.2 (http://research.nki.nl/fornerodlab/NES-Finder.htm) and LocNES (http://prodata.swmed.edu/LocNES/LocNES.php), with default software parameters [43,44].
3. Results
3.1. Subcellular Distribution of BioID Fusion Proteins and Biotinylation Induction
To validate the subcellular localization of the BioID fusion proteins, we first examined the localization of NHA9-BioID, SN214-BioID, and control BirAR118G, after biotinylation was induced. As shown in Figure 1A, NHA9-BioID localized to the nucleus in a distinctive punctate pattern, whereas SN214-BioID formed intranuclear foci and localized to the nuclear rim. The localization of the two BioID fusion proteins is similar to the respective GFP-fusion proteins of NHA9 and SN214 (Figure 1B; [22,45]). BirAR118G was found throughout the entire cell (Figure 1A). Streptavidin labeling revealed the same distribution pattern as HA- and GFP-NHA9 and -SN214, indicating the presence of biotin (Figure 1C). Western blot analysis furthermore showed that endogenous proteins were biotinylated in whole cellular extracts of NHA9-BioID and SN214-BioID transfected cells (Figure 1D, bound fraction), in contrast to BirAR118G transfected cells. Of note, the NHA9-BioID fraction showed a strong enrichment of a specific band above 100 kDa, likely corresponding to the fusion protein itself. Due to the lower intensity of the SN214-BioID fraction, biotinylated SET-NUP214 cannot reliably be allocated, but might correspond to the band appearing below the 250-kDa band. For BirAR118G no specific enrichment was observed, given the unspecific nature of biotinylation mediated by the biotin ligase alone.
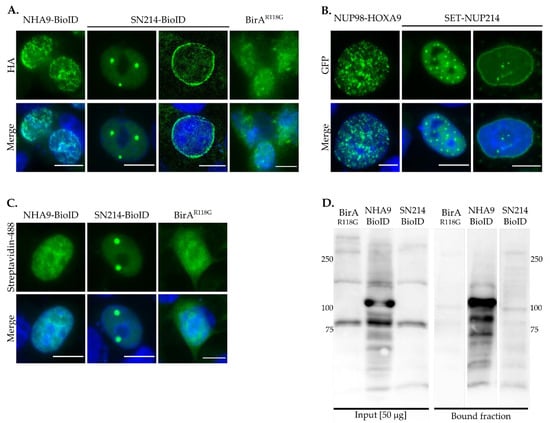
Figure 1.
Proximity-dependent biotin identification (BioID) fusion protein expression and biotinylation of endogenous proteins. (A) Localization of NHA9-BioID, SN214-BioID, and BirAR118G was evaluated by immunostaining with anti-HA antibody. (B) Localization of GFP-tagged NUP98-HOXA9 and SET-NUP214 was evaluated by green fluorescent protein (GFP) fluorescence. NHA9-BioID and SN214-BioID exhibit the same distribution pattern in nuclear foci and at the nuclear envelope as their GFP-tagged counterparts. (C) Detection of protein biotinylation by Streptavidin-488. Cells were transfected with NHA9-BioID, SN214-BioID, and BirAR118G and probed with Streptavidin-Alexa Fluor™ 488 conjugate. Shown are representative confocal images. DNA was visualized with DAPI (blue). Scale bars, 10 µm. (D) For detection of protein biotinylation by NHA9-BioID, SN214-BioID, and BirAR118G, corresponding cell lysates were enriched on Streptavidin-coated magnetic beads and whole protein lysates and the bound fractions were analyzed by immunoblotting. Note, virtually no specific bands were detected in the bound fraction of BirAR118G, in contrast to the bound fractions of NHA9-BioID and SN214-BioID, which exhibited patterns of differentially biotinylated proteins.
3.2. Identification of Known Proximal Interactors of NUP98-HOXA9 and SET-NUP214
Having validated the correct subcellular localization and the reliability of NHA9-BioID and SN214-BioID, we then carried out a mass-spectrometric analysis of the biotinylated proteins co-purifying with the respective BioID fusion protein. We first screened for candidate proximal interactors and identified several known binding partners of NUP98-HOXA9 and SET-NUP214. These were the protein nuclear export factor CRM1 [24,29], the mRNA export factor RAE1 [46], the transcription factor NFAT5 [24], the histone methyltransferase MLL1 [28], and the histone deacetylase HDAC1 [47] for NUP98-HOXA9 (Table 1). We did not find the acetyltransferase CREB binding protein/p300 (CBP/p300; [48]) nor WDR5, a component of the non-lethal specific (NSL)-histone modifying complex [27], co-purifying with NHA9-BioID, which might be due to the fact that both genes are mutated in HCT-116 cells [49,50]. Among known SET-NUP214 interactors (Table 2), we identified the nuclear RNA export factor 1 (NXF1/TAP; [25]), CRM1 [23,25], and nucleoporin NUP62 [23,45].

Table 1.
List of validated NUP98-HOXA9 interaction partners found in the biotinylated fraction of NHA9-BioID.

Table 2.
List of validated SET-NUP214 interaction partners found in the biotinylated fraction of SN214-BioID.
A major limitation of our BioID approach is the use of transient transfection instead of stable, inducible expression of the BioID fusion proteins. Due to its smaller size, the transfection efficiency of BirAR118G is much higher as for the larger NHA9-BioID and SN214-BioID constructs. We therefore considered the rate of biotinylation by BirAR118G higher than by the fusion proteins. Moreover, BirAR118G is a promiscuous biotin ligase, which also likely results in higher biotinylation levels in the control. To control for these differences in biotinylation, we employed a two-step normalization strategy that accounted for differences between BirAR118G, NHA9-BioID, and SN214-BioID protein expression (Figure S1). To further validate our BioID approach, next we performed Gene ontology (GO) analysis of proteins exclusively co-purifying with the BirAR118G control to determine unspecific interactors of BirAR118G-mediated biotinylation [29], as described in the Methods section. Table S1 summarizes the list of proteins that were exclusively biotinylated by BirAR118G and their respective subcellular localization. No significant enrichment in any of the GO categories (i) biological processes (GOBP), (ii) molecular function (GOMF), and (iii) cellular components (GOCC) was found, suggesting that biotinylation of these proteins is of an unspecific nature, as previously reported [29].
Moreover, given the differences in transfection efficiency between the BirAR118G, NHA9-BioID, and SN214-BioID constructs, some potential candidates might have disappeared, because known interactors, such as CRM1 and others, were more strongly biotinylated by BirAR118G as compared to NHA9-BioID and SN214-BioID. This in consequence explains the negative fold change (F.C.) values (Table 1). High normalized label-free quantification values (LFQNORMALIZED; see Data Analysis, Section 2.6) indicated that these proteins, after pull-down, are more enriched with NHA9-BioID or SN214-BioID, albeit being biotinylated by BirAR118G, meaning that their interaction with the fusion proteins is of a rather specific nature.
