The Transcription Factor EB Reduces the Intraneuronal Accumulation of the Beta-Secretase-Derived APP Fragment C99 in Cellular and Mouse Alzheimer’s Disease Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Viral Infection
2.2. Cell Culture and Treatments
2.3. Immunolabeling of Tissue and Cultured Cells
2.4. Immunofluorescence Quantification
2.5. Western Blot Analysis
2.6. mRNA Extraction and Quantitative RT-PCR
2.7. Statistics
3. Results
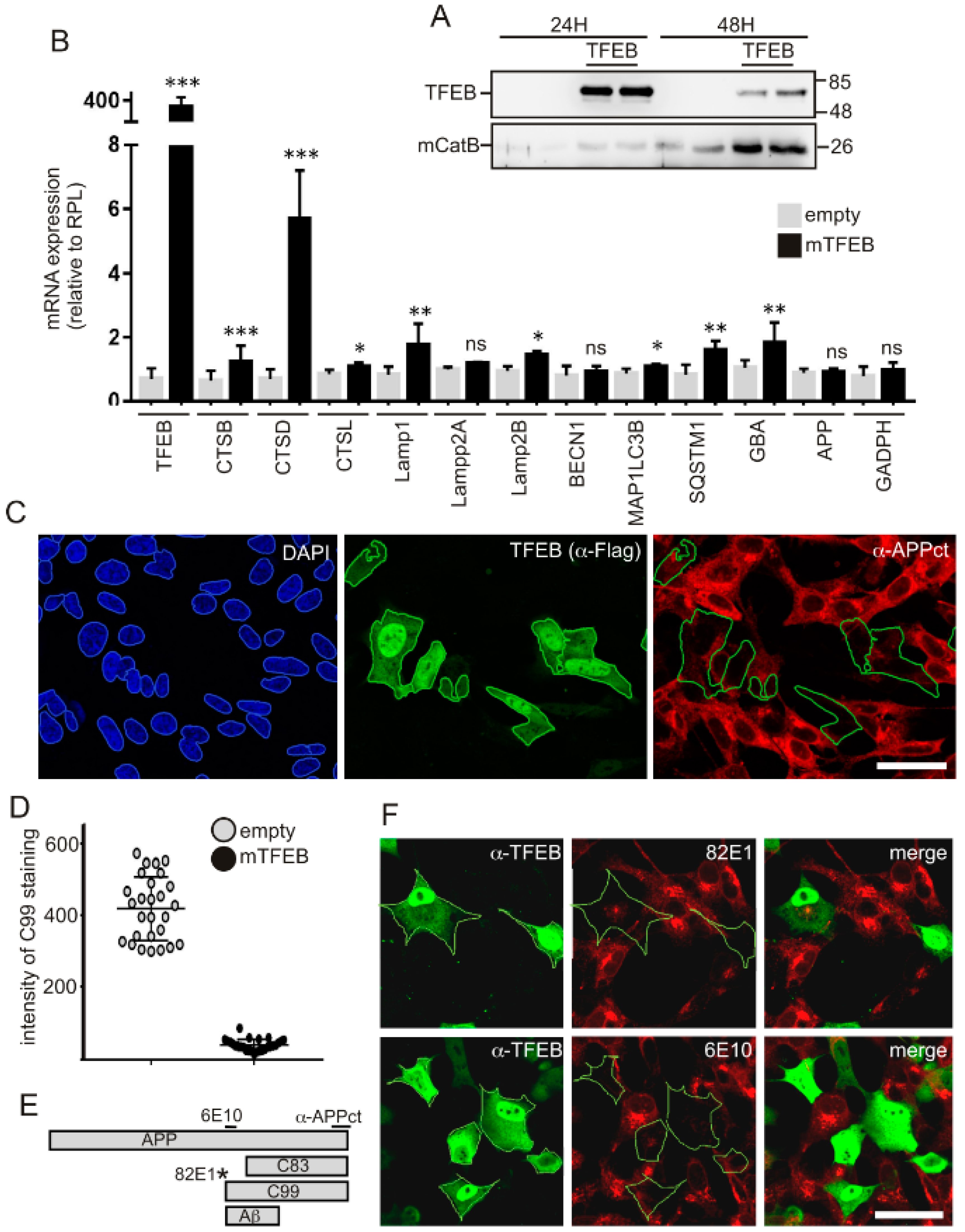
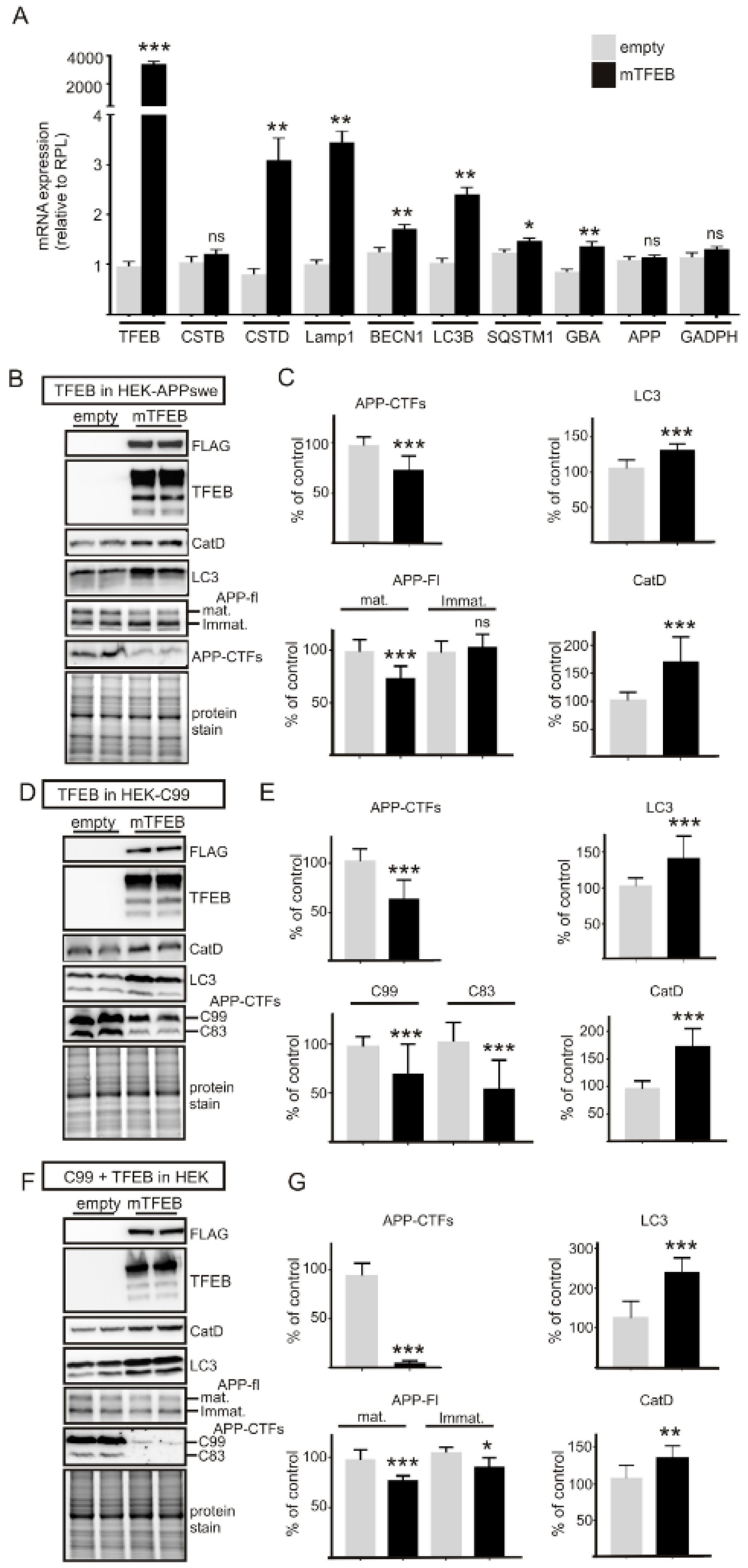
3.1. TFEB Overexpression Reduces APP-CTF Levels in both APPswe and C99 Expressing Cells
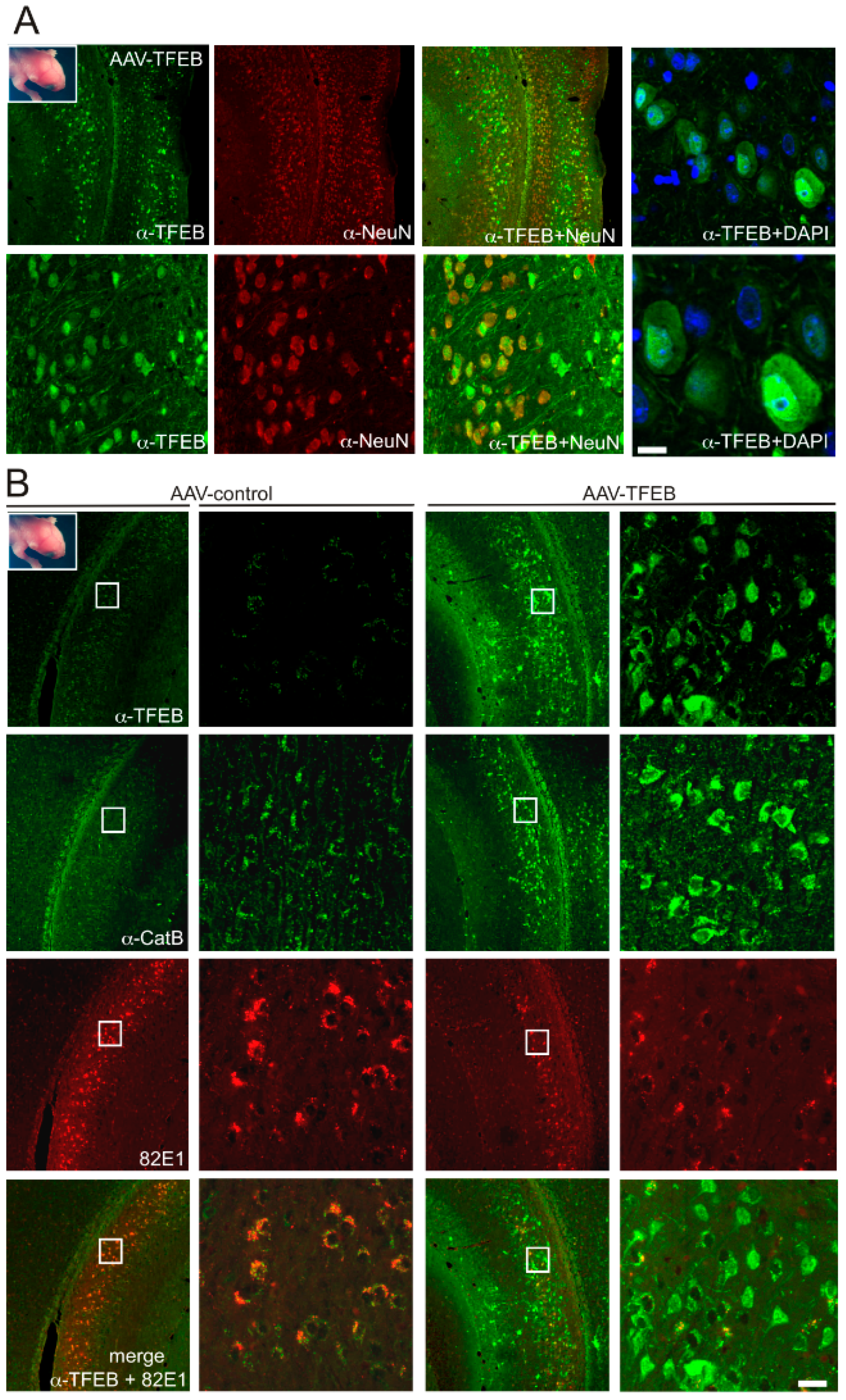
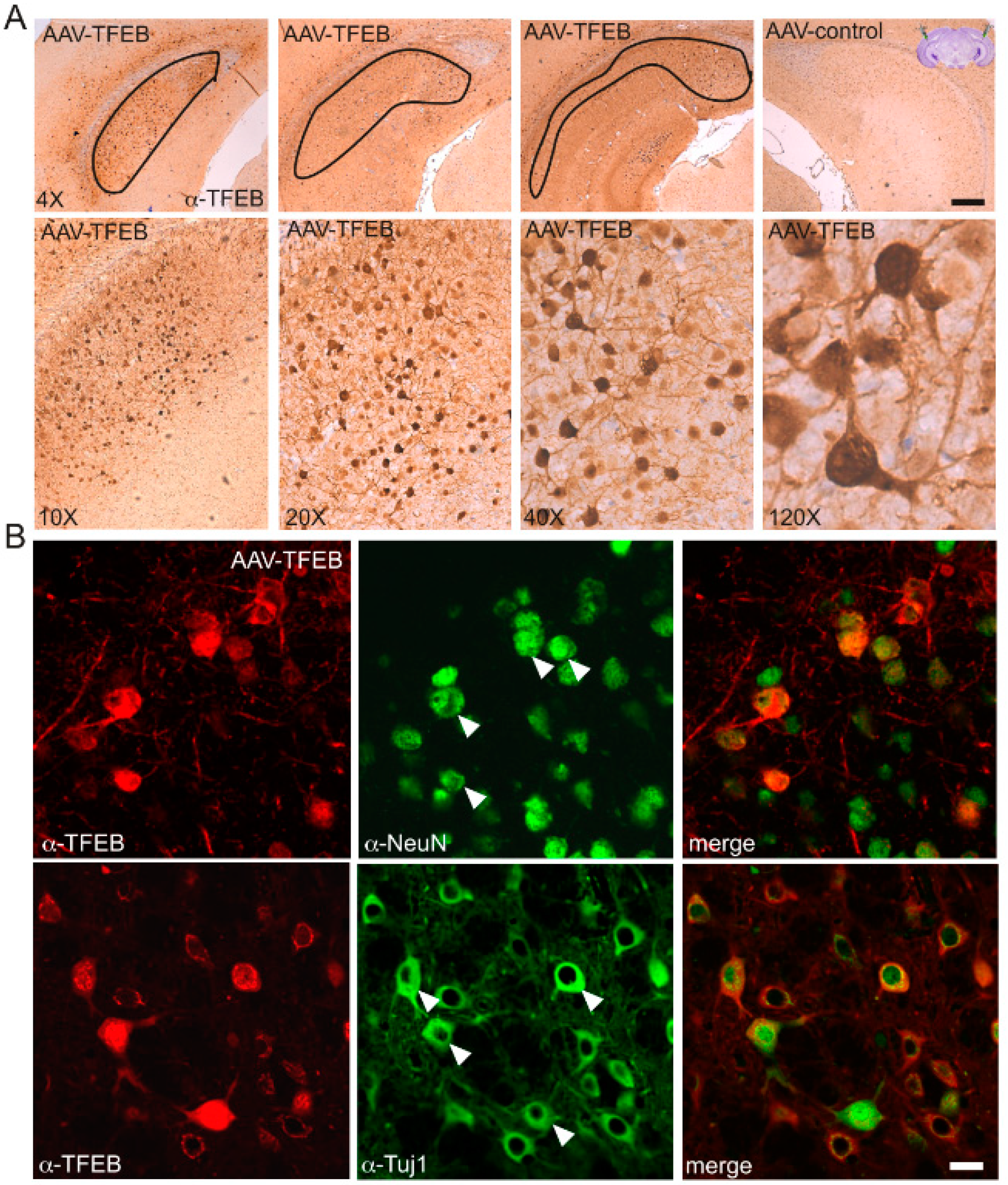
3.2. TFEB Overexpression Lowers C99 Accumulation in the Triple-Transgenic Mouse Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
List of Abbreviations
References
- Checler, F. Processing of the beta-amyloid precursor protein and its regulation in Alzheimer’s disease. J. Neurochem. 1995, 65, 1431–1444. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Mullaney, K.A.; Peterhoff, C.M.; Che, S.; Schmidt, S.D.; Boyer-Boiteau, A.; Ginsberg, S.D.; Cataldo, A.M.; Mathews, P.M.; Nixon, R.A. Alzheimer’s-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc. Natl. Acad. Sci. USA 2010, 107, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Rigoglioso, A.; Peterhoff, C.M.; Pawlik, M.; Sato, Y.; Bleiwas, C.; Stavrides, P.; Smiley, J.F.; Ginsberg, S.D.; Mathews, P.M.; et al. Partial BACE1 reduction in a Down syndrome mouse model blocks Alzheimer-related endosomal anomalies and cholinergic neurodegeneration: Role of APP-CTF. Neurobiol. Aging 2016, 39, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Sato, Y.; Mohan, P.S.; Peterhoff, C.; Pensalfini, A.; Rigoglioso, A.; Jiang, Y.; Nixon, R.A. Evidence that the rab5 effector APPL1 mediates APP-betaCTF-induced dysfunction of endosomes