YRNAs: New Insights and Potential Novel Approach in Head and Neck Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. HNSCC Cell Lines
2.2. Patient Samples
2.3. RNA Isolation and Quantitative RT-PCR
2.4. TCGA Data
2.5. Interaction between YRNA1 Expression and Other Genes
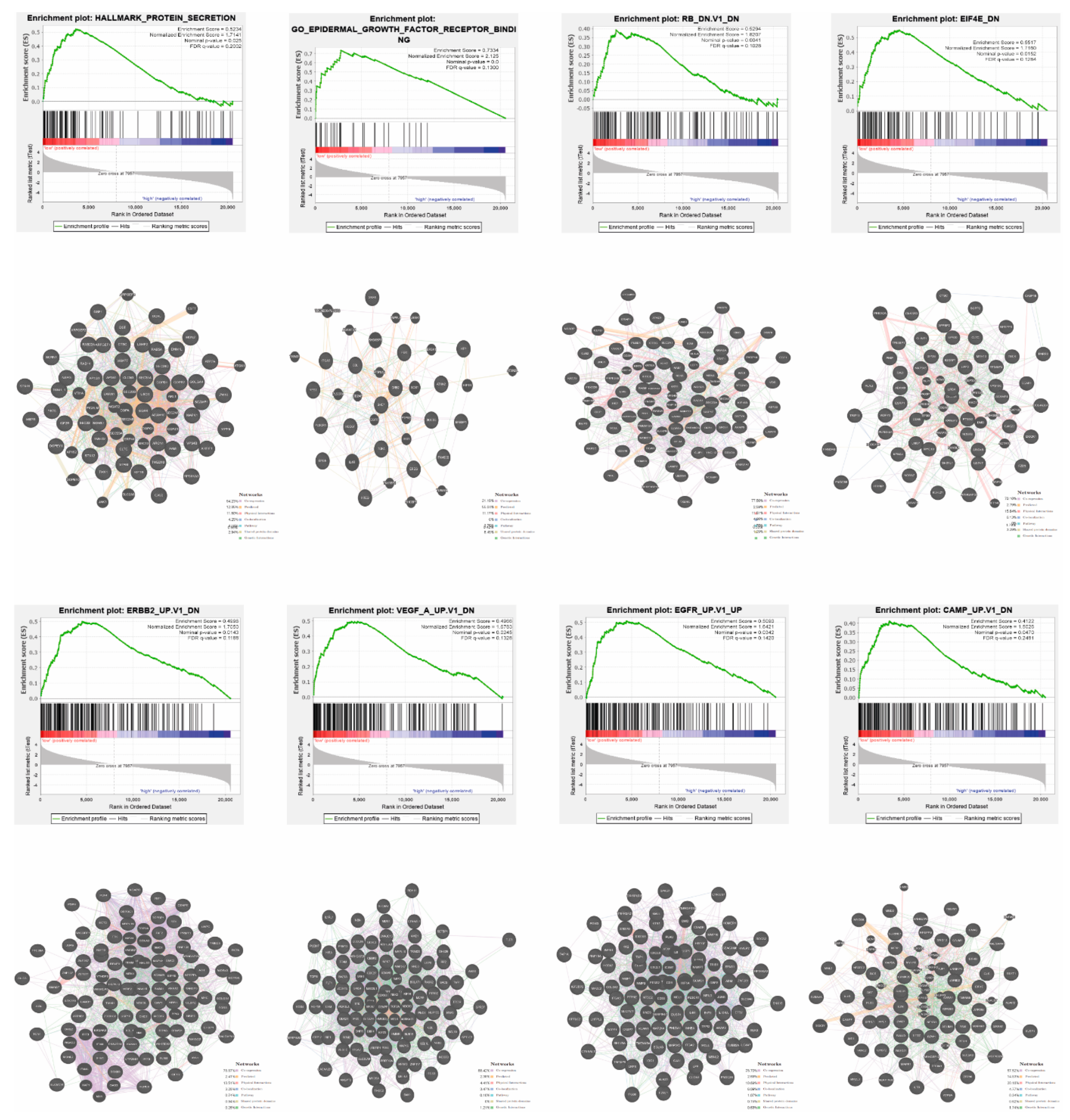
2.6. Functional Enrichment Analysis and Prediction of Gene Function
2.7. Statistical Analysis
3. Results
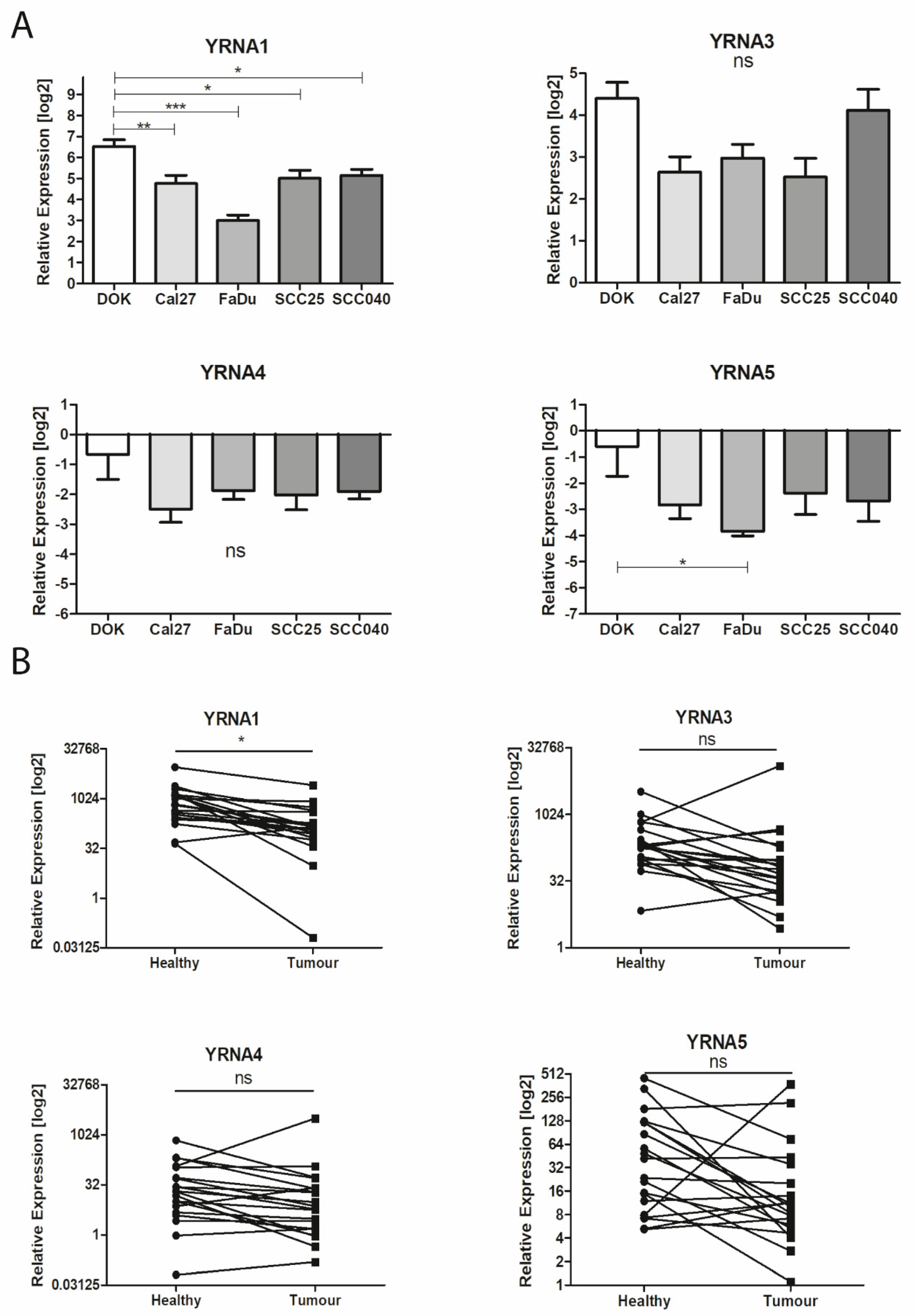
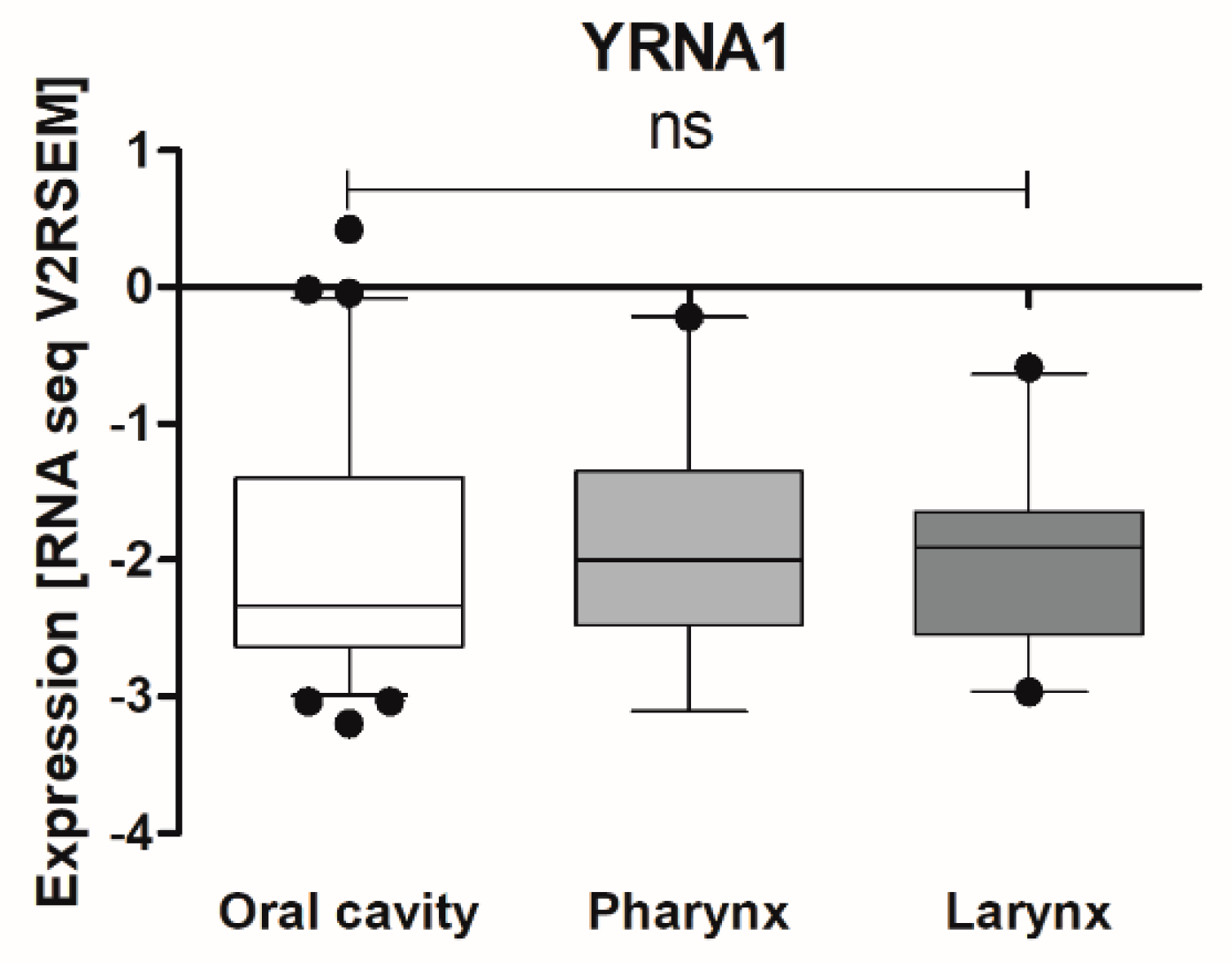
3.1. Expression of YRNAs Is Changed in HNSCC Cell Lines and in Patients’ Tumours
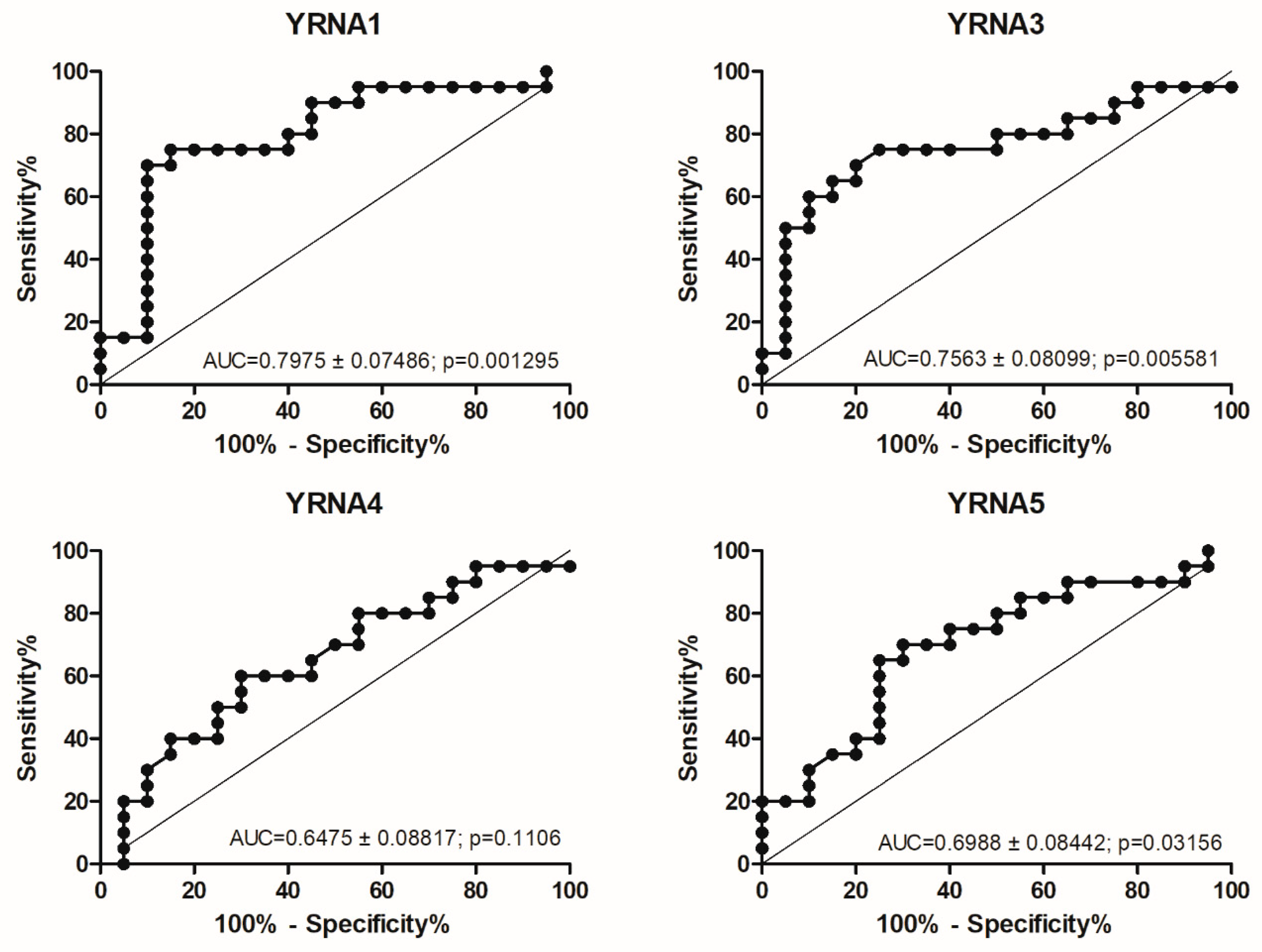
3.2. TCGA Analysis Indicates That YRNA1 Levels Differ According to HPV Status
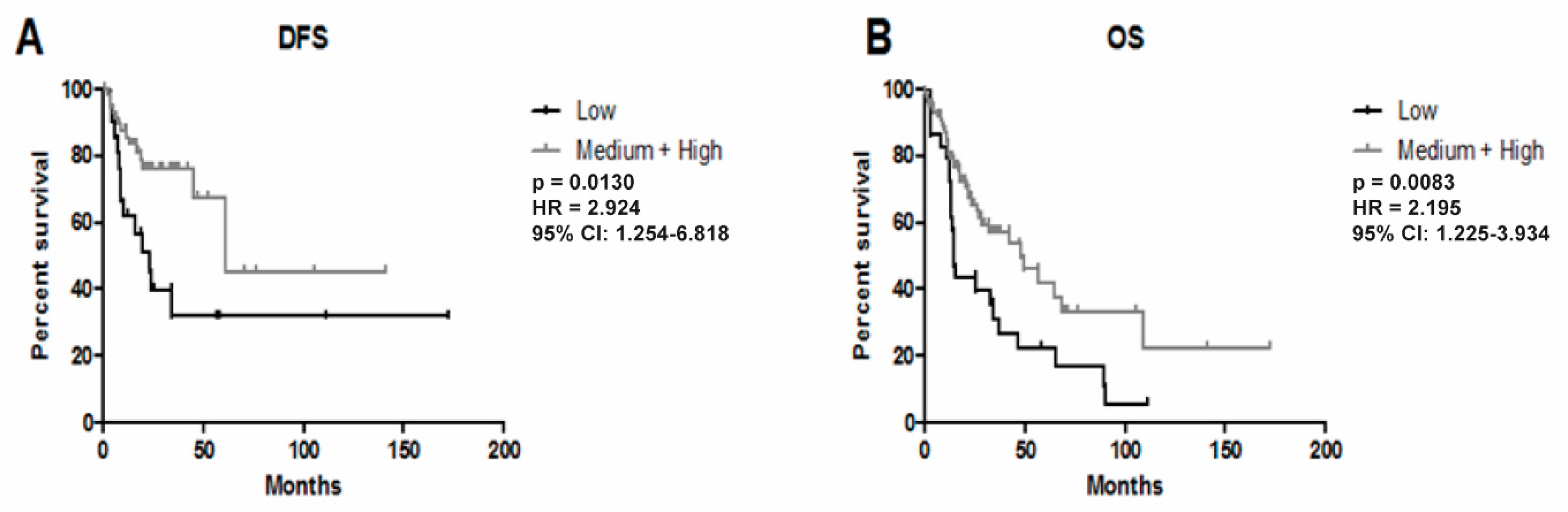
3.3. Higher Expression of YRNA1 Is Associated with Longer DFS and OS
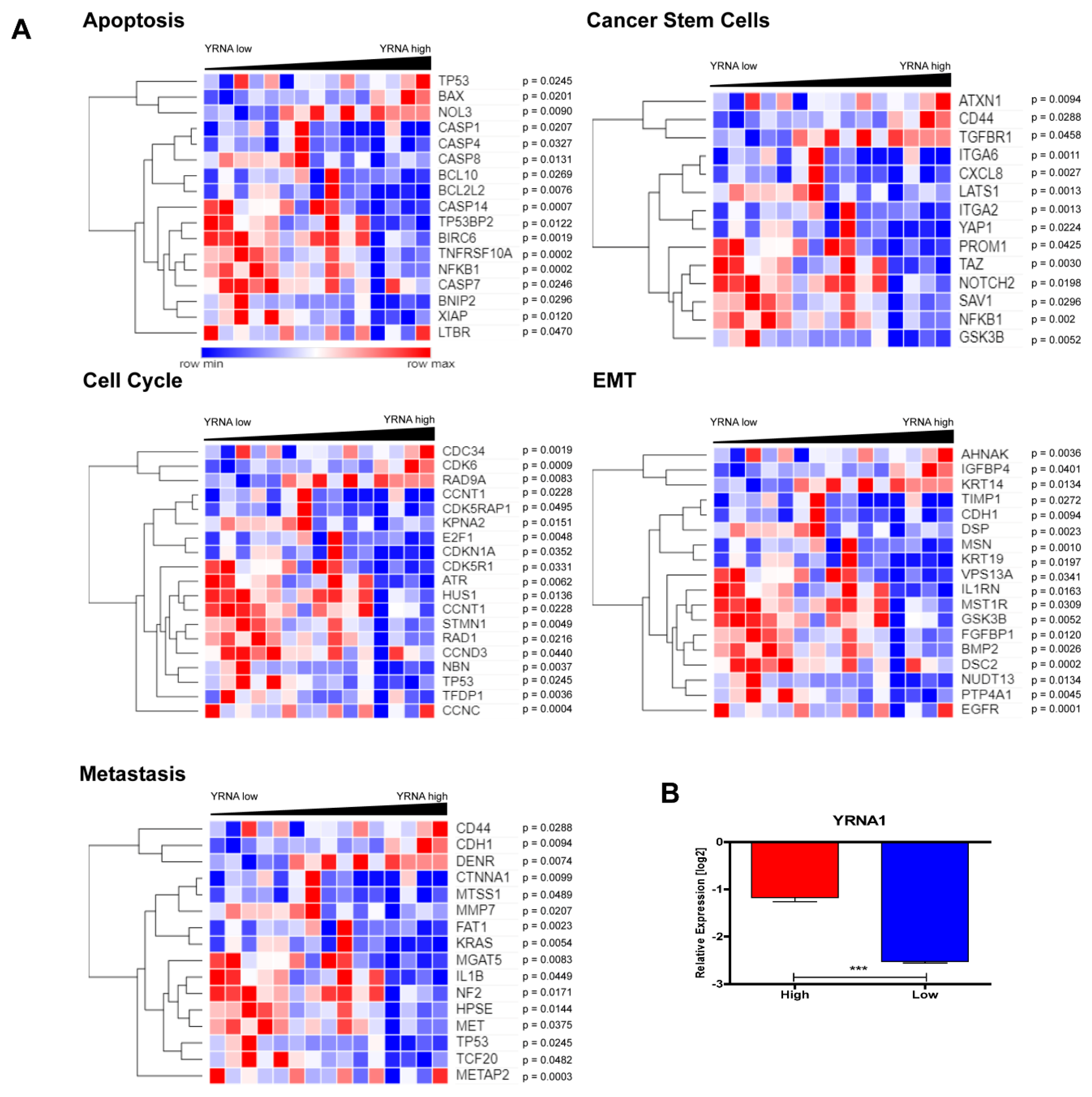
3.4. YRNA1 Expression Is Associated with Changes in the Expression of Many Important Genes
3.5. Patients with High and Low Expression of YRNA1 Have a Different Pattern of Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethics Statement
Abbreviations
| HNSCC | head and neck squamous cell carcinoma |
| TSCC | tongue squamous cell carcinoma |
| OSCC | oral squamous cell carcinoma |
| LSCC | laryngeal squamous cell carcinoma |
| NSCC | nasopharyngeal squamous cell carcinoma |
| lncRNA | long non-coding RNA |
| qRT-PCR | quantitative reverse transcriptase PCR |
| PCR | polymerase chain reaction |