3.3. Identification of Novel Proximal Interactors of NUP98-HOXA9
Next, we employed a two-step normalization strategy to evaluate NHA9-BioID and SN214-BioID proximal interactors relative to BirAR118G to account for differences in protein expression levels due to the transient expression of the BioID fusion proteins (Figure S1). In doing so, we obtained 131 proteins that were at least 50% more abundant for NHA9-BioID when compared to BirAR118G (Table S2, fold change ≥0.6). GOCC analysis revealed that these NHA9-BioID proximal interactors located to both cytoplasmic and nuclear compartments with a general enrichment of chromatin-associated proteins and particularly the transcription factor AP-1 complex (Figure 2A). Cytoskeleton-related proteins were also significantly overrepresented in the pool of NHA9-BioID proximal interactors, namely proteins from the spindle pole, actin stress fibers, and centrosome (Figure 2A,B). GOMF analysis showed that NUP98-HOXA9 proximal interactors are significantly enriched in DNA binding proteins that target the promoters of genes transcribed by RNA polymerase II (RNAPII; Figure 2C).
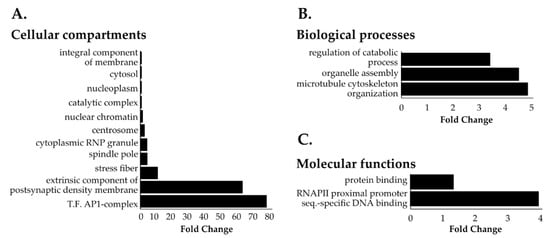
Figure 2.
Gene ontology (GO) of NHA9-BioID proximal interactors. Most represented (A) cellular compartments (GOCC), (B) biological processes (GOBP), and (C) molecular functions (GOMF) among NHA9-BioID proximal interactors. Statistical analysis of the overrepresented proteins in the NHA9-BioID fraction (total 131 proteins) was performed with the PANTHER classification online software (v14.1) using Fisher’s exact test. Results are displayed for FDR p < 0.05.
For a systematic analysis of the landscape of the NUP98-HOXA9 proximal interactome, next we performed clustered pathway analysis (Figure 3). Here, the most significantly enriched functional clusters included proteins involved in RNA processing and the RNAPII transcriptional machinery (Figure 3, Figure S2). Moreover, proximal NHA9-BioID interactors clustered in functional groups associated with the expression of genes from key signaling pathways frequently dysregulated in cancer. These included estrogen receptor (ER) and MAPK signaling pathways, and p53-mediated transcription of DNA damage repair genes (Figure 3A, Figure S2; [51,52,53]). Further, β-catenin-mediated transcription may be negatively regulated by NUP98-HOXA9, suggesting a dysregulation of the Wnt signaling pathway. Interestingly, some members of the Wnt signaling pathway, i.e., the segment polarity protein disheveled homolog DVL-1, the trinucleotide repeat-containing gene 6A protein (TNRC6A), and the transducing-like enhancer protein, isoforms 1-4 (TLE 1-4), were exclusively biotinylated by NHA9-BioID, further supporting the hypothesis that NUP98-HOXA9 affects this signaling pathway. Altogether, GO and clustered pathway analyses of NUP98-HOXA9 suggest that transcription dysregulation is a major defect in NUP98-HOXA9 driven leukemia.
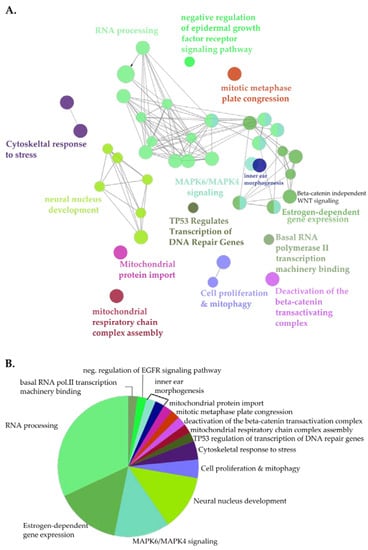
Figure 3.
Clustered pathway analysis of NHA9-BioID proximal interactors. (A) Functionally grouped network of NHA9-BioID proximal interactors. Nodes correspond to functional clusters, resulting from term grouping based on their overlapping level. (B) Overview pie-chart showing functional groups. Statistical analysis was performed using the Cytoscape plugin ClueGo (v.2.5.5) using the hypergeometric test and the following parameters: p < 0.01, kappa-score (k) = 0.4; (min/%) genes = 3/4%, GO tree levels: 3-11. Ontology databases: GOBP, GOCC, and GOMF, Reactome Pathways and KEGG.
3.4. NUP98-HOXA9 Proximal Interactors are Enriched in Nuclear Export Signal (NES) Containing Proteins
NUP98 is essential for CRM1-mediated nuclear export and NUP98 chimeras have been shown to affect CRM1-mediated nuclear export, to an extent, however, that is not fully understood [7,54]. To obtain a deeper insight into the impact of NUP98-HOXA9 on CRM1-mediated nuclear export, we screened the list of NUP98-HOXA9 proximal interactors for the presence of classical NESs using two algorithms: NES finder 0.2 and LocNES [43,44]. Both algorithms revealed that the majority of NHA9-BioID proximal interactors exhibit at least one classical NES motif (NES+ proteins, Figure 4A, Table S3). Given that the LocNES software has previously been shown to identify a higher rate of false positive NESs, we applied GO analysis for predicted NES+ interactors obtained by the NES Finder 0.2 software [55]. As shown in Figure 4B, this revealed a significant enrichment in proteins from cytoplasmic ribonuclear granules and in proteins from the microtubule organizing center. Conversely, NES- proximal interactors of NUP98-HOXA9 were significantly enriched in proteins implicated in transcription regulation, namely the ß-catenin/TCF complex and the transcription factor AP-1 complex (Figure 4B).
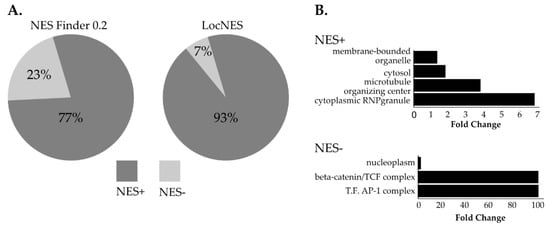
Figure 4.
Screening of nuclear export signals (NESs) in NHA9-BioID proximal interactors. The amino acid sequences of the 131 NHA9-BioID proximal interactors were analyzed by two different algorithms (NES Finder 0.2 and LocNES) to evaluate the presence of classical NES. (A) The results from both algorithms show that the majority of NHA9-BioID proximal interactors have at least one putative classical NES, with a higher percentage of NES+ proteins determined by the LocNES algorithm. (B) GO analysis of NES+ and NES- proteins identified by NES Finder 0.2 shows an overrepresentation of proteins from cytoplasmic RNP granules and the microtubule organizing center. NES- proteins are mostly nucleoplasmic and are associated with the ß-catenin and the AP-1 transcription factor complexes. Statistical analysis was performed with the PANTHER classification online software (v14.1), using Fisher’s exact test. Results are displayed for FDR p < 0.05.