in Down syndrome and Alzheimer’s disease. Mol. Psychiatry 2016, 21, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Pawlik, M.; Gandy, S.E.; Ehrlich, M.E.; Smiley, J.F.; Levy, E. Lysosomal dysfunction in the brain of a mouse model with intraneuronal accumulation of carboxyl terminal fragments of the amyloid precursor protein. Mol. Psychiatry 2016, 22, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Lauritzen, I.; Pardossi-Piquard, R.; Bourgeois, A.; Pagnotta, S.; Biferi, M.G.; Barkats, M.; Lacor, P.; Klein, W.; Bauer, C.; Checler, F. Intraneuronal aggregation of the beta-CTF fragment of APP (C99) induces Abeta-independent lysosomal-autophagic pathology. Acta Neuropathol. 2016, 132, 257–276. [Google Scholar] [CrossRef]
- Xu, W.; Weissmiller, A.M.; White, J.A., II; Fang, F.; Wang, X.; Wu, Y.; Pearn, M.L.; Zhao, X.; Sawa, M.; Chen, S.; et al. Amyloid precursor protein-mediated endocytic pathway disruption induces axonal dysfunction and neurodegeneration. J. Clin. Investig. 2016, 126, 1815–1833. [Google Scholar] [CrossRef]
- Hung, C.O.Y.; Livesey, F.J. Altered gamma-Secretase Processing of APP Disrupts Lysosome and Autophagosome Function in Monogenic Alzheimer’s Disease. Cell Rep. 2018, 25, 3647–3660. [Google Scholar] [CrossRef]
- Kwart, D.; Gregg, A.; Scheckel, C.; Murphy, E.; Paquet, D.; Duffield, M.; Fak, J.; Olsen, O.; Darnell, R.; Tessier-Lavigne, M. A Large Panel of Isogenic APP and PSEN1 Mutant Human iPSC Neurons Reveals Shared Endosomal Abnormalities Mediated by APP beta-CTFs, Not Abeta. Neuron 2019, 104, 256–270. [Google Scholar] [CrossRef]
- Ying, J.; Sato, Y.; Im, E.; Berg, M.; Bordi, M.; Darji, S.; Kumar, A.; Mohan, P.S.; Bandyopadhyay, U.; Diaz, A.; et al. Lysosomal dysfunction in Down syndrome is APP-dependent and mediated by APP-betaCTF (C99). J. Neurosci. 2019, 39, 5255–5268. [Google Scholar]