| FFPET | Formalin-Fixed Paraffin-Embedded Tissue |
| TCGA | The Cancer Genome Atlas |
| SEM | standard error of the mean |
| EMT | epithelial to mesenchymal transition |
| HPRT1 | hypoxanthine phosphoribosyltransferase 1 |
| B2M | beta-2 microglobulin |
| FDR | false discovery rate |
| ROC | receiver operating characteristic |
| AUC | area under the curve |
| DFS | disease-free survival |
| OS | overall survival |
| RB | retinoblastoma protein |
| EIF4E | eukaryotic translation initiation factor 4E |
| ERBB2 | erb-B2 receptor tyrosine kinase 2 |
| VEGF | vascular endothelial growth factor |
| EGFR | epidermal growth factor receptor |
| cAMP | cyclic adenosine monophosphate |
| NGS | next generation sequencing |
References
- Guglas, K.; Bogaczyńska, M.; Kolenda, T.; Ryś, M.; Teresiak, A.; Bliźniak, R.; Łasińska, I.; Mackiewicz, J.; Lamperska, K. lncRNA in HNSCC: Challenges and potential. Współczesna Onkol. 2017, 21, 259–266. [Google Scholar] [CrossRef] [Green Version]
- Guglas, K.; Kolenda, T.; Teresiak, A.; Kopczyńska, M.; Łasińska, I.; Mackiewicz, J.; Mackiewicz, A.; Lamperska, K. LncRNA Expression after Irradiation and Chemoexposure of HNSCC Cell lines. NonCoding RNA 2018, 4, 33. [Google Scholar] [CrossRef] [Green Version]
- Kolenda, T.; Guglas, K.; Ryś, M.; Bogaczyńska, M.; Teresiak, A.; Bliźniak, R.; Łasińska, I.; Mackiewicz, J.; Lamperska, K. Biological role of long non-coding RNA in head and neck cancers. Rep. Pr. Oncol. Radiother. 