3.5. Identification of Novel Proximal Interactors of SET-NUP214
Our LFQ normalization strategy produced a list of 1125 proteins enriched in SN214-BioID relative to the BirAR118G control (Table S4). To disclose the SET-NUP214 proximal interactome, we performed GO enrichment analysis of the SN214-BioID fraction using the same fold-change threshold of ≥0.6 and statistical parameters as for NHA9-BioID (Figure 5, Figures S3 and S4). For clustered pathway analysis, we applied a conservative approach to reduce redundancy of the functional network given the elevated number of potential SN214-BioID interactors. Figure 6 shows the functional groups that were significantly represented, obtained with the following statistical parameters: p <0.01, GO levels: 13-15 (ClueGO default: 3-8), threshold kappa score of 0.5 (ClueGO default 0.4), thus increasing the threshold for group overlapping, and the minimum gene number corresponding to percentage of genes of 40/4% (ClueGO default 3/4%).
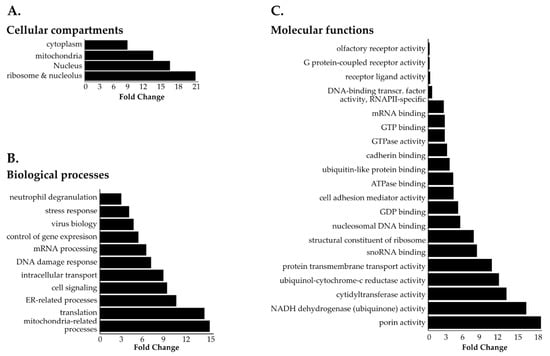
Figure 5.
Gene ontology (GO) of SN214-BioID proximal interactors. Most represented (A) cellular compartments (GOCC), (B) biological processes (GOBP), and (C) molecular functions (GOMF) among SN214-BioID proximal interactors. Statistical analysis of the overrepresented proteins in the SN214-BioID fraction (total 1125 proteins) was performed with the PANTHER classification online software (v14.1) using Fisher’s exact test. Results are displayed for FDR p < 0.05. Summarized graphic representation of the significantly enriched GO terms among SN214-BioID proximal interactors. The detailed results of GOBP and GOCC analysis can be consulted in Figures S3 and S4.
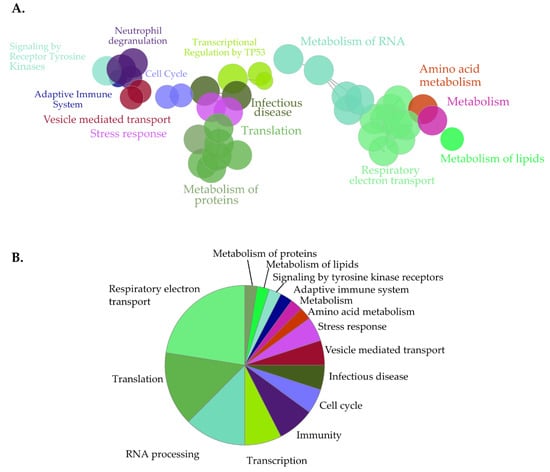
Figure 6.
Clustered pathway analysis of SN214-BioID proximal interactors. (A) Functionally grouped network of SN214-BioID proximal interactors. Nodes correspond to functional clusters resulting from term grouping based on their overlapping level. (B) Overview pie-chart with functional groups. Statistical analysis was performed with the Cytoscape plugin ClueGo (v2.5.5) using the hypergeometric test and the following parameters: p < 0.01, kappa-score (k) = 0.5; (min/%) genes = 40/4%, GO tree levels: 13-15. Ontology databases: GOBP, GOCC, and GOMF, Reactome Pathways and KEGG.
GOCC (Figure 5A and Figure S3) and GOBP (Figure 5B and Figure S4) analysis produced a list of cytoplasmic and nuclear structures and processes likely to be affected by SET-NUP214, such as mRNA processing, intracellular transport, viral transport, and transcription (Figure 5B, Figure S4) [48,50,51,52,53]. Consistently GOMF analysis (Figure 5C) unveiled an enrichment in proteins with GTP- and GDP-binding activity, as well as mRNA and DNA binding proteins. Moreover, proteins involved in neutrophil degranulation were enriched with SET-NUP214 (Figure 5B), establishing a direct link between SET-NUP214 and immune regulation. GO also showed an enrichment in nucleolar and ribosomal proteins (Figure 5A and Figure S3), suggesting an effect of the fusion protein on translation. It further revealed that mitochondrial proteins were enriched with SN214-BioID, especially proteins from the respiratory chain complexes and proteins involved in mitochondrial translation (Figure 5A, Figures S3 and S4), suggesting involvement of the fusion protein in cell metabolism through an effect on mitochondria. In line with the GO results, clustered pathway analysis of proximal SET-NUP214 interactors revealed an enrichment in proteins involved in amino acid metabolism, translation and infection, and proteins involved in virus biology, such as transcription, transport, and interaction with host cells (Figure 6, Figures S4 and S6, Table S5). Clustered pathway analysis further supports GO findings of an interplay between SET-NUP214 and mitochondrial proteins (Figure 6B, Figures S3 and S4, Table S5). Moreover, the results suggest an association of SET-NUP214 with several transcription factors and point to a possible effect on TP53-mediated transcription (Figure 6A,B, Table S5, Figure S5). Finally, our clustered pathway analysis reinforces the link between SET-NUP214 and immunity, with two distinct, yet overlapping, immunity-related clusters that link the fusion protein to immune system regulation, and more specifically to neutrophil activation (Figure 6A, Table S5).
We identified a total of 47 proteins that were enriched with both NHA9-BioID and SN214-BioID, of which a vast majority presented at least one putative classic NES (Figure S7A and Table S6). Furthermore, GO analysis showed that the pool of shared proximal interactors is enriched in proteins associated with microtubule cytoskeleton, suggesting that SET-NUP214, like NUP98-HOXA9, might be involved in microtubule organization (Figure S7B).
4. Discussion
We used a modified BioID approach to study the proximal interactome of NUP98-HOXA9 and SET-NUP214. We compared the pool of proteins biotinylated by NHA9-BioID and SN214-BioID, respectively, with the pool of proteins biotinylated by BirAR118G, using a normalization strategy that accounts for differences in NHA9-BioID, SN214-BioID and BirAR118G expression due to their transient transfection. After validating the expression, cellular distribution, and the capacity of protein biotinylation of all three BioID proteins, we performed gene ontology (GO) and clustered pathway enrichment analysis of the proximal interactors of NHA9-BioID and SN214-BioID. As expected, we observed that BirAR118G alone biotinylates endogenous proteins in an unspecific manner [31]. The respective proximal interactomes of NHA9-BioID and SN214-BioID were, somewhat surprisingly, enriched in cytoplasmic proteins, which appears inconsistent with the nuclear localization of the fusion proteins. This might be explained by the fact that mitosis in vertebrates involves nuclear envelope breakdown and mixing of the cytoplasmic and nuclear protein content [56]. At the end of mitosis, proteins must be appropriately segregated between the nucleus and the cytosol. This process depends on transport factors, including CRM1, which alone is predicted to transport one fourth of the entire proteome [57]. As shown by us and others, NUP98-HOXA9 and SET-NUP214 sequester CRM1 nuclear export complexes, resulting in the nuclear accumulation of proteins and RNPs [23,24,25]. Thus, it is possible that in cells expressing NUP98-HOXA9 and SET-NUP214 fusion proteins protein segregation after mitosis is affected, and that otherwise exclusively cytoplasmic proteins are retained in the nucleus.
