An Innovative Lipidomic Workflow to Investigate the Lipid Profile in a Cystic Fibrosis Cell Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagent and Chemicals
2.2. Cell Culture
2.3. Lipids Extraction
2.4. LC-MS/MS Untargeted Method
2.5. Lipidomic Data Processing
2.6. Statistical and Data Analysis
3. Results
3.1. Pre-Analytical Optimization
3.2. Optimization of the Analytical Conditions for Lipidomics Analysis
3.3. Performances of the Untargeted Lipidomics Analysis
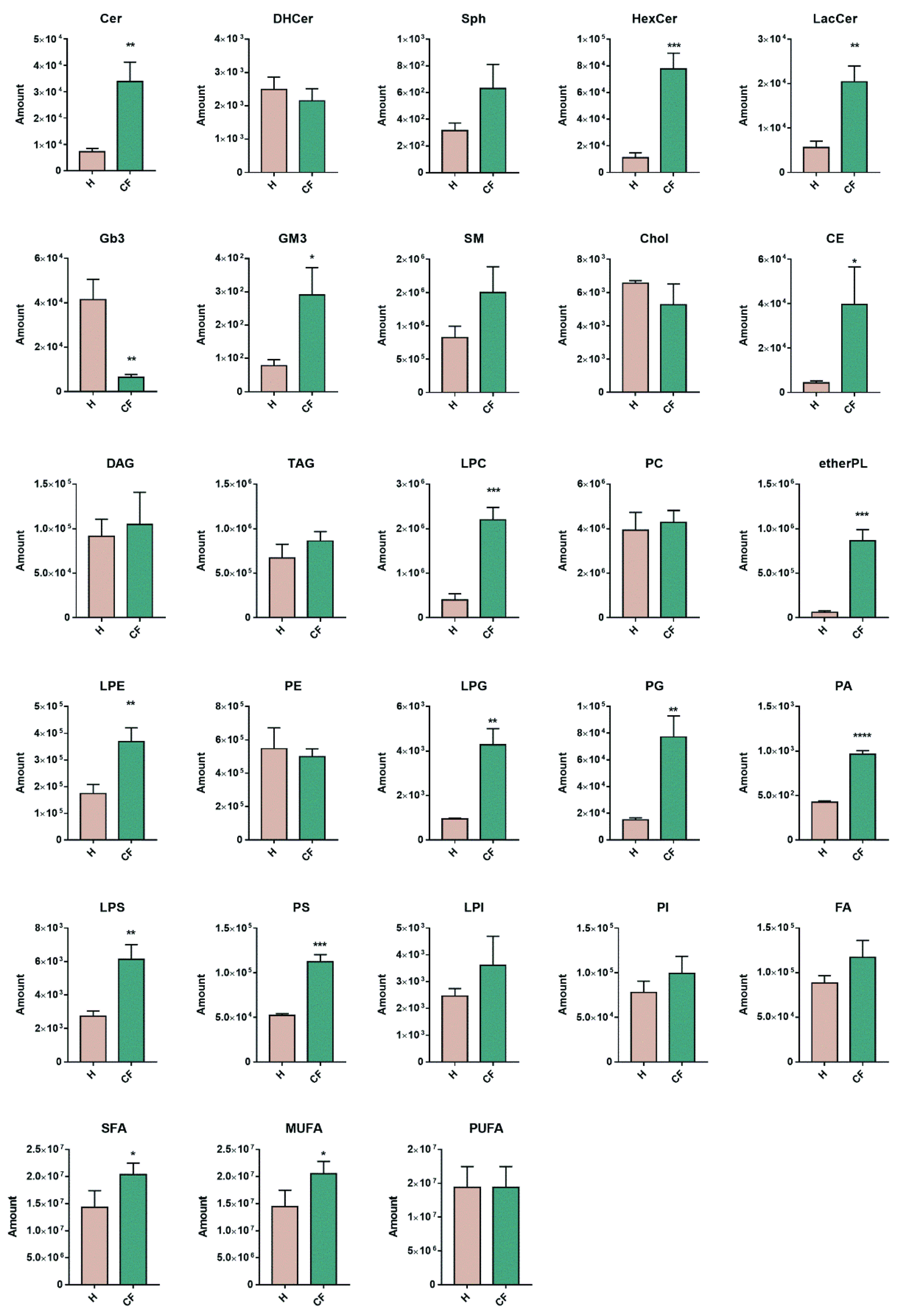
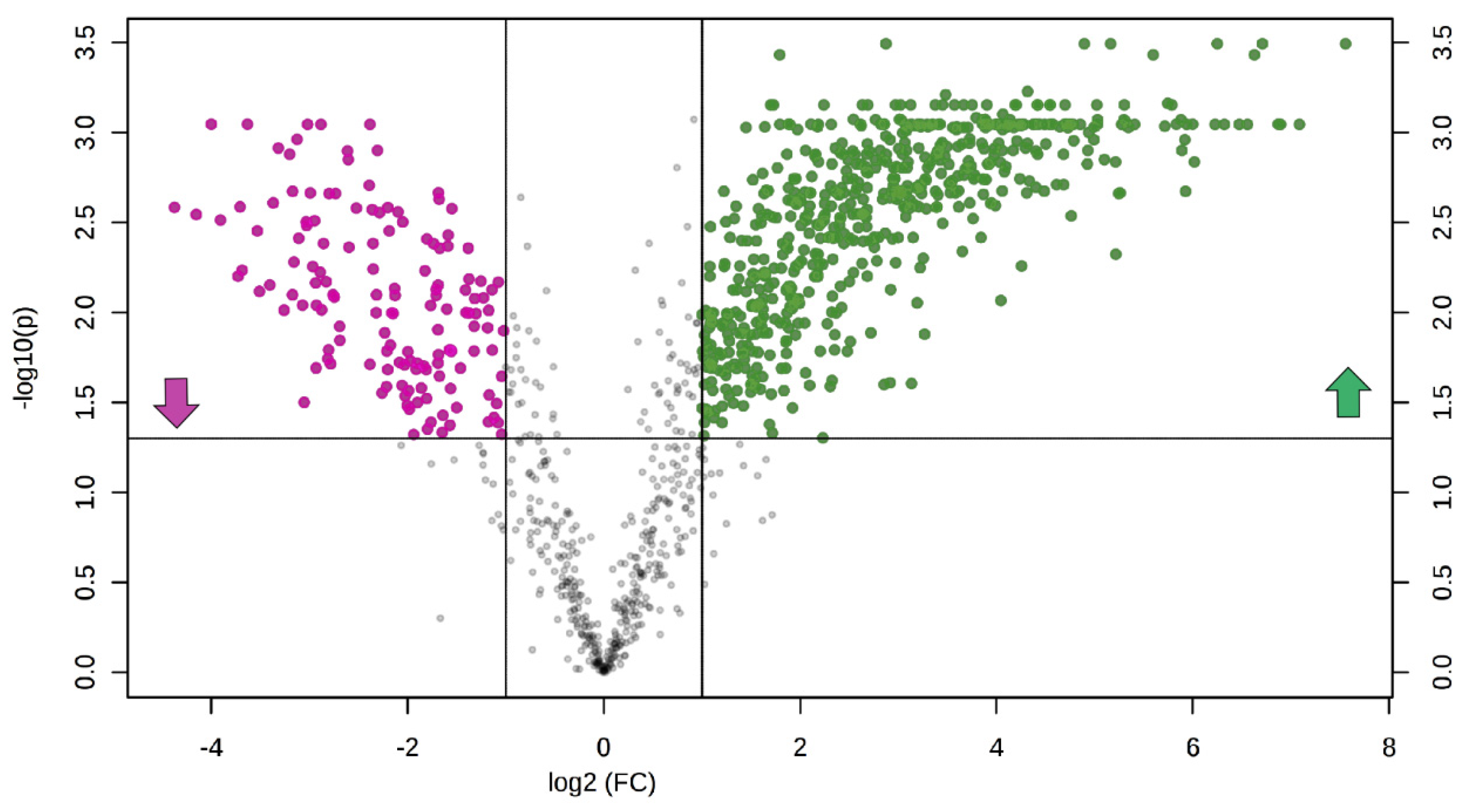
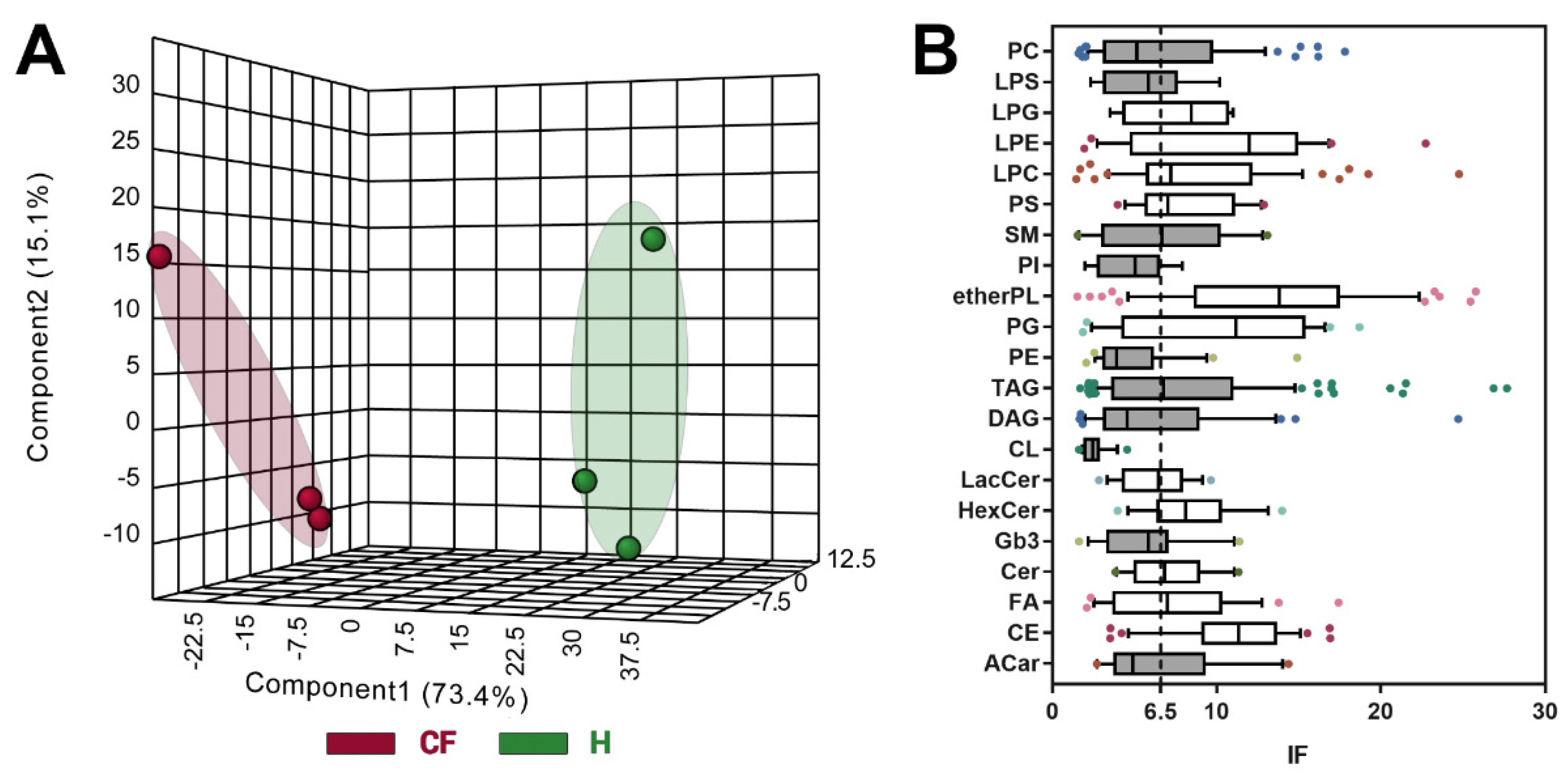
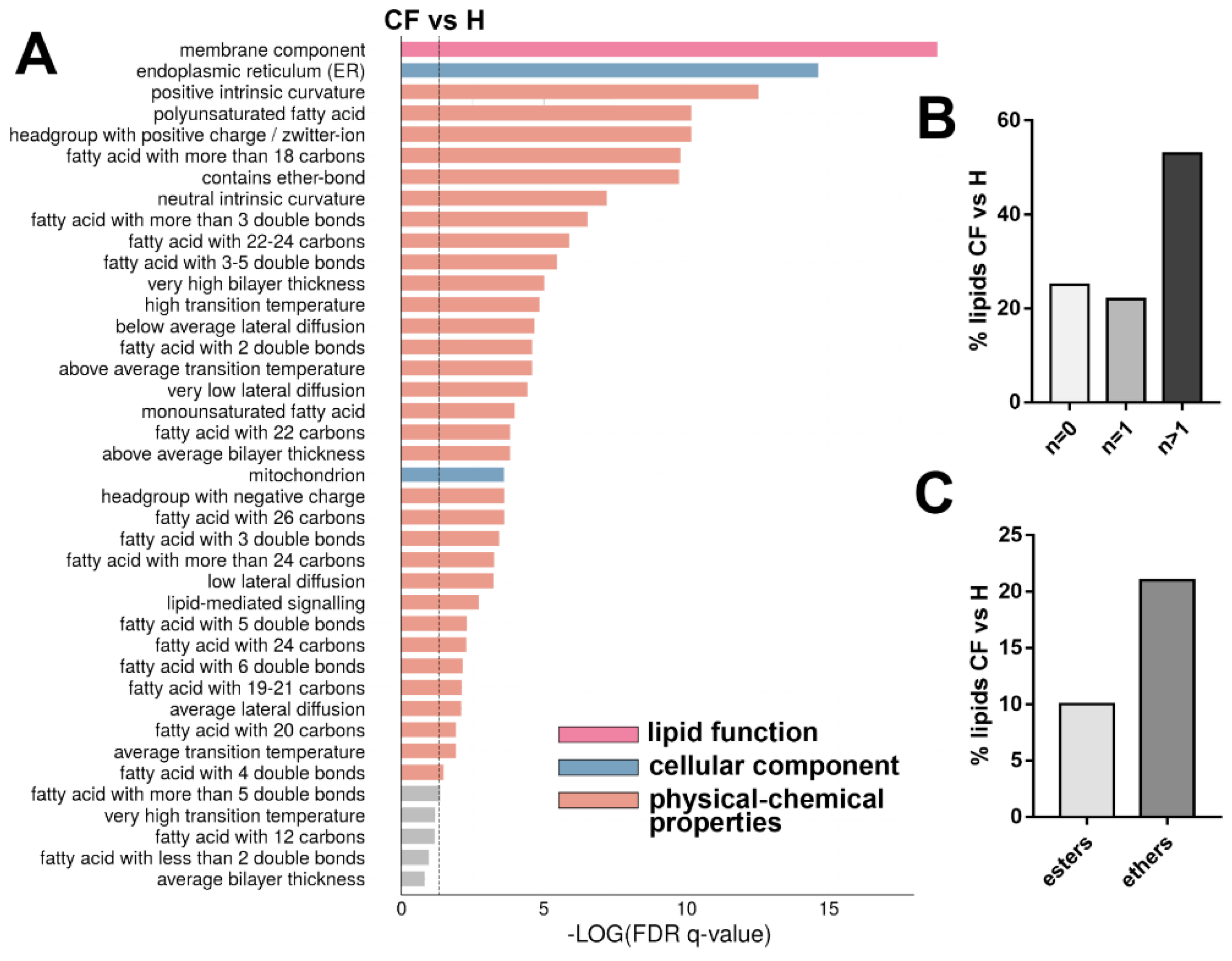