2017, 22, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Tolkach, Y.; Niehoff, E.-M.; Stahl, A.F.; Zhao, C.; Kristiansen, G.; Müller, S.C.; Ellinger, J. YRNA expression in prostate cancer patients: Diagnostic and prognostic implications. World J. Urol. 2018, 36, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Nientiedt, M.; Schmidt, D.; Kristiansen, G.; Müller, S.C.; Ellinger, J. YRNA expression profiles are altered in clear cell renal cell carcinoma. Eur. Urol. Focus 2018, 4, 260–266. [Google Scholar] [CrossRef]
- Tolkach, Y.; Stahl, A.F.; Niehoff, E.-M.; Zhao, C.; Kristiansen, G.; Müller, S.C.; Ellinger, J. YRNA expression predicts survival in bladder cancer patients. BMC Cancer 2017, 17, 749. [Google Scholar] [CrossRef] [Green Version]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Boil. 2015, 66, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-M.; Tseng, C.-H.; Chen, Y.-C.; Yu, W.-Y.; Ho, M.-Y.; Ho, C.-Y.; Lai, M.M.C.; Su, W.-C. Exosome-delivered and Y RNA-derived small RNA suppresses influenza virus replication. J. Biomed. Sci. 2019, 26, 58. [Google Scholar] [CrossRef] [Green Version]
- Hizir, Z.; Bottini, S.; Grandjean, V.; Trabucchi, M.; Repetto, E. RNY (YRNA)-derived small RNAs regulate cell death and inflammation in monocytes/macrophages. Cell Death Dis. 2017, 8, e2530. [Google Scholar] [CrossRef] [Green Version]