The results from GO and pathway enrichment analysis suggest that both NUP98-HOXA9 and SET-NUP214 interact with major regulatory proteins, such as members of the RNAPII complex, ribosomal proteins, and transcription factor complexes, suggesting a widespread effect on gene expression (Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6). NUP98-HOXA9 and SET-NUP214 form dynamic nuclear foci that accumulate endogenous proteins [23,46]. These structures may represent factories that bring transcriptional and epigenetic regulators into close proximity to promote gene expression changes [29,30,58]. Interestingly, our analyses suggest that p53-mediated transcription is a common target of NUP98-HOXA9 and SET-NUP214. The association of the fusion proteins with p53 might occur via their NUP98 and SET portions, respectively. Both proteins regulate TP53-mediated transcription, namely of the cyclin-dependent kinase inhibitor p21 (p21CDKN1A), a DNA damage response and cell cycle regulator [59,60,61]. p53 dysregulation is frequent in both ALL and AML, and loss of p53 function was previously reported to promote AML progression in a NUP98-HOXD13 mouse model, suggesting that it might contribute to NUP98-related leukemia [62,63,64]. Nevertheless, despite the direct binding between SET and p53, the status of p53 signaling in SET-NUP214 leukemia has not been studied.
Changes in gene expression by NUP98-HOXA9 might result from its interaction with major transcription factors, such as TFAP2A (Table S2) and the AP-1 transcription factor complex (Figure 2 and Figure 3), and proteins from cytoplasmic RNP granules (Figure 2 and Figure 3), where mRNA is processed for translation or targeted for degradation [47,65,66,67,68]. Chromatin immunoprecipitation (ChIP) experiments showed that NUP98-HOXA9 (but not NUP98 nor HOXA9) binds at genomic regions that are adjacent to the TFAP2A sequence recognition motif, further supporting a potential interaction between the fusion protein and this transcription factor [47]. TFAP2A regulates the transcription of several developmental genes and has been reported to promote HOX gene upregulation of clustered HOX genes in AML [69]. Moreover, members of Wnt, MAPK, and ER signaling pathways are enriched with NHA9-BioID (Figure 3). Among them, the DVL-1, TLE, isoforms 1–4, and TNRC6A proteins, which are common to Wnt and ER signaling, were exclusively found in the NHA9-BioID pool of biotinylated proteins, reinforcing the idea of a specific effect of NUP98-HOXA9 in these two signaling pathways. In line with our findings, previous work showed that in NUP98-HOXA-transduced hematopoietic stem cells, Wnt and ER signaling pathways were dysregulated, which correlated with cellular transformation [67].
Most of the NHA9-BioID proximal interactors are predicted to have at least one NES, supporting the idea that the biological consequences of CRM1 inhibition by the fusion protein might depend on the type of cargoes that become trapped in the nucleus. NES+ NHA9-BioID proximal interactors were enriched in cytoplasmic RNP granules and microtubule organizing center (MTOC)-associated proteins (Figure 4B). As a MTOC docking protein, CRM1 facilitates microtubule nucleation at the NPC periphery [70]. Several cell-death-related processes, such as autophagy and apoptosis, which are activated under stress, rely on the microtubule system [71,72]. Still, the biological implications of the nuclear retention of MTOC-related proteins, which has been reported in cancer cells, have remained unclear [73,74].
Among SET-NUP214 proximal interactors, our results revealed an unexpected enrichment of mitochondrial proteins. This enrichment might be explained by changes in the communication between the mitochondria and the nucleus (anterograde/retrograde transport) that may be imposed by SET-NUP214 at the NPC. The molecular determinants of anterograde and retrograde transport are still unclear, but current evidence shows that some mitochondrial proteins also have functions in the nucleus, and that nuclear translocation of mitochondrial proteins is an emerging mitochondrial signaling pathway [75,76]. This system relies on the microtubule network that proteins and even entire organelles use to move from and towards the vicinity of the NE, respectively [77]. Given the recently reported MTOC function of CRM1, it is tempting to hypothesize that SET-NUP214 might dock CRM1 at the cytoplasmic side of the NPC and support its MTOC function, thereby promoting the accumulation of mitochondrial proteins (or even entire mitochondria) in the periphery of the NPC. The possibility that SET-NUP214 is related to mitochondria dysregulation is supported by the observation that patients with SET-NUP214 T-related ALL are resistant to glucocorticoid (GC) therapy, which induces a metabolic shift from glycolysis to oxidative phosphorylation (OXPHOS; [20,78]). The AML-associated DEK-NUP214 fusion protein, which has the same NUP214 portion as SET-NUP214, also promotes a shift from glycolysis to OXPHOS [79]. Yet, in a recent proteomics report, no mitochondrial proteins were reported in the DEK-NUP214 interactome and dysregulation of the mammalian target of rapamycin (mTOR) was assumed as the main cause of metabolic shift [80].
Among SN214-BioID interactors, eight proteins (i.e., RAB6A, PCBD1, RPTOR, RIN1, NRAS, GCC2, LYN, and CRKL) were common to the DEK-NUP214 interactome (Table S4; [80]). The association of these proteins with DEK-NUP214 was proposed to promote activation of several cancer associated pathways, such as AKT/mTOR, Src family kinase (SFK), ABL1, and c-MYC pathways [81,82,83,84]. Further studies will be necessary to confirm the same pathway profile activation by SET-NUP214. Nevertheless, given the structural similarities of the two chimeras, it is reasonable to expect a significant overlap in the biological processes that are affected by both fusion proteins [12,14,85].
Despite the identification of the above-mentioned numerous NUP98-HOXA9 SN214-BioID interactors, our approach possibly falls short of others due to the promiscuous biotinylation activity of BirAR118G. Although our normalization strategy accounts for differences in the expression of the BioID fusion proteins, we observed that some known NUP98-HOXA9 and SET-NUP214 binding partners, such as CRM1 (for both fusion proteins) and MLL1 (for NUP98-HOXA9), were not strongly enriched in the NHA9-BioID and SN214-BioID fractions relative to BirAR118G alone. We hypothesize that the same may occur with other NUP98-HOXA9 and SET-NUP214 interactors and for that reason we recognize that our BioID approach might miss relevant candidate binding partners. Nevertheless, the unspecific protein biotinylation by BirAR118G alone, and the identification of several known binding partners enriched with NHA9-BioID and SN214-BioID, reinforce our approach in the study of NUP98-HOXA9 and SET-NUP214 proximal interactome.
