Circulating Tumor Cells Characterization Revealed TIMP1 as a Potential Therapeutic Target in Ovarian Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients Inclusion and Samples Collection
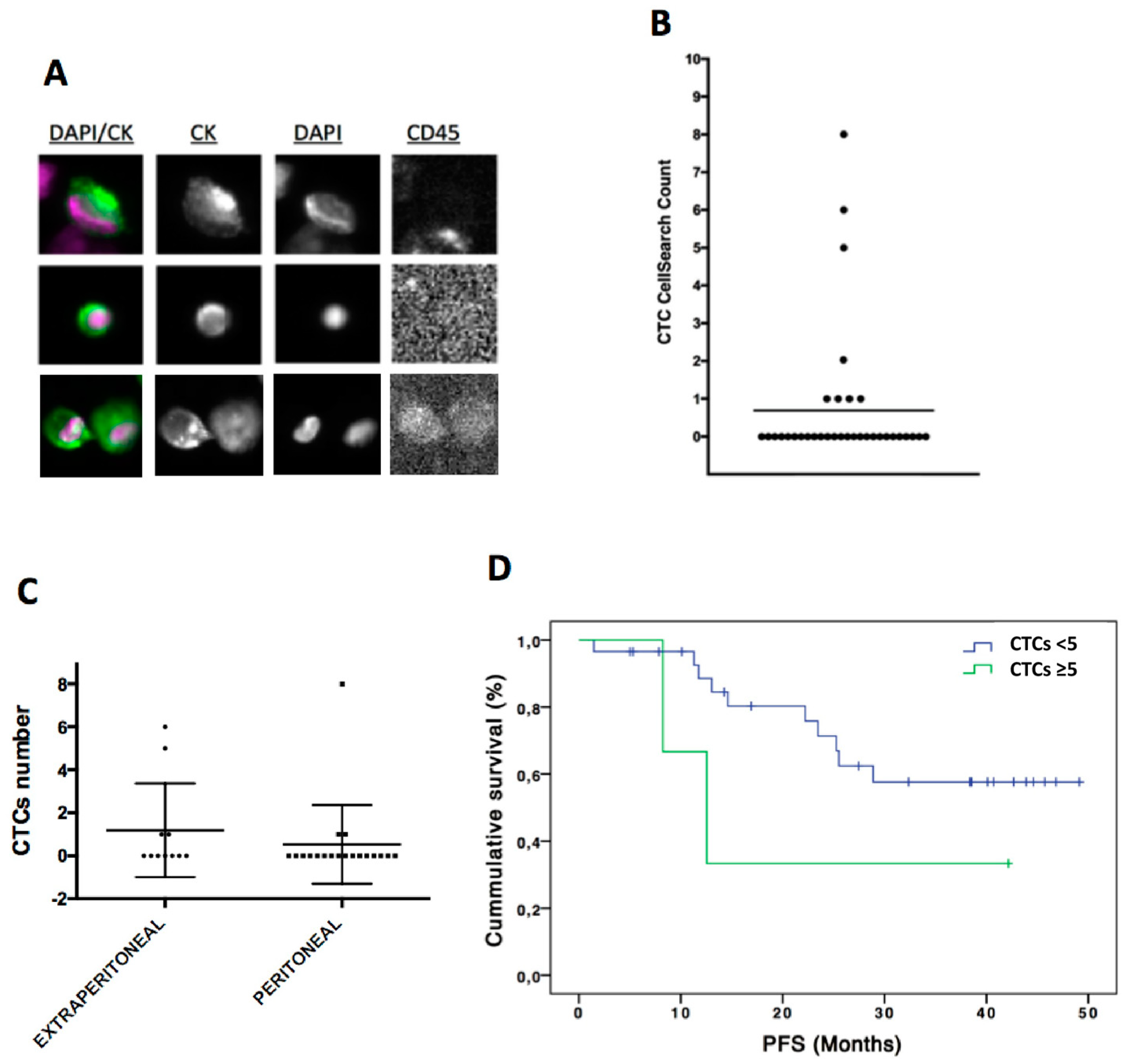
2.2. CTCs Enumeration and Characterization
2.3. Cell Lines
2.4. TIMP1 Silencing
2.5. Gene Expression Assays in Cell Lines
2.6. Western Blot
2.7. In vitro Functional Assays
2.7.1. Transwell Migration Assay
2.7.2. Proliferation Assay
2.7.3. Colony Formation Assay
2.7.4. Collagen Adhesion Assay
2.8. In Vivo Functional Assays Using Zebrafish Xenograft
2.8.1. Zebrafish Care and Breeding
2.8.2. Xenograft Assays and Image Analysis
2.8.3. Quantification of Cell Proliferation in Zebrafish Assays
2.9. In Vivo Functional Assays Using Mouse Xenografts
2.10. Statistical Analysis
3. Results
3.1. CTCs Enumeration by Cellsearch System was Higher in Patients with a More Aggressive Disease
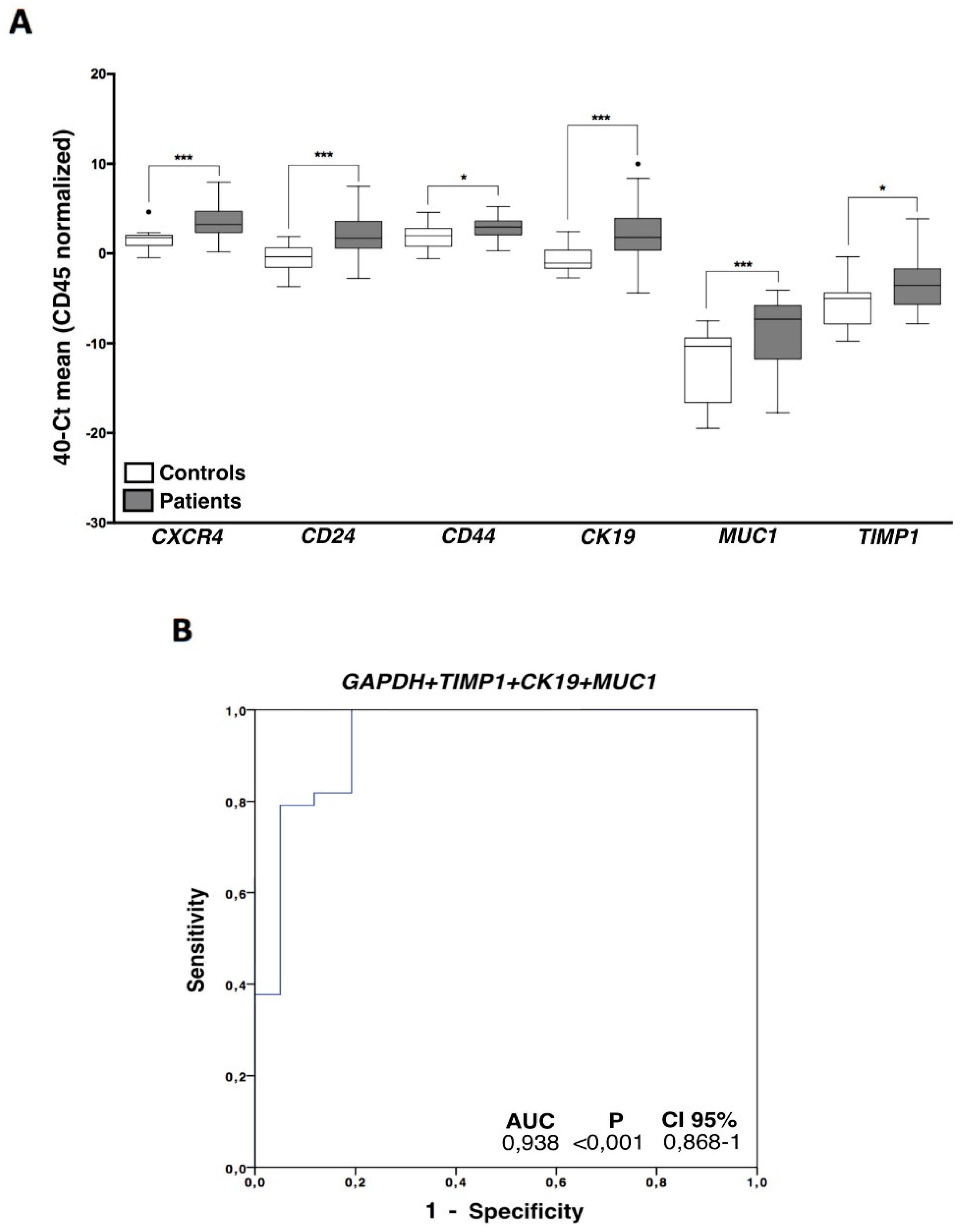
3.2. Indirect Quantification of CTCS by RT-qPCR Showed High Accuracy to Detect Cancer Patients
3.3. CTCs Characterization in Ovarian Cancer Patients Evidenced an Important Cell Plasticity
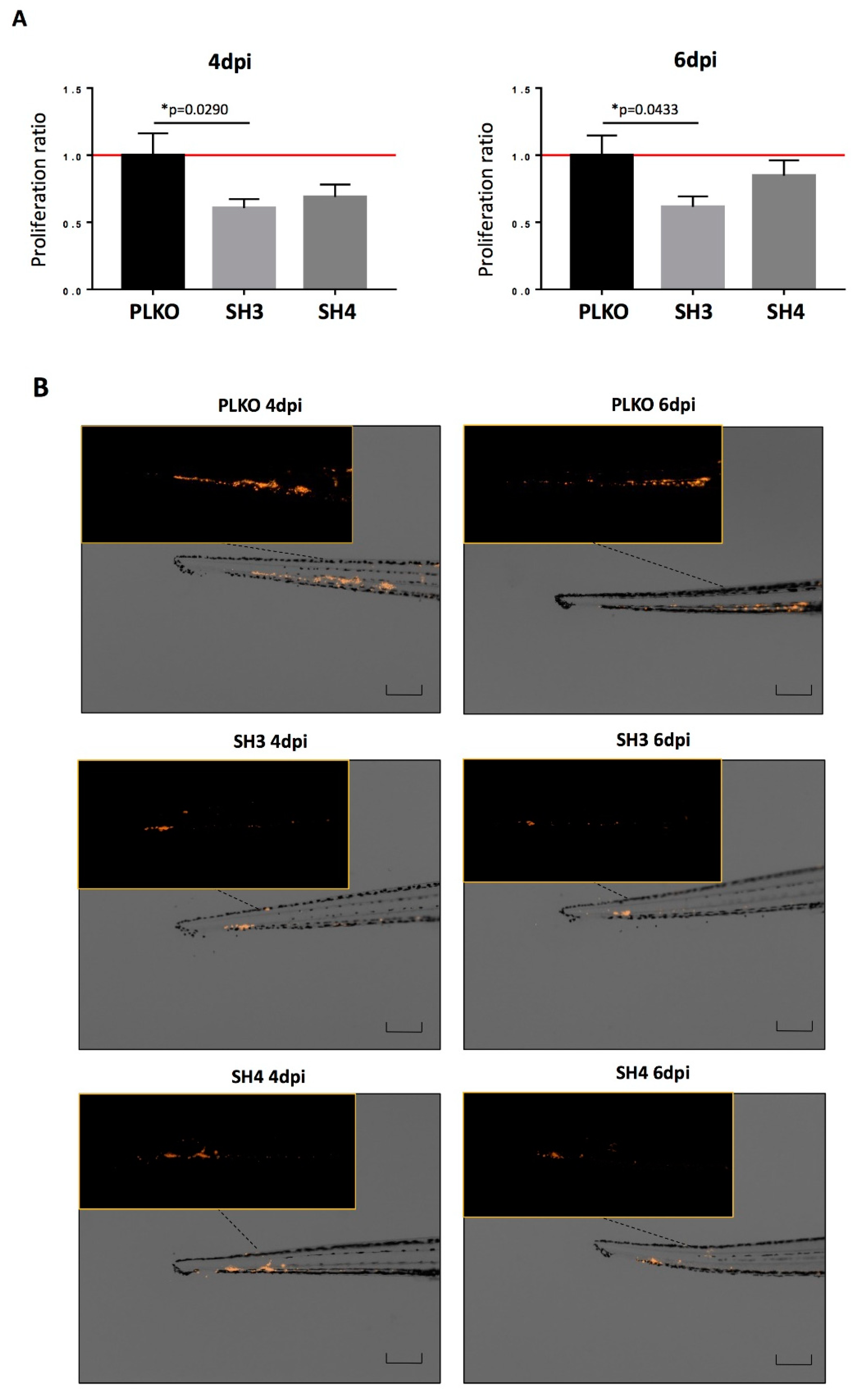
3.4. TIMP1 Down-Regulation Decreased Tumor Growth In Vitro and In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Colombo, N.; Peiretti, M.; Parma, G.; Lapresa, M.; Mancari, R.; Carinelli, S.; Sessa, C.; Castiglione, M. ESMO Guidelines Working Group Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010, 21 (Suppl. 5), v23–v30. [Google Scholar] [CrossRef] [PubMed]
- Worzfeld, T.; Pogge von Strandmann, E.; Huber, M.; Adhikary, T.; Wagner, U.; Reinartz, S.; Müller, R. The Unique Molecular and Cellular Microenvironment of Ovarian Cancer. Front. Oncol. 2017, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Chebouti, I.; Kasimir-Bauer, S.; Buderath, P.; Wimberger, P.; Hauch, S.; Kimmig, R.; Kuhlmann, J.D. EMT-like circulating tumor cells in ovarian cancer patients are enriched by platinum-based chemotherapy. Oncotarget 2017, 8, 48820–48831. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, E. Ovarian cancer development and metastasis. Am. J. Pathol. 2010, 177, 1053–1064. [Google Scholar] [CrossRef]