- Repetto, E.; Lichtenstein, L.; Hizir, Z.; Tekaya, N.; Benahmed, M.; Ruidavets, J.-B.; Zaragosi, L.-E.; Perret, B.; Bouchareychas, L.; Genoux, A.; et al. RNY-derived small RNAs as a signature of coronary disease. BMC Med. 2015, 13, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.E.; Foster, S.; Betts, D.; Marnock, W.E. DOK, a cell line established from human dysplastic oral mucosa, shows a partially transformed non-malignant phenotype. Int. J. Cancer 1992, 52, 896–902. [Google Scholar] [CrossRef]
- Lamperska, K.; Kolenda, T.; Teresiak, A.; Kowalik, A.; Kruszyna-Mochalska, M.; Jackowiak, W.; Bliźniak, R.; Przybyła, W.; Kapałczyńska, M.; Kozlowski, P. Different levels of let-7d expression modulate response of FaDu cells to irradiation and chemotherapeutics. PLoS ONE 2017, 12, e0180265. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Pavlovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Konwledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehár, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive Genes Involved in Oxidative Phosphorylation Are Coordinately Downregulated in Human Diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Franz, M.; Rodríguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.; Morris, Q. GeneMANIA Update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef] [Green Version]
- Kolenda, T.; Guglas, K.; Kopczyńska, M.; Teresiak, A.; Bliźniak, R.; Mackiewicz, A.; Lamperska, K.; Mackiewicz, J. Oncogenic role of ZFAS1 lncRNA in Head and Neck Squamous Cell Carcinomas. Cells 2019, 8, 366. [Google Scholar] [CrossRef] [Green Version]