- Lauritzen, I.; Pardossi-Piquard, R.; Bauer, C.; Brigham, E.; Abraham, J.D.; Ranaldi, S.; Fraser, P.; St-George-Hyslop, P.; Le Thuc, O.; Espin, V.; et al. The beta-secretase-derived C-terminal fragment of betaAPP, C99, but not Abeta, is a key contributor to early intraneuronal lesions in triple-transgenic mouse hippocampus. J. Neurosci. 2012, 32, 16243–162550. [Google Scholar] [CrossRef]
- Wisniewski, K.E.; Wisniewski, H.M.; Wen, G.Y. Occurrence of neuropathological changes and dementia of Alzheimer’s disease in Down’s syndrome. Ann. Neurol. 1985, 17, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, A.M.; Barnett, J.L.; Berman, S.A.; Li, J.; Quarless, S.; Bursztajn, S.; Lippa, C.; Nixon, R.A. Gene expression and cellular content of cathepsin D in Alzheimer’s disease brain: Evidence for early up-regulation of the endosomal-lysosomal system. Neuron 1995, 14, 671–680. [Google Scholar] [CrossRef]
- Goutagny, R.; Gu, N.; Cavanagh, C.; Jackson, J.; Chabot, J.G.; Quirion, R.; Krantic, S.; Williams, S. Alterations in hippocampal network oscillations and theta-gamma coupling arise before Abeta overproduction in a mouse model of Alzheimer’s disease. Eur. J. Neurosci. 2013, 37, 1896–1902. [Google Scholar] [CrossRef] [PubMed]
- Tamayev, R.; Matsuda, S.; Arancio, O.; D’Adamio, L. beta- but not gamma-secretase proteolysis of APP causes synaptic and memory deficits in a mouse model of dementia. EMBO Mol. Med. 2012, 4, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, A.; Lauritzen, I.; Lorivel, T.; Bauer, C.; Checler, F.; Pardossi-Piquard, R. Intraneuronal accumulation of C99 contributes to synaptic alterations, apathy-like behavior, and spatial learning deficits in 3xTgAD and 2xTgAD mice. Neurobiol. Aging 2018, 71, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Pulina, M.V.; Hopkins, M.; Haroutunian, V.; Greengard, P.; Bustos, V. C99 selectively accumulates in vulnerable neurons in Alzheimer’s disease. Alzheimers Dement. 2020, 16, 273–282. [Google Scholar]