- Zhou, Y.; Bian, B.; Yuan, X.; Xie, G.; Ma, Y.; Shen, L. Prognostic Value of Circulating Tumor Cells in Ovarian Cancer: A Meta-Analysis. PLoS ONE 2015, 10, e0130873. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, L.; Chen, Y.; Qing, C. Circulating cell-free DNA and circulating tumor cells, the “liquid biopsies” in ovarian cancer. J. Ovarian Res. 2017, 10, 75. [Google Scholar] [CrossRef]
- Gupta, G.P.; Massagué, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef]
- Guo, Y.-X.; Neoh, K.H.; Chang, X.-H.; Sun, Y.; Cheng, H.-Y.; Ye, X.; Ma, R.-Q.; Han, R.P.S.; Cui, H. Diagnostic value of HE4+ circulating tumor cells in patients with suspicious ovarian cancer. Oncotarget 2018, 9, 7522–7533. [Google Scholar] [CrossRef]
- Aktas, B.; Kasimir-Bauer, S.; Heubner, M.; Kimmig, R.; Wimberger, P. Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int. J. Gynecol. Cancer 2011, 21, 822–830. [Google Scholar] [CrossRef]
- Kuhlmann, J.D.; Wimberger, P.; Bankfalvi, A.; Keller, T.; Schöler, S.; Aktas, B.; Buderath, P.; Hauch, S.; Otterbach, F.; Kimmig, R.; et al. ERCC1-positive circulating tumor cells in the blood of ovarian cancer patients as a predictive biomarker for platinum resistance. Clin. Chem. 2014, 60, 1282–1289. [Google Scholar] [CrossRef]
- Barbazán, J.; Alonso-Alconada, L.; Muinelo-Romay, L.; Vieito, M.; Abalo, A.; Alonso-Nocelo, M.; Candamio, S.; Gallardo, E.; Fernández, B.; Abdulkader, I.; et al. Molecular characterization of circulating tumor cells in human metastatic colorectal cancer. PLoS ONE 2012, 7, e40476. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Alconada, L.; Muinelo-Romay, L.; Madissoo, K.; Diaz-Lopez, A.; Krakstad, C.; Trovik, J.; Wik, E.; Hapangama, D.; Coenegrachts, L.; Cano, A.; et al. Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Mol. Cancer 2014, 13, 223. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Xu, S.; Zhang, H.; Wang, Y.; Xiao, C.; Jiang, T.; Wu, L.; Zhang, T.; Sun, X.; Zhong, L.; et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J. Exp. Clin. Cancer Res. 2016, 35, 148. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Scott, E.; Lu, R.; Xu, Y.; Oh, W.K.; Yu, Q. TIMP-1 promotes accumulation of cancer associated fibroblasts and cancer progression. PLoS ONE 2013, 8, e77366. [Google Scholar] [CrossRef]