- Martinez, B.V.; Dhahbi, J.M.; Lopez, Y.N.; Lamperska, K.; Golusinski, P.; Luczewski, L.; Kolenda, T.; Atamna, H.; Spindler, S.R.; Golusinski, W.; et al. Circulating small non-coding RNA signature in Head and Neck Squamous Cell Carcinoma. Oncotarget 2015, 6, 19246–19263. [Google Scholar] [CrossRef]
- Hess, J. Predictive factors for outcome and quality of life in HPV-positive and HPV-negative HNSCC. Methods Mol. Biol. 2016, 206, 233–242. [Google Scholar]
- Koneva, L.A.; Zhang, Y.; Virani, S.; Hall, P.B.; McHugh, J.B.; Chepeha, D.B.; Wolf, G.T.; Carey, T.E.; Rozek, L.S.; Sartor, M.A. HPV integration in HNSCC correlates with survival outcomes, immune response signatures and candidate drivers. Mol. Cancer Res. 2017, 16, 90–102. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.-C.; Lin, L.-H.; Yu, S.-Y.; Kao, S.-Y.; Chang, K.-W.; Cheng, H.-W.; Liu, C.-J. FAT1 somatic mutations in head and neck carcinoma are associated with tumor progression and survival. Carcinogenesis 2018, 39, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wu, Y.; Guo, W.; Yu, F.; Kong, L.; Ren, Y.; Wang, Y.; Yao, X.; Jing, C.; Zhang, C.; et al. STAT3/HOTAIR Signaling Axis Regulates HNSCC Growth in an EZH2-dependent Manner. Clin. Cancer Res. 2018, 24, 2665–2677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, T.N.; Georgopoulos, R.; Shagisultanova, E.I.; Sarcu, D.; Handorf, E.A.; Dubyk, C.; Lango, M.N.; Ridge, J.A.; Astsaturov, I.; Serebriiskii, I.G.; et