- Lauritzen, I.; Pardossi-Piquard, R.; Bourgeois, A.; Bécot, A.; Checler, F. Does intraneuronal accumulation of caboxyl-terminal fragment Amyloid Precursor Protein trigger early neurotoxicity in Alzheimers disease? Curr. Alzheimer Res. 2019, 16, 453–457. [Google Scholar] [CrossRef]
- Vingtdeux, V.; Hamdane, M.; Begard, S.; Loyens, A.; Delacourte, A.; Beauvillain, J.C.; Buee, L.; Marambaud, P.; Sergeant, N. Intracellular pH regulates amyloid precursor protein intracellular domain accumulation. Neurobiol. Dis. 2007, 25, 686–696. [Google Scholar] [CrossRef]
- Boland, B.; Smith, D.A.; Mooney, D.; Jung, S.S.; Walsh, D.M.; Platt, F.M. Macroautophagy is not directly involved in the metabolism of amyloid precursor protein. J. Biol. Chem. 2010, 285, 37415–37426. [Google Scholar] [CrossRef]
- Asai, M.; Yagishita, S.; Iwata, N.; Saido, T.C.; Ishiura, S.; Maruyama, K. An alternative metabolic pathway of amyloid precursor protein C-terminal fragments via cathepsin B in a human neuroglioma model. FASEB J. 2011, 25, 3720–3730. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A.; Yang, D.S. Autophagy failure in Alzheimer’s disease--locating the primary defect. Neurobiol. Dis. 2011, 43, 38–45. [Google Scholar] [CrossRef]
- Sardiello, M.; Palmieri, M.; di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A gene network regulating lysosomal biogenesis and function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef]
- Sardiello, M. Transcription factor EB: From master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases. Ann. N. Y. Acad. Sci. 2016, 1371, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, L.; Lotfi, P.; Pal, R.; Ronza, A.D.; Sharma, J.; Sardiello, M. Lysosome biogenesis in health and disease. J. Neurochem. 2019, 148, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S.; Caccamo, A.; Shepherd, J.D.; Murphy, M.P.; Golde, T.E.; Kayed, R.; Metherate, R.; Mattson, M.P.; Akbari, Y.; LaFerla, F.M. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: Intracellular Abeta and synaptic dysfunction. Neuron 2003, 39, 409–421. [Google Scholar] [CrossRef]