5. Conclusions
Overall, our report provides new data on the landscape of potential binding partners of nucleoporin fusion proteins, and suggests novel cellular processes and signaling pathways that may be affected by NUP98-HOXA9 and SET-NUP214. Although we identified several previously validated binding partners of both fusion proteins, experimental validation by gold-standard experimental methods, such as protein immunoprecipitation, is necessary to confirm the predicted interaction candidates. Our work unveils, for the first time, putative new players in nucleoporin-related leukemia, and provides the basis for a new understanding of the biological actions of nucleoporin fusion proteins. Our findings may be of particular relevance in the search for new druggable targets, such as the MAPK and ER pathways, that might be explored in the development of specific therapies for NUP98 and NUP214 leukemia.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/7/1666/s1, Figure S1: Normalization strategy of BioID results, Figure S2: Clustered Pathway Analysis of NUP98-HOXA9-BioID proximal interactors, Figure S3: Most represented cellular compartments (GOCC) among SN214-BioID proximal interactors, Figure S4: Most represented biological processes (GOBP) among SN214-BioID proximal interactors., Figure S5: Network of direct p53 binding proteins in SN214-BioID and NHA9-BioID proximal interactors., Figure S6: Clustered Pathway Analysis of SET-NUP2147-BioID proximal interactors, Table S1: List of biotinylated proteins found exclusively in the biotinylated fraction of BirAR118G, Table S2: List of overrepresented proteins in the biotinylated fraction of NHA9-BioID (NHA9-BioID proximal interactors) and respective fold-change relative to control BirAR118G, Table S3: Presence of classical nuclear export signals (NES) in NHA9-BioID proximal interactors, Table S5: List of functional groups with corresponding genes of SN214-BioID proximal interactors generated by the ClueGO Cytoscape plugin.
Author Contributions
Conceptualization, A.M. and B.F.; methodology, A.M., S.B.; software, A.M., R.J.; formal analysis, A.M. and R.J.; investigation, A.M., S.B.; resources, R.J.; writing—original draft preparation, A.M.; writing—review and editing, B.F.; supervision, B.F.; project administration, B.F.; funding acquisition, B.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the Fonds National de la Recherche Scientifique (F.R.S.–FNRS–FRIA, FC 16752) and Fondation Rose et Jean Hoguet to A.M., as well as by research grants from the FNRS (T.0082.14 and J.013616F) and the Fédération Wallonie-Bruxelles (ARC 4.110. F.000092F) to B.F.
Acknowledgments
We are grateful to Francis Impens and Evy Timmerman from the VIB Proteomics Core Facility (VIB, University of Gent, Belgium) for the mass spectrometry service. We thank yle Roux (University of South Dakota, Vermillon, SD, USA), Karl Kramer (Technical University of Munich, Germany), and Denis Lafontaine (Université Libre de Bruxelles, Belgium) for sharing reagents. Confocal images were acquired at the CMMI (Gosselies, Belgium), which is supported by the European Regional Development Fund (ERDF).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Appendix A.1. Cloning of SET-NUP214 into the pcDNA3.1 MCS-BirA(R118G)-HA Vector
For the cloning of SET-NUP214-GFP, total RNA (RNAT) was extracted from LOUCY cells, which carry del(9)(q34) resulting in the SET-NUP214 fusion transcript, using the High Pure RNA Isolation Kit (Roche Life Sciences, Basel, Switzerland) according to the manufacturer’s instructions and stored at –80 °C for further use. Synthesis of cDNA was performed by reverse transcriptase polymerase chain reaction (RT-PCR) as follows (all reagents were kept on ice for manipulation): 100 ng of RNAT were mixed with 3.4 µg of deoxy-thymidine oligomer (OligodT)12-18 primer and 10 nmol of desoxyribonucleotides (dNTPs) in a final volume of 20 µl. Samples were first incubated at 65 °C for 5 min and the RT-PCR reaction was initiated by the addition of 2 µl of 0.1 µM DTT, 4 µl of 5x First Strand Buffer, 40 units (U) of RNase OUTTM and 200 U of SuperScriptTM Reverse Transcriptase II (SSII-ThermoFisher Scientific, Merelbek, Belgium). The final mixture was incubated at the following thermal program: 42 °C for 2 min, 25 °C for 10 min and 42 °C for 1 h. The resulting cDNA was immediately used for PCR amplification or stored at −20 °C for further applications. Next, the converted cDNA was used as template for the amplification of the SET-NUP214 coding sequence by PCR using specific primers (forward 5′- ATGTCGGCGCCGGCGGCCAA-AGTCAGTAAA-3’; reverse 5’- TATCCCGGGCTTCGCCAGCCACCA-AAACC-3’). The resulting PCR product was purified using the High Pure PCR Product Purification Kit (Roche Life Sciences) and inserted into the pGEM®-T intermediate vector backbone (Promega, Leiden, Netherlands). The ligation product was used to transform E. coli XL1-blue competent cells and positive colonies were selected by blue/white screening. Plasmid DNA was isolated using the GenElute™ Plasmid MidiPrep Kit (Sigma–Aldrich) according to the manufacturer’s instructions. To generate the SN214-BioID construct, the SET-NUP214 coding sequence was subcloned from the pGEM-T backbone into the pcDNA3.1 MCS-BirA(R118G)-HA using AgeI and EcoRI restriction enzymes (New England Biolabs—NEB, Ipswich, MA, USA). To insert AgeI and EcoRI restriction sites into the PGEM-T-SET-NUP214 construct the following primers were used for the PCR reaction: forward 5′ -TACCGGTATGTCGGCGCCGGCGGCCAAAGTCAGTAAA-3′; reverse 5′- ACTGGAATTCGCTTCGCCAGCCACCAAAACC – 3′. Digestion products were purified using the High Pure PCR Product Purification Kit (Roche Life Sciences). Ligation was performed for 2 h using T4 DNA ligase (Thermo Fisher Scientific) and the purified ligation product was transformed into XL1-blue competent cells. Cells were then seeded on LB-agar plates supplemented with ampicillin (100 µg/ml) and grown overnight at 37 °C. Colonies were screened by colony-PCR and positive clones were expanded in LB/ampicillin medium. The SET-NUP214-BirAR118G plasmid was fully sequenced to verify the correct sequence.
Appendix A.2. Incubation and Elution of Biotinylated Proteins from Streptavidin-Coated Magnetic Beads
Protein-bead mixtures were incubated for 1 h with slow end-to-end agitation. Next, beads were magnetically precipitated and washed twice in wash buffer (2 M urea,150 mM NaCl, 50 mM Tris, pH 7.5). After washing, proteins were eluted by boiling for 7 min in excess biotin, as described in [86]. Laemmli buffer (2% SDS, 10% glycerol, 60 mM Tris-HCl, pH 6.8) was then added to each eluate and boiled for 5 min. Detection of biotinylated proteins with horseradish-conjugated streptavidin (HRP-Strep, Thermo Fischer Scientific, Merelbeke, Belgium) was performed as previously described in [86]. As input, 50 µg of whole protein extract was used.
Appendix A.3. Sample Injection and Mass Spectrometer Operation
Trapping was performed at 10 μl/min for 4 min in loading solvent A on a 20 mm trapping column (made in-house, 100 μm internal diameter (I.D.), 5 μm beads, C18 Reprosil-HD, Ammerbuch, Germany) and the sample was loaded on a 20 cm analytical column (made in-house, 75 μm I.D. × 400 mm, 1.9 μm beads C18 Reprosil-HD, Ammerbuch, Germany), kept at a constant temperature of 50 °C. Peptides were eluted by a non-linear gradient reaching 56% MS solvent B (0.1% FA in water/acetonitrile (2:8, v/v)) in 87 min. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the 12 most abundant ion peaks per MS spectrum. Full-scan MS spectra (375–1500 m/z) were acquired at a resolution of 60,000 in the orbitrap analyzer after accumulation to a target value of 1E5. The 12 most intense ions above a threshold value of 1.3E4 were isolated for fragmentation at a normalized collision energy of 30% after filling the trap at a target value of 1E3 for maximum 80 MS. MS/MS spectra (200–2000 m/z) were acquired at a resolution of 15,000 in the orbitrap analyzer.