al. EGFR and RB1 as Dual Biomarkers in HPV-Negative Head and Neck Cancer. Mol. Cancer Ther. 2016, 15, 2486–2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macha, M.A.; Rachagani, S.; Pai, P.; Gupta, S.; Lydiatt, W.M.; Smith, R.B.; Johansson, S.L.; Lele, S.M.; Kakar, S.S.; Farghaly, H.; et al. MUC4 regulates cellular senescence in head and neck squamous cell carcinoma through p16/Rb pathway. Oncogene 2015, 34, 1698–1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkeland, A.C.; Yanik, M.; Tillman, B.N.; Scott, M.V.; Foltin, S.; Mann, J.E.; Michmerhuizen, N.L.; Ludwig, M.L.; Sandelski, M.M.; Komarck, C.M.; et al. Identification of Targetable ERBB2 Aberrations in Head and Neck Squamous Cell Carcinoma. JAMA Otolaryngol. Neck Surg. 2016, 142, 559–567. [Google Scholar] [CrossRef] [Green Version]
- Schlüter, A.; Weller, P.; Kanaan, O.; Nel, I.; Heusgen, L.; Höing, B.; Haßkamp, P.; Zander, S.; Mandapathil, M.; Dominas, N.; et al. CD31 and VEGF are prognostic biomarkers in early-stage, but not in late-stage, laryngeal squamous cell carcinoma. BMC Cancer 2018, 18, 272. [Google Scholar] [CrossRef] [Green Version]
| Parameter | Group | Mean ± SEM | n | p-Value |
|---|---|---|---|---|
| Age | ≤70 | 0.03773 ± 0.009145 | 35 | 0.4873 |
| >70 | 0.06560 ± 0.0444 | 29 | ||
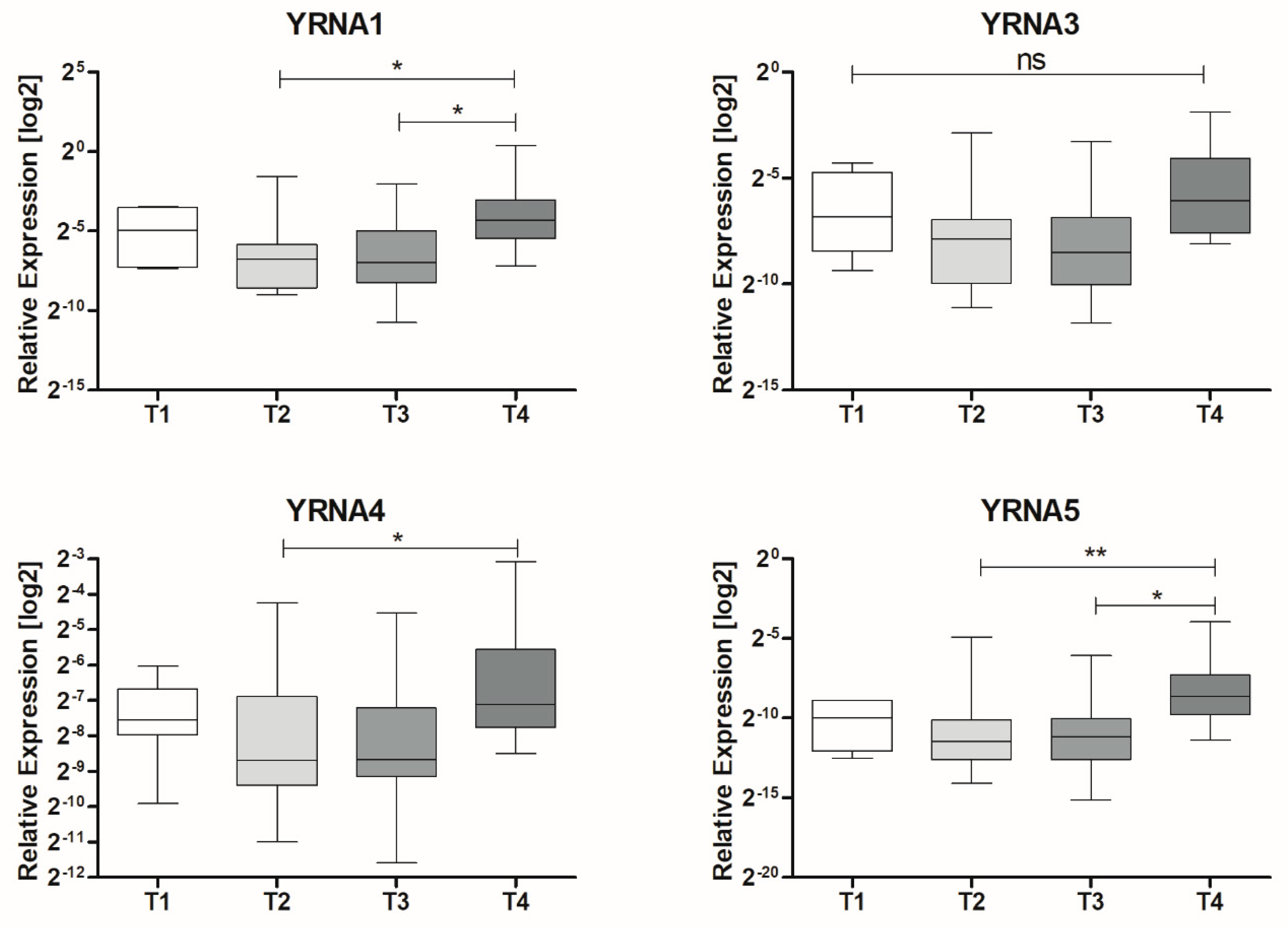
| T Stage | T1 | 0.04212 ± 0.01625 | 6 | 0.0211 |
| T2 | 0.03231 ± 0.01764 | 19 | ||
| T3 | 0.03344 ± 0.01369 | 20 | ||