- Xiao, Q.; Yan, P.; Ma, X.; Liu, H.; Perez, R.; Zhu, A.; Gonzales, E.; Tripoli, D.L.; Czerniewski, L.; Ballabio, A.; et al. Neuronal-Targeted TFEB Accelerates Lysosomal Degradation of APP, Reducing Abeta Generation and Amyloid Plaque Pathogenesis. J. Neurosci. 2015, 35, 12137–12151. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Ash, R.T.; Ceballos-Diaz, C.; Levites, Y.; Golde, T.E.; Smirnakis, S.M.; Jankowsky, J.L. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. Eur. J. Neurosci. 2013, 37, 1203–1220. [Google Scholar] [CrossRef]
- Oules, B.; Del Prete, D.; Greco, B.; Zhang, X.; Lauritzen, I.; Sevalle, J.; Moreno, S.; Paterlini-Brechot, P.; Trebak, M.; Checler, F.; et al. Ryanodine receptor blockade reduces amyloid-beta load and memory impairments in Tg2576 mouse model of Alzheimer disease. J. Neurosci. 2012, 32, 11820–11834. [Google Scholar] [CrossRef]
- Chevallier, N.; Jiracek, J.; Vincent, B.; Baur, C.P.; Spillantini, M.G.; Goedert, M.; Dive, V.; Checler, F. Examination of the role of endopeptidase 3.4.24.15 in A beta secretion by human transfected cells. Br. J. Pharmacol. 1997, 121, 556–562. [Google Scholar] [CrossRef]
- Lauritzen, I.; Becot, A.; Bourgeois, A.; Pardossi-Piquard, R.; Biferi, M.G.; Barkats, M.; Checler, F. Targeting gamma-secretase triggers the selective enrichment of oligomeric APP-CTFs in brain extracellular vesicles from Alzheimer cell and mouse models. Transl. Neurodegener. 2019, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarty, P.; Rosario, A.; Cruz, P.; Siemienski, Z.; Ceballos-Diaz, C.; Crosby, K.; Jansen, K.; Borchelt, D.R.; Kim, J.Y.; Jankowsky, J.L.; et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS ONE 2013, 8, e67680. [Google Scholar] [CrossRef]