- Cheng, G.; Fan, X.; Hao, M.; Wang, J.; Zhou, X.; Sun, X. Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer. Mol. Cancer 2016, 15, 30. [Google Scholar] [CrossRef]
- Abreu, M.; Cabezas-Sainz, P.; Pereira-Veiga, T.; Falo, C.; Abalo, A.; Morilla, I.; Curiel, T.; Cueva, J.; Rodríguez, C.; Varela-Pose, V.; et al. Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells. J. Clin. Med. 2020, 9, 353. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Poveda, A.; Vergote, I.; Tjulandin, S.; Kong, B.; Roy, M.; Chan, S.; Filipczyk-Cisarz, E.; Hagberg, H.; Kaye, S.B.; Colombo, N.; et al. Trabectedin plus pegylated liposomal doxorubicin in relapsed ovarian cancer: Outcomes in the partially platinum-sensitive (platinum-free interval 6-12 months) subpopulation of OVA-301 phase III randomized trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011, 22, 39–48. [Google Scholar] [CrossRef]
- Obermayr, E.; Sanchez-Cabo, F.; Tea, M.-K.M.; Singer, C.F.; Krainer, M.; Fischer, M.B.; Sehouli, J.; Reinthaller, A.; Horvat, R.; Heinze, G.; et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer 2010, 10, 666. [Google Scholar] [CrossRef]
- Liu, J.F.; Kindelberger, D.; Doyle, C.; Lowe, A.; Barry, W.T.; Matulonis, U.A. Predictive value of circulating tumor cells (CTCs) in newly-diagnosed and recurrent ovarian cancer patients. Gynecol. Oncol. 2013, 131, 352–356. [Google Scholar] [CrossRef]
- Pradeep, S.; Kim, S.W.; Wu, S.Y.; Nishimura, M.; Chaluvally-Raghavan, P.; Miyake, T.; Pecot, C.V.; Kim, S.-J.; Choi, H.J.; Bischoff, F.Z.; et al. Hematogenous metastasis of ovarian cancer: Rethinking mode of spread. Cancer Cell 2014, 26, 77–91. [Google Scholar] [CrossRef]
- Santamaria, P.G.; Moreno-Bueno, G.; Portillo, F.; Cano, A. EMT: Present and future in clinical oncology. Mol. Oncol. 2017, 11, 718–738. [Google Scholar] [CrossRef] [PubMed]