| T4 | 0.1829 ± 0.1251 | 10 | ||
| N Stage | N0 + N1 | 0.0435 ± 0.01017 | 44 | 0.1747 |
| N2 + N3 | 0.1319 ± 0.1172 | 11 | ||
| Grade | G1 + G2 | 0.06342 ± 0.02687 | 50 | 0.5764 |
| G3 + G4 | 0.03474 ± 0.01272 | 9 | ||
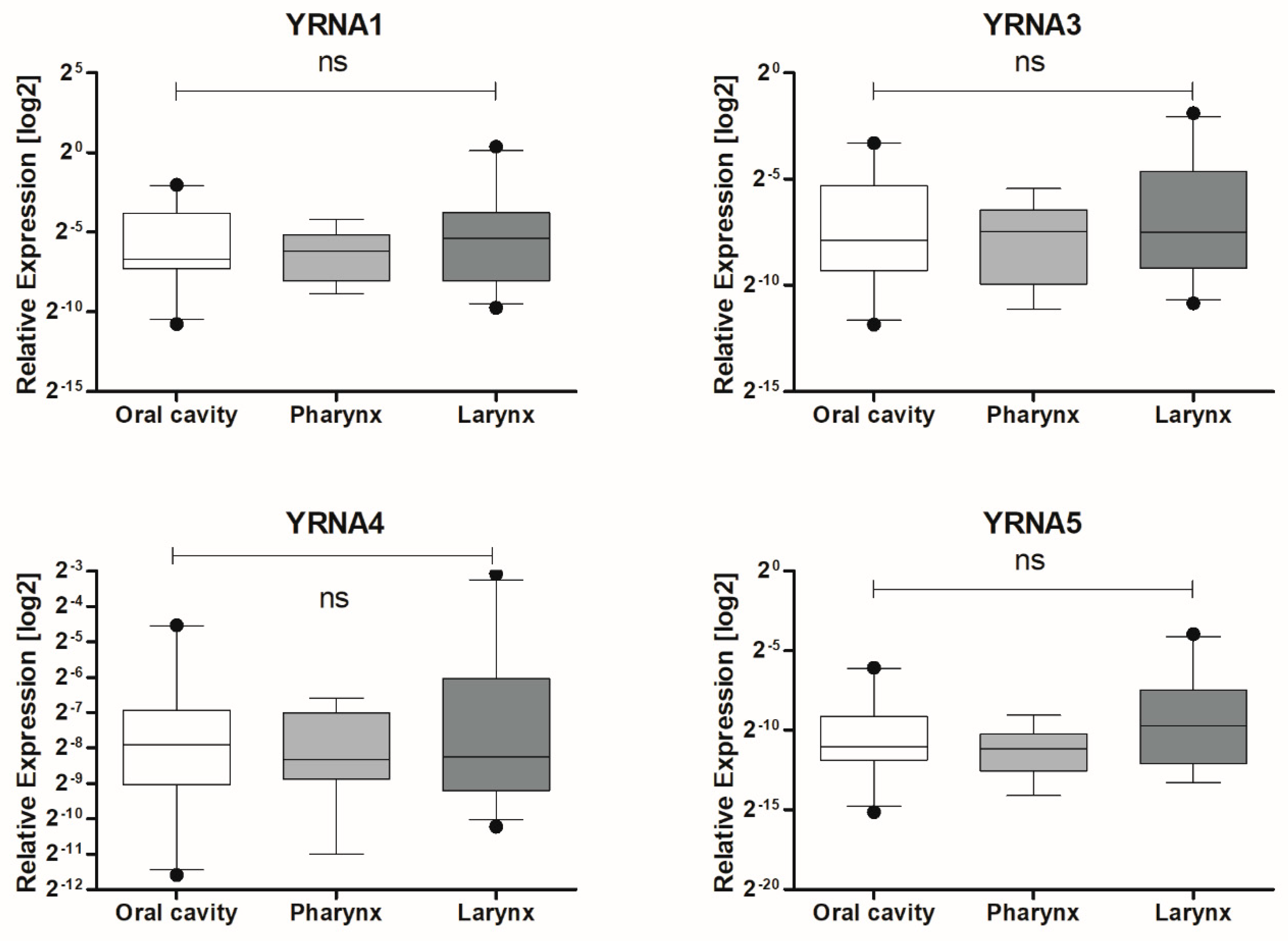
| Localization | Oral Cavity | 0.04271 ± 0.01368 | 21 | 0.4274 |
| Pharynx | 0.01781 ± 0.004761 | 13 | ||
| Larynx | 0.1049 ± 0.05659 | 23 |
| Parameter | Group | Mean ± SEM | n | p-Value |
|---|---|---|---|---|
| Age | ≤62 | −1.931 ± 0.1019 | 65 | 0.7491 |
| >62 | −2.001 ± 0.1126 | 51 | ||
| Sex | Female | −2.134 ± 0.1562 | 27 | 0.0932 |
| Male | −1.910 ± 0.0857 | 89 | ||
| Alcohol | Yes | −1.977 ± 0.0957 | 81 | 0.3472 |
| No | −1.887 ± 0.1258 | 32 | ||
| Smoking | Yes | −1.983 ± 0.1104 | 38 | 0.7405 |
| No/Ex | −1.950 ± 0.1022 | 75 | ||
| Cancer Stage | I + II | −1.967 ± 0.1774 | 15 | 0.8773 |
| III + IV | −1.973 ± 0.0864 | 85 | ||
| T Stage | T1 | −2.064 ± 0.2855 | 6 | 0.9603 |
| T2 | −1.963 ± 0.1214 | 32 | ||
| T3 | −2.030 ± 0.1783 | 21 | ||
| T4 | −1.947 ± 0.1265 | 44 | ||
| N Stage | N0 + N1 | −2.129 ± 0.1038 | 45 | 0.1597 |
| N2 + N3 | −1.896 ± 0.1124 | 47 | ||
| Grade | G1 + G2 | −1.972 ± 0.0955 | 71 | 0.9464 |
| G3 + G4 | −1.974 ± 0.1222 | 43 | ||
| Perineural Invasion | Positive | −1.969 ± 0.1331 | 40 | 0.3927 |
| Negative | −1.855 ± 0.1322 | 38 | ||
| Lymph Node Dissection | Positive | −1.800 ± 0.1763 | 90 | 0.2735 |
| Negative | −2.010 ± 0.0836 | 25 | ||