- Wang, H.; Wang, R.; Xu, S.; Lakshmana, M.K. Transcription Factor EB Is Selectively Reduced in the Nuclear Fractions of Alzheimer’s and Amyotrophic Lateral Sclerosis Brains. Neurosci. J. 2016, 2016, 4732837. [Google Scholar] [CrossRef] [PubMed]
- Decressac, M.; Mattsson, B.; Weikop, P.; Lundblad, M.; Jakobsson, J.; Bjorklund, A. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc. Natl. Acad. Sci. USA 2013, 110, E1817–E1826. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Cusack, C.L.; Nnah, I.C.; Khayati, K.; Saqcena, C.; Huynh, T.B.; Noggle, S.A.; Ballabio, A.; Dobrowolski, R. Dysregulation of Nutrient Sensing and CLEARance in Presenilin Deficiency. Cell Rep. 2016, 14, 2166–2179. [Google Scholar] [CrossRef]
- Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.; George-Hyslop, P.H.; Pericak-Vance, M.A.; Joo, S.H.; Rosi, B.L.; Gusella, J.F.; Crapper-MacLachlan, D.R.; Alberts, M.J.; et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 1993, 43, 1467–1472. [Google Scholar] [CrossRef]
- Parcon, P.A.; Balasubramaniam, M.; Ayyadevara, S.; Jones, R.A.; Liu, L.; Shmookler Reis, R.J.; Barger, S.W.; Mrak, R.E.; Griffin, W.S.T. Apolipoprotein E4 inhibits autophagy gene products through direct, specific binding to CLEAR motifs. Alzheimers Dement. 2018, 14, 230–242. [Google Scholar] [CrossRef]
- Bordi, M.; Berg, M.J.; Mohan, P.S.; Peterhoff, C.M.; Alldred, M.J.; Che, S.; Ginsberg, S.D.; Nixon, R.A. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy 2016, 12, 2467–2483. [Google Scholar] [CrossRef]
- Landel, V.; Baranger, K.; Virard, I.; Loriod, B.; Khrestchatisky, M.; Rivera, S.; Benech, P.; Feron, F. Temporal gene profiling of the 5XFAD transgenic mouse model highlights the importance of microglial activation in Alzheimer’s disease. Mol. Neurodegener. 2014, 9, 33. [Google Scholar] [CrossRef]