- Mariscal, J.; Alonso-Nocelo, M.; Muinelo-Romay, L.; Barbazan, J.; Vieito, M.; Abalo, A.; Gomez-Tato, A.; Maria de Los Angeles, C.d.C.; Garcia-Caballero, T.; Rodriguez, C.; et al. Molecular Profiling of Circulating Tumour Cells Identifies Notch1 as a Principal Regulator in Advanced Non-Small Cell Lung Cancer. Sci. Rep. 2016, 6, 37820. [Google Scholar] [CrossRef] [PubMed]
- León-Mateos, L.; Casas, H.; Abalo, A.; Vieito, M.; Abreu, M.; Anido, U.; Gómez-Tato, A.; López, R.; Abal, M.; Muinelo-Romay, L. Improving circulating tumor cells enumeration and characterization to predict outcome in first line chemotherapy mCRPC patients. Oncotarget 2017, 8, 54708. [Google Scholar]
- Kolostova, K.; Pinkas, M.; Jakabova, A.; Pospisilova, E.; Svobodova, P.; Spicka, J.; Cegan, M.; Matkowski, R.; Bobek, V. Molecular characterization of circulating tumor cells in ovarian cancer. Am. J. Cancer Res. 2016, 6, 973–980. [Google Scholar]
- Nakamura, K.; Terai, Y.; Tanabe, A.; Ono, Y.J.; Hayashi, M.; Maeda, K.; Fujiwara, S.; Ashihara, K.; Nakamura, M.; Tanaka, Y.; et al. CD24 expression is a marker for predicting clinical outcome and regulates the epithelial-mesenchymal transition in ovarian cancer via both the Akt and ERK pathways. Oncol. Rep. 2017, 37, 3189–3200. [Google Scholar] [CrossRef]
- Zhang, J.; Chang, B.; Liu, J. CD44 standard form expression is correlated with high-grade and advanced-stage ovarian carcinoma but not prognosis. Hum. Pathol. 2013, 44, 1882–1889. [Google Scholar] [CrossRef]
- Mao, M.; Zheng, X.; Jin, B.; Zhang, F.; Zhu, L.; Cui, L. Effects of CD44 and E-cadherin overexpression on the proliferation, adhesion and invasion of ovarian cancer cells. Exp. Ther. Med. 2017, 14, 5557–5563. [Google Scholar] [CrossRef]
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef]
- Blassl, C.; Kuhlmann, J.D.; Webers, A.; Wimberger, P.; Fehm, T.; Neubauer, H. Gene expression profiling of single circulating tumor cells in ovarian cancer - Establishment of a multi-marker gene panel. Mol. Oncol. 2016, 10, 1030–1042. [Google Scholar] [CrossRef]