Appendix A.4. Raw Data Analysis Using MaxQuant
For the MaxQuant analysis of raw data, the mass tolerance for precursor and fragment ions was set to 20 and 4.5 ppm, respectively, during the main search. Enzyme specificity was set to C-terminal to arginine and lysine, also allowing cleavage at arginine/lysine-proline bonds with a maximum of two missed cleavages. Variable modifications were set to oxidation of methionine (to sulfoxides) and acetylation of protein N termini. A minimum of one peptide in total was required for identification. Matching was allowed between runs using a 1.5 min match and a 20 min alignment time window. Proteins were quantified by the MaxLFQ algorithm integrated in the MaxQuant software. A minimum ratio count of two unique or razor peptides was required for quantification.
References
- Terry, L.J.; Wente, S. Flexible Gates: Dynamic Topologies and Functions for FG Nucleoporins in Nucleocytoplasmic Transport. Eukaryot. Cell 2009, 8, 1814–1827. [Google Scholar] [CrossRef]
- Paulillo, S.M.; Powers, M.A.; Ullman, K.S.; Fahrenkrog, B. Changes in Nucleoporin Domain Topology in Response to Chemical Effectors. J. Mol. Biol. 2006, 363, 39–50. [Google Scholar] [CrossRef]
- Chatel, G.; Desai, S.H.; Mattheyses, A.L.; Powers, M.A.; Fahrenkrog, B. Domain topology of nucleoporin Nup98 within the nuclear pore complex. J. Struct. Biol. 2012, 177, 81–89. [Google Scholar] [CrossRef]
- Yang, J.; Lyu, X.; Zhu, X.; Meng, X.-G.; Zuo, W.; Ai, H.; Deng, M. Chromosome t(7;11)(p15;p15) translocation in acute myeloid leukemia coexisting with multilineage dyspoiesis and mutations in NRAS and WT1: A case report and literature review. Oncol. Lett. 2017, 13, 3066–3070. [Google Scholar] [CrossRef][Green Version]
- Gough, S.M.; Slape, C.; Aplan, P.D. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef]
- Fahrenkrog, B. Nucleoporin Gene Fusions and Hematopoietic Malignancies. New J. Sci. 2014, 2014, 1–18. [Google Scholar] [CrossRef]
- Martins, N.; Mendes, A.; Fahrenkrog, B. On the Effects of Leukemogenic Nucleoporin Fusion Proteins on Nucleocytoplasmic Transport and Gene Expression; Springer Science and Business Media LLC: New York, NY, USA, 2018; pp. 223–248. [Google Scholar]
- Lam, D.H.; Aplan, P.D. NUP98 gene fusions in hematologic malignancies. Leukemia 2001, 15, 1689–1695. [Google Scholar] [CrossRef] [PubMed]
- Ghannam, G.; Takeda, A.; Camarata, T.; Moore, M.A.; Viale, A.; Yaseen, N.R. The OncogeneNup98-HOXA9Induces Gene Transcription in Myeloid Cells. J. Biol. Chem. 2003, 279, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.T.; Hess, J.L. Role of HOXA9 in leukemia: Dysregulation, cofactors and essential targets. Oncogene 2015, 35, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, G.; Lansdorp, P.M.; Eaves, C.J.; Hogge, D.E.; Dragowska, W.H.; Reid, D.S.; Largman, C.; Lawrence, H.J.; Humphries, R.K. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. USA 1994, 91, 12223–12227. [Google Scholar] [CrossRef] [PubMed]
- von Lindern, M.; van Baal, S.; Wiegant, J.; Raap, A.; Hagemeijer, A.; Grosveld, G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3’ half to different genes: Characterization of the set gene. Mol. Cell. Biol. 1992, 12, 3346–3355. [Google Scholar] [CrossRef] [PubMed]
- Mendes, A.; Fahrenkrog, B. NUP214 in Leukemia: It’s More than Transport. Cells 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.-B.; McNamara, P.; Heo, S.; Turner, A.; Lane, W.S.; Chakravarti, D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 2001, 104, 119–130. [Google Scholar] [CrossRef]
- Kutney, S.N.; Hong, R.; Macfarlan, T.S.; Chakravarti, D. A Signaling Role of Histone-binding Proteins and INHAT Subunits pp32 and Set/TAF-Iβ in Integrating Chromatin Hypoacetylation and Transcriptional Repression. J. Biol. Chem. 2004, 279, 30850–30855. [Google Scholar] [CrossRef] [PubMed]
- Switzer, C.H.; Cheng, R.Y.; Vitek, T.M.; Christensen, D.J.; Wink, D.A.; Vitek, M.P. Targeting SET/I2PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene 2011, 30, 2504–2513. [Google Scholar] [CrossRef]
- Ichijo, T.; Chrousos, G.P.; Kino, T. Activated glucocorticoid receptor interacts with the INHAT component Set/TAF-Iβ and releases it from a glucocorticoid-responsive gene promoter, relieving repression: Implications for the pathogenesis of glucocorticoid resistance in acute undifferentiated leukemia with Set-Can translocation. Mol. Cell. Endocrinol. 2008, 283, 19–31. [Google Scholar] [CrossRef]
- Enjoji, S.; Yabe, R.; Tsuji, S.; Yoshimura, K.; Kawasaki, H.; Sakurai, M.; Sakai, Y.; Takenouchi, H.; Yoshino, S.; Hazama, S.; et al. Stemness Is Enhanced in Gastric Cancer by a SET/PP2A/E2F1 Axis. Mol. Cancer Res. 2018, 16, 554–563. [Google Scholar] [CrossRef]
- Fahrenkrog, B.; Martinelli, V.; Nilles, N.; Fruhmann, G.; Chatel, G.; Juge, S.; Sauder, U.; Di Giacomo, D.; Mecucci, C.; Schwaller, J. Expression of Leukemia-Associated Nup98 Fusion Proteins Generates an Aberrant Nuclear Envelope Phenotype. PLoS One 2016, 11, e0152321. [Google Scholar] [CrossRef][Green Version]
- Port, S.A.; Mendes, A.; Valkova, C.; Spillner, C.; Fahrenkrog, B.; Kaether, C.; Kehlenbach, R.H. The Oncogenic Fusion Proteins SET-Nup214 and Sequestosome-1 (SQSTM1)-Nup214 Form Dynamic Nuclear Bodies and Differentially Affect Nuclear Protein and Poly(A)+ RNA Export*. J. Biol. Chem. 2016, 291, 23068–23083. [Google Scholar] [CrossRef]
- Saito, S.; Cigdem, S.; Okuwaki, M.; Nagata, K. Leukemia-Associated Nup214 Fusion Proteins Disturb the XPO1-Mediated Nuclear-Cytoplasmic Transport Pathway and Thereby the NF-κB Signaling Pathway. Mol. Cell. Biol. 2016, 36, 1820–1835. [Google Scholar] [CrossRef]
- Mendes, A.; Juehlen, R.; Martinelli, V.; Fahrenkrog, B. Targeted CRM1-inhibition perturbs leukemogenic NUP214 fusion proteins and exerts anti-cancer effects in leukemia cell lines with NUP214 rearrangements. bioRxiv 2020. [Google Scholar] [CrossRef]