- Xiao, Q.; Yan, P.; Ma, X.; Liu, H.; Perez, R.; Zhu, A.; Gonzales, E.; Burchett, J.M.; Schuler, D.R.; Cirrito, J.R.; et al. Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 2014, 34, 9607–9620. [Google Scholar] [CrossRef]
- Bao, J.; Zheng, L.; Zhang, Q.; Li, X.; Zhang, X.; Li, Z.; Bai, X.; Zhang, Z.; Huo, W.; Zhao, X.; et al. Deacetylation of TFEB promotes fibrillar Abeta degradation by upregulating lysosomal biogenesis in microglia. Protein. Cell 2016, 7, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Song, J.X.; Malampati, S.; Zeng, Y.; Durairajan, S.S.K.; Yang, C.B.; Tong, B.C.; Iyaswamy, A.; Shang, W.B.; Sreenivasmurthy, S.G.; Zhu, Z.; et al. A small molecule transcription factor EB activator ameliorates beta-amyloid precursor protein and Tau pathology in Alzheimer’s disease models. Aging Cell 2019, 19, e13069. [Google Scholar] [CrossRef] [PubMed]

| Gene name | Forward Primer | Reverse Primer |
|---|---|---|
| Human | ||
| TFEB | 5′-CCAGAAGCGAGAGCTCACAGAT-3′ | 5′-TGTGATTGTCTTTCTTCTGCCG-3′ |
| CSTB | 5′-TCATGGCCGAGATCTACAAA-3′ | 5′-TGACGTGTTGGTACACTCCTG-3′ |
| CSTD | 5′-GAAAGGCCTACTGGCAGGT-3′ | 5′TCCCTGTGTCCACAATAGCC-3′ |
| CSTL | 5′-TCCTATCCATATGAGGCAACAG-3′ | 5′-GTTGCAACTGCCTTCATCAG-3′ |
| LAMP1 | 5′-ACGTTACAGCGTCCAGCTCAT-3′ | 5′-TCTTTGGAGCTCGCATTGG-3′ |
| LAMP2a | 5′-CAACAAAGAGCAGACTGTTTCAG-3′ | 5′-GCACTGCAGTCTTGAGCTGT-3′ |
| LAMP2b | 5′-AAGGGTTCAGCCTTTCAATG-3′ | 5′-CAACTATAATTGGGATTAGAATGGTGT-3 |
| MAP1LC3B | 5′-GAGAAGCAGCTTCCTGTTCTGG-3′ | 5′-GTGTCCGTTCACCAACAGGAAG-3′ |
| BECN1 | 5′-GGCTGAGAGACTGGATCAGG-3′ | 5′-CTGCGTCTGGGCATAACG-3′ |
| SQSTM1 | 5′-GCACCCCAATGTGATCTGC-3′ | 5′-CGCTACACAAGTCGTAGTCTGG-3′ |
| GBA | 5′-TCCTACCCTCGCCAACAGTA-3′ | 5′-GCTGCTTCTGGGTCTGTCA-3′ |
| APP | 5′-TTCATCATGGTGTGGTGGAG-3′ | 5′-GTTCTGCTGCATCTTGGACA-3′ |
| GAPDH | 5′-AGCCACATCGCTCAGACAC-3′ | 5′-GCCCAATACGACCAAATCC-3′ |