- Figueras, A.; Alsina-Sanchís, E.; Lahiguera, A.; Abreu, M.; Muinelo-Romay, L.; Moreno-Bueno, G.; Casanovas, O.; Graupera, M.; Matias-Guiu, X.; Vidal, A.; et al. A Role for Cxcr4 in Peritoneal and Hematogenous Ovarian Cancer Dissemination. Mol. Cancer Ther. 2017, 17, 532–543. [Google Scholar] [CrossRef] [PubMed]
- Będkowska, G.E.; Gacuta, E.; Zajkowska, M.; Głażewska, E.K.; Osada, J.; Szmitkowski, M.; Chrostek, L.; Dąbrowska, M.; Ławicki, S. Plasma levels of MMP-7 and TIMP-1 in laboratory diagnostics and differentiation of selected histological types of epithelial ovarian cancers. J. Ovarian Res. 2017, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Määtta, M.; Talvensaari-Mattila, A.; Turpeenniemi-Hujanen, T.; Santala, M. Matrix metalloproteinase-2 (MMP-2) and -9 (MMP-9) and their tissue inhibitors (TIMP-1 and TIMP-2) in differential diagnosis between low malignant potential (LMP) and malignant ovarian tumours. Anticancer Res. 2007, 27, 2753–2758. [Google Scholar] [CrossRef]
- Manenti, L.; Paganoni, P.; Floriani, I.; Landoni, F.; Torri, V.; Buda, A.; Taraboletti, G.; Labianca, R.; Belotti, D.; Giavazzi, R. Expression levels of vascular endothelial growth factor, matrix metalloproteinases 2 and 9 and tissue inhibitor of metalloproteinases 1 and 2 in the plasma of patients with ovarian carcinoma. Eur. J. Cancer 2003, 39, 1948–1956. [Google Scholar] [CrossRef]
- Steffensen, K.D.; Waldstrøm, M.; Christensen, R.K.; Bartels, A.; Brünner, N.; Jakobsen, A. Lack of relationship between TIMP-1 tumour cell immunoreactivity, treatment efficacy and prognosis in patients with advanced epithelial ovarian cancer. BMC Cancer 2010, 10, 185. [Google Scholar] [CrossRef][Green Version]
- Brun, J.L.; Cortez, A.; Lesieur, B.; Uzan, S.; Rouzier, R.; Daraï, E. Expression of MMP-2, -7, -9, MT1-MMP and TIMP-1 and -2 has no prognostic relevance in patients with advanced epithelial ovarian cancer. Oncol. Rep. 2012, 27, 1049–1057. [Google Scholar] [CrossRef]
- Mahner, S.; Woelber, L.; Eulenburg, C.; Schwarz, J.; Carney, W.; Jaenicke, F.; Milde-Langosch, K.; Mueller, V. TIMP-1 and VEGF-165 serum concentration during first-line therapy of ovarian cancer patients. BMC Cancer 2010, 10, 139. [Google Scholar] [CrossRef]