- Takeda, A.; Sarma, N.J.; Abdul-Nabi, A.M.; Yaseen, N.R. Inhibition of CRM1-mediated Nuclear Export of Transcription Factors by Leukemogenic NUP98 Fusion Proteins. J. Biol. Chem. 2010, 285, 16248–16257. [Google Scholar] [CrossRef] [PubMed]
- Van Vlierberghe, P.; Van Grotel, M.; Tchinda, J.; Lee, C.; Beverloo, H.B.; Van Der Spek, P.J.; Stubbs, A.; Cools, J.; Nagata, K.; Fornerod, M.; et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood 2008, 111, 4668–4680. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Valerio, D.G.; Eisold, M.E.; Sinha, A.; Koche, R.P.; Hu, W.; Chen, C.-W.; Chu, S.H.; Brien, G.L.; Park, C.Y.; et al. NUP98 Fusion Proteins Interact with the NSL and MLL1 Complexes to Drive Leukemogenesis. Cancer Cell 2016, 30, 863–878. [Google Scholar] [CrossRef] [PubMed]
- Shima, Y.; Yumoto, M.; Katsumoto, T.; Kitabayashi, I. MLL is essential for NUP98-HOXA9-induced leukemia. Leukemia 2017, 31, 2200–2210. [Google Scholar] [CrossRef]
- Oka, M.; Mura, S.; Yamada, K.; Sangel, P.; Hirata, S.; Maehara, K.; Kawakami, K.; Tachibana, T.; Ohkawa, Y.; Kimura, H.; et al. Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes. eLife 2016, 5, 1195. [Google Scholar] [CrossRef]
- Oka, M.; Mura, S.; Otani, M.; Miyamoto, Y.; Nogami, J.; Maehara, K.; Harada, A.; Tachibana, T.; Yoneda, Y.; Ohkawa, Y. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. eLife 2019, 8. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Galon, J.; Bindea, G. Automated exploration of gene ontology term and pathway networks with ClueGO-REST. Bioinformatics 2019, 35, 3864–3866. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef]
- La Cour, T.; Kiemer, L.; Gupta, R.; Skriver, K.; Brunak, S.; Mølgaard, A. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Marquis, K.; Pei, J.; Fu, S.-C.; Cağatay, T.; Grishin, N.V.; Chook, Y.M. LocNES: A computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 2014, 31, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Powers, M.A. Nup98-Homeodomain Fusions Interact with Endogenous Nup98 during Interphase and Localize to Kinetochores and Chromosome Arms during Mitosis. Mol. Biol. Cell 2010, 21, 1585–1596. [Google Scholar] [CrossRef]
- Rio-Machin, A.; Gómez-López, G.; Muñoz, J.; Garcia, F.; Maiques-Diaz, A.; Alvarez, S.; Salgado, R.N.; Shrestha, M.; Torres-Ruiz, R.; Haferlach, C.; et al. The molecular pathogenesis of the NUP98-HOXA9 fusion protein in acute myeloid leukemia. Leukemia 2017, 31, 2000–2005. [Google Scholar] [CrossRef][Green Version]
- Kasper, L.H.; Brindle, P.; Schnabel, C.A.; Pritchard, C.E.J.; Cleary, M.L.; Van Deursen, J.M.A. CREB Binding Protein Interacts with Nucleoporin-Specific FG Repeats That Activate Transcription and Mediate NUP98-HOXA9 Oncogenicity. Mol. Cell. Biol. 1999, 19, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2016, 45, D777–D783. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2018, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Aguilera, A.; Arranz, L.; Pérez, D.M.; García-García, A.; Stavropoulou, V.; Kubovcakova, L.; Isern, J.; Martín-Salamanca, S.; Langa, X.; Skoda, R.C.; et al. Estrogen Signaling Selectively Induces Apoptosis of Hematopoietic Progenitors and Myeloid Neoplasms without Harming Steady-State Hematopoiesis. Cell Stem Cell 2014, 15, 791–804. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2016, 36, 1461–1473. [Google Scholar] [CrossRef]
- Zolotukhin, A.S.; Felber, B.K. Nucleoporins Nup98 and Nup214 Participate in Nuclear Export of Human Immunodeficiency Virus Type 1 Rev. J. Virol. 1999, 73, 120–127. [Google Scholar] [CrossRef]
- Lee, Y.; Pei, J.; Baumhardt, J.M.; Chook, Y.M.; Grishin, N.V. Structural prerequisites for CRM1-dependent nuclear export signaling peptides: Accessibility, adapting conformation, and the stability at the binding site. Sci. Rep. 2019, 9, 6627. [Google Scholar] [CrossRef]
- Schmitt, C.; Von Kobbe, C.; Bachi, A.; Panté, N.; Rodrigues, J.P.; Boscheron, C.; Rigaut, G.; Wilm, M.; Seraphin, B.; Carmo-Fonseca, M.; et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999, 18, 4332–4347. [Google Scholar] [CrossRef]
- Trotman, L.C.; Mosberger, N.; Fornerod, M.; Stidwill, R.P.; Greber, U.F. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001, 3, 1092–1100. [Google Scholar] [CrossRef]
- Von Moeller, H.; Basquin, C.; Conti, E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009, 16, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Bernad, R.; van der Velde, H.; Fornerod, M.; Pickersgill, H. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell. Biol. 2004, 24, 2373–2384. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, B.; Barral, Y. The cell biology of open and closed mitosis. Nucleus 2013, 4, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Kırlı, K.; Karaca, S.; Dehne, H.J.; Samwer, M.; Pan, K.-T.; Lenz, C.; Urlaub, H.; Görlich, D. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. eLife 2015, 4. [Google Scholar] [CrossRef]
- Griffis, E.; Altan, N.; Lippincott-Schwartz, J.; Powers, M.A. Nup98 Is a Mobile Nucleoporin with Transcription-dependent Dynamics. Mol. Biol. Cell 2002, 13, 1282–1297. [Google Scholar] [CrossRef]
- Singer, S.; Zhao, R.; Barsotti, A.M.; Ouwehand, A.; Fazollahi, M.; Coutavas, E.; Breuhahn, K.; Neumann, O.; Longerich, T.; Pusterla, T.; et al. Nuclear Pore Component Nup98 Is a Potential Tumor Suppressor and Regulates Posttranscriptional Expression of Select p53 Target Genes. Mol. Cell 2012, 48, 799–810. [Google Scholar] [CrossRef]
- Cazzalini, O.; Scovassi, A.I.; Savio, M.; Stivala, L.A.; Prosperi, E. Multiple roles of the cell cycle inhibitor p21CDKN1A in the DNA damage response. Mutat. Res. Mutat. Res. 2010, 704, 12–20. [Google Scholar] [CrossRef]
- Wang, N.; Kon, N.; Lasso, G.; Jiang, L.; Leng, W.; Zhu, W.-G.; Qin, J.; Honig, B.; Gu, W. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 2016, 538, 118–122. [Google Scholar] [CrossRef]
- Prokocimer, M.; Molchadsky, A.; Rotter, V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: Projections on diagnostic workup and therapy. Blood 2017, 130, 699–712. [Google Scholar] [CrossRef]