| RPL19 | 5′GGGCATAGGTAAGCGGAAGG-3′ | 5′TCAGGTACAGGCTGTGATACA-3 |
| Mouse | ||
| TFEB | 5′-GAGCTGGGAATGCTGATCC-3′ | 5′-GGGACTTCTGCAGGTCCTT-3′ |
| CSTB | 5′-CACACTGAAACCAGGCCTTT-3′ | 5′-CTTGCTGTGGTATCCAGTGTG-3′ |
| CSTD | 5′-GCCTACTGGCAGGTCCAC-3′ | 5′-GTGTCCACAATGGCCTGAC-3′ |
| LAMP1 | 5′-GTGGCAACTTCAGCAAGGA-3′ | 5′-GTGGGCACAAGTGGTGGT-3′ |
| GAPDH | 5′-TGTCCGTCGTGGATCTGAC-3′ | 5′-CCTGCTTCACCACCTTCTTG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bécot, A.; Pardossi-Piquard, R.; Bourgeois, A.; Duplan, E.; Xiao, Q.; Diwan, A.; Lee, J.-M.; Lauritzen, I.; Checler, F. The Transcription Factor EB Reduces the Intraneuronal Accumulation of the Beta-Secretase-Derived APP Fragment C99 in Cellular and Mouse Alzheimer’s Disease Models. Cells 2020, 9, 1204. https://doi.org/10.3390/cells9051204
Bécot A, Pardossi-Piquard R, Bourgeois A, Duplan E, Xiao Q, Diwan A, Lee J-M, Lauritzen I, Checler F. The Transcription Factor EB Reduces the Intraneuronal Accumulation of the Beta-Secretase-Derived APP Fragment C99 in Cellular and Mouse Alzheimer’s Disease Models. Cells. 2020; 9(5):1204. https://doi.org/10.3390/cells9051204
Chicago/Turabian StyleBécot, Anaïs, Raphaëlle Pardossi-Piquard, Alexandre Bourgeois, Eric Duplan, Qingli Xiao, Abhinav Diwan, Jin-Moo Lee, Inger Lauritzen, and Frédéric Checler. 2020. "The Transcription Factor EB Reduces the Intraneuronal Accumulation of the Beta-Secretase-Derived APP Fragment C99 in Cellular and Mouse Alzheimer’s Disease Models" Cells 9, no. 5: 1204. https://doi.org/10.3390/cells9051204
APA StyleBécot, A., Pardossi-Piquard, R., Bourgeois, A., Duplan, E., Xiao, Q., Diwan, A., Lee, J.-M., Lauritzen, I., & Checler, F. (2020). The Transcription Factor EB Reduces the Intraneuronal Accumulation of the Beta-Secretase-Derived APP Fragment C99 in Cellular and Mouse Alzheimer’s Disease Models. Cells, 9(5), 1204. https://doi.org/10.3390/cells9051204