- Hartland, W.J.; Defamie, W.; Waterhouse, P.; Khokha, R. TIMPs: Versatile Extracellular Regulators in Cancer. Nat. Rev. Cancer 2017, 17, 38–53. [Google Scholar]
- Sonego, M.; Poletto, M.; Pivetta, E.; Nicoloso, S.M.; Pellicani, R.; Rampioni, V.G.L.; Citron, F.; Sorio, R.; Mongiat, M.; Baldassarre, G. TIMP-1 is overexpressed and secreted by platinum resistant epithelial ovarian cancer. Cells 2019, 9, 6. [Google Scholar] [CrossRef]
- Omar, O.M.; Soutto, M.; Bhat, N.S.; Bhat, A.A.; Lu, H.; Chen, Z.; El-Rifai, W. TFF1 antagonizes TIMP-1 mediated proliferative functions in gastric cancer. Mol. Carcinog. 2018, 57, 1577–1587. [Google Scholar] [CrossRef]
- Forte, D.; Salvestrini, V.; Corradi, G.; Rossi, L.; Catani, L.; Lemoli, R.M.; Cavo, M.; Curti, A. The tissue inhibitor of metalloproteinases-1 (TIMP-1) promotes survival and migration of acute myeloid leukemia cells through CD63/PI3K/Akt/p21 signaling. Oncotarget 2017, 8, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Fridman, R.; Kim, H.R. Tissue inhibitor of metalloproteinase-1 inhibits apoptosis of human breast epithelial cells. Cancer Res. 1999, 59, 6267–6275. [Google Scholar] [PubMed]



| Features | ||
|---|---|---|
| Age | Mean (range) | 58 (23–83) |
| n (%) | ||
| Stage at diagnosis | I | 1 (2.6%) |
| II | 5 (13.2%) | |
| III | 21 (55.2%) | |
| IV | 5 (13.2%) | |
| Unknown | 6 (15.8%) | |
| Status at sample collection | First diagnosis | 25 (65.8%) |
| Recurrence | 11 (28.9%) | |
| Unknown | 2 (5.3%) | |
| Metastasis at sample collection | Peritoneal | 19 (50%) |
| Extraperitoneal | 11 (28.9%) | |
| Unknown | 8 (21%) | |
| Histology | Serous | 22 (57.9%) |
| Endometrioid | 3 (7.9%) | |
| Other | 10 (26.3%) | |
| Unknown | 3 (7.9%) | |
| Grade | High | 36 (94.7%) |
| Unknown | 2 (5.3%) | |
| BRCA status | Mutant | 10 (26.3%) |
| Wt | 26 (68.4%) | |
| Unknown | 2 (5.3%) | |
| Under treatment at sample collection | Yes | 9 (23.7%) |
| No | 29 (76.3%) | |
| CA125 levels at diagnosis (units/mL) | >35 | 24 (63.2%) |