- Chiaretti, S.; Brugnoletti, F.; Tavolaro, S.; Bonina, S.; Paoloni, F.; Marinelli, M.; Patten, N.; Bonifacio, M.; Kropp, M.G.; Sica, S.; et al. TP53 mutations are frequent in adult acute lymphoblastic leukemia cases negative for recurrent fusion genes and correlate with poor response to induction therapy. Haematologica 2013, 98, e59–e61. [Google Scholar] [CrossRef]
- Xu, H.; Menéndez, S.; Schlegelberger, B.; Bae, N.; Aplan, P.D.; Göhring, G.; DeBlasio, T.R.; Nimer, S.D. Loss of p53 accelerates the complications of myelodysplastic syndrome in a NUP98-HOXD13–driven mouse model. Blood 2012, 120, 3089–3097. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Goolsby, C.; Yaseen, N.R. NUP98-HOXA9 Induces Long-term Proliferation and Blocks Differentiation of Primary Human CD34+ Hematopoietic. Cells Cancer Res. 2006, 66, 6628–6637. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Nabi, A.M.; Yassin, E.R.; Varghese, N.; Deshmukh, H.; Yaseen, N.R. In Vitro Transformation of Primary Human CD34+ Cells by AML Fusion Oncogenes: Early Gene Expression Profiling Reveals Possible Drug Target in AML. PLoS One 2010, 5, e12464. [Google Scholar] [CrossRef]
- Yassin, E.R.; Sarma, N.J.; Abdul-Nabi, A.M.; Dombrowski, J.; Han, Y.; Takeda, A.; Yaseen, N.R. Dissection of the Transformation of Primary Human Hematopoietic Cells by the Oncogene NUP98-HOXA9. PLoS One 2009, 4, e6719. [Google Scholar] [CrossRef][Green Version]
- Alberti, S.; Matějů, D.; Mediani, L.; Carra, S. Granulostasis: Protein Quality Control of RNP Granules. Front. Mol. Neurosci. 2017, 10, 45920. [Google Scholar] [CrossRef]
- Pushpalatha, K.V.; Besse, F. Local Translation in Axons: When Membraneless RNP Granules Meet Membrane-Bound Organelles. Front. Mol. Biosci. 2019, 6, 129. [Google Scholar] [CrossRef]
- Ding, X.; Yang, Z.; Zhou, F.; Wang, F.; Li, X.; Chen, C.; Li, X.; Hu, X.; Xiang, S.; Zhang, J. Transcription factor AP-2α regulates acute myeloid leukemia cell proliferation by influencing Hoxa gene expression. Int. J. Biochem. Cell Biol. 2013, 45, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.X.; Spanos, C.; Kojidani, T.; Lynch, E.; Rappsilber, J.; Hiraoka, Y.; Haraguchi, T.; Sawin, K.E. Exportin Crm1 is repurposed as a docking protein to generate microtubule organizing centers at the nuclear pore. eLife 2018, 7, e33465. [Google Scholar] [CrossRef]
- Arduíno, D.M.; Esteves, A.R.; Cardoso, S.M. Mitochondria drive autophagy pathology via microtubule disassembly. Autophagy 2013, 9, 112–114. [Google Scholar] [CrossRef]
- Mackeh, R.; Perdiz, D.; Lorin, S.; Codogno, P.; Poüs, C. Autophagy and microtubules - new story, old players. J. Cell Sci. 2013, 126, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Schwarzerová, K.; Bellinvia, E.; Martinek, J.; Sikorová, L.; Dostál, V.; Libusová, L.; Bokvaj, P.; Fischer, L.; Schmit, A.C.; Nick, P. Tubulin is actively exported from the nucleus through the Exportin1/CRM1 pathway. Sci. Rep. 2019, 9, 5725. [Google Scholar] [CrossRef]
- Akoumianaki, T.; Kardassis, D.; Polioudaki, H.; Georgatos, S.; Theodoropoulos, P.A. Nucleocytoplasmic shuttling of soluble tubulin in mammalian cells. J. Cell Sci. 2009, 122, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, R.; Whitmarsh, A.J. Mitochondrial Proteins Moonlighting in the Nucleus. Trends Biochem. Sci. 2015, 40, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Kotiadis, V.; Duchen, M.R.; Osellame, L.D. Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim. Biophys. Acta (BBA) Bioenerg. 2013, 1840, 1254–1265. [Google Scholar] [CrossRef]
- Schnapp, B.J.; Reese, T.S. Dynein is the motor for retrograde axonal transport of organelles. Proc. Natl. Acad. Sci. USA 1989, 86, 1548–1552. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.; Morita, M.; Hirao, T.; Yamaguchi, M.; Shiratori, R.; Kikuya, M.; Chibana, H.; Ito, K. Shift in energy metabolism caused by glucocorticoids enhances the effect of cytotoxic anti-cancer drugs against acute lymphoblastic leukemia cells. Oncotarget 2017, 8, 94271–94285. [Google Scholar] [CrossRef][Green Version]
- Sandén, C.; Ageberg, M.; Petersson, J.; Lennartsson, A.; Gullberg, U. Forced expression of the DEK-NUP214 fusion protein promotes proliferation dependent on upregulation of mTOR. BMC Cancer 2013, 13, 440. [Google Scholar] [CrossRef]
- Guillen, N.; Wieske, M.; Otto, A.; Mian, A.A.; Rokicki, M.; Guy, C.; Alvares, C.; Hole, P.; Held, H.; Ottmann, O.G.; et al. Subtractive Interaction Proteomics Reveal a Network of Signaling Pathways Activated by an Oncogenic Transcription Factor in Acute Myeloid Leukemia. SSRN Electron. J. 2018. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Zhang, J. mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int. J. Mol. Sci. 2019, 20, 755. [Google Scholar] [CrossRef] [PubMed]
- Sen, B.; Johnson, F.M. Regulation of Src Family Kinases in Human Cancers. J. Signal Transduct. 2011, 2011, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Greuber, E.K.; Smith-Pearson, P.; Wang, J.; Pendergast, A.M. Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat. Rev. Cancer 2013, 13, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018, 3, 5. [Google Scholar] [CrossRef]
- Von Lindern, M.; Poustka, A.; Lerach, H.; Grosveld, G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol. Cell. Biol. 1990, 10, 4016–4026. [Google Scholar] [CrossRef] [PubMed]
- Von Lindern, M.; Breems, D.; Van Baal, S.; Adriaansen, H.; Grosveld, G. Characterization of the translocation breakpoint sequences of twoDEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and aSET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosom. Cancer 1992, 5, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Cheah, J.S.; Yamada, S. A simple elution strategy for biotinylated proteins bound to streptavidin conjugated beads using excess biotin and heat. Biochem. Biophys. Res. Commun. 2017, 493, 1522–1527. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).