| Angiolymphatic Invasion | Positive | −1.906 ± 0.1416 | 32 | 0.3307 |
| Negative | −2.057 ± 0.1207 | 41 | ||
| Disease Surgical Margin Status | Positive | −1.879 ± 0.1469 | 28 | 0.369 |
| Negative | −1.990 ± 0.0947 | 75 | ||
| HPV p16 Status | Positive | −1.150 ± 0.2755 | 11 | 0.0002 |
| Negative | −2.330 ± 0.1236 | 16 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guglas, K.; Kolenda, T.; Stasiak, M.; Kopczyńska, M.; Teresiak, A.; Ibbs, M.; Bliźniak, R.; Lamperska, K. YRNAs: New Insights and Potential Novel Approach in Head and Neck Squamous Cell Carcinoma. Cells 2020, 9, 1281. https://doi.org/10.3390/cells9051281
Guglas K, Kolenda T, Stasiak M, Kopczyńska M, Teresiak A, Ibbs M, Bliźniak R, Lamperska K. YRNAs: New Insights and Potential Novel Approach in Head and Neck Squamous Cell Carcinoma. Cells. 2020; 9(5):1281. https://doi.org/10.3390/cells9051281
Chicago/Turabian StyleGuglas, Kacper, Tomasz Kolenda, Maciej Stasiak, Magda Kopczyńska, Anna Teresiak, Matthew Ibbs, Renata Bliźniak, and Katarzyna Lamperska. 2020. "YRNAs: New Insights and Potential Novel Approach in Head and Neck Squamous Cell Carcinoma" Cells 9, no. 5: 1281. https://doi.org/10.3390/cells9051281
APA StyleGuglas, K., Kolenda, T., Stasiak, M., Kopczyńska, M., Teresiak, A., Ibbs, M., Bliźniak, R., & Lamperska, K. (2020). YRNAs: New Insights and Potential Novel Approach in Head and Neck Squamous Cell Carcinoma. Cells, 9(5), 1281. https://doi.org/10.3390/cells9051281