| <35 | 3 (7.9%) | |
| Unknown | 11 (28.9%) | |
| Recurrence | PD | 12 (31.5%) |
| PFS (median months, CI) | 22.8 (0.39–49.1) | |
| Survival | Death | 6 (15.8%) |
| OS (median months, CI) | 29.9 (0.39–49.1) | |
| Marker | AUC 1 | p | CI 95% |
|---|---|---|---|
| MUC1 | 0.812 | <0.001 | 0.703–0.920 |
| CXCR4 | 0.880 | <0.001 | 0.783–0.976 |
| CK19 | 0.826 | <0.001 | 0.718–0.933 |
| CD24 | 0.845 | <0.001 | 0.746–0.945 |
| CD44 | 0.686 | 0.021 | 0.540–0.831 |
| TIMP1 | 0.696 | 0.015 | 0.552–0.840 |
| GAPDH | 0.812 | <0.001 | 0.695–0.929 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abreu, M.; Cabezas-Sainz, P.; Alonso-Alconada, L.; Ferreirós, A.; Mondelo-Macía, P.; Lago-Lestón, R.M.; Abalo, A.; Díaz, E.; Palacios-Zambrano, S.; Rojo-Sebastian, A.; et al. Circulating Tumor Cells Characterization Revealed TIMP1 as a Potential Therapeutic Target in Ovarian Cancer. Cells 2020, 9, 1218. https://doi.org/10.3390/cells9051218
Abreu M, Cabezas-Sainz P, Alonso-Alconada L, Ferreirós A, Mondelo-Macía P, Lago-Lestón RM, Abalo A, Díaz E, Palacios-Zambrano S, Rojo-Sebastian A, et al. Circulating Tumor Cells Characterization Revealed TIMP1 as a Potential Therapeutic Target in Ovarian Cancer. Cells. 2020; 9(5):1218. https://doi.org/10.3390/cells9051218
Chicago/Turabian StyleAbreu, Manuel, Pablo Cabezas-Sainz, Lorena Alonso-Alconada, Alba Ferreirós, Patricia Mondelo-Macía, Ramón Manuel Lago-Lestón, Alicia Abalo, Eva Díaz, Sara Palacios-Zambrano, Alejandro Rojo-Sebastian, and et al. 2020. "Circulating Tumor Cells Characterization Revealed TIMP1 as a Potential Therapeutic Target in Ovarian Cancer" Cells 9, no. 5: 1218. https://doi.org/10.3390/cells9051218
APA StyleAbreu, M., Cabezas-Sainz, P., Alonso-Alconada, L., Ferreirós, A., Mondelo-Macía, P., Lago-Lestón, R. M., Abalo, A., Díaz, E., Palacios-Zambrano, S., Rojo-Sebastian, A., López-López, R., Sánchez, L., Moreno-Bueno, G., & Muinelo-Romay, L. (2020). Circulating Tumor Cells Characterization Revealed TIMP1 as a Potential Therapeutic Target in Ovarian Cancer. Cells, 9(5), 1218. https://doi.org/10.3390/cells9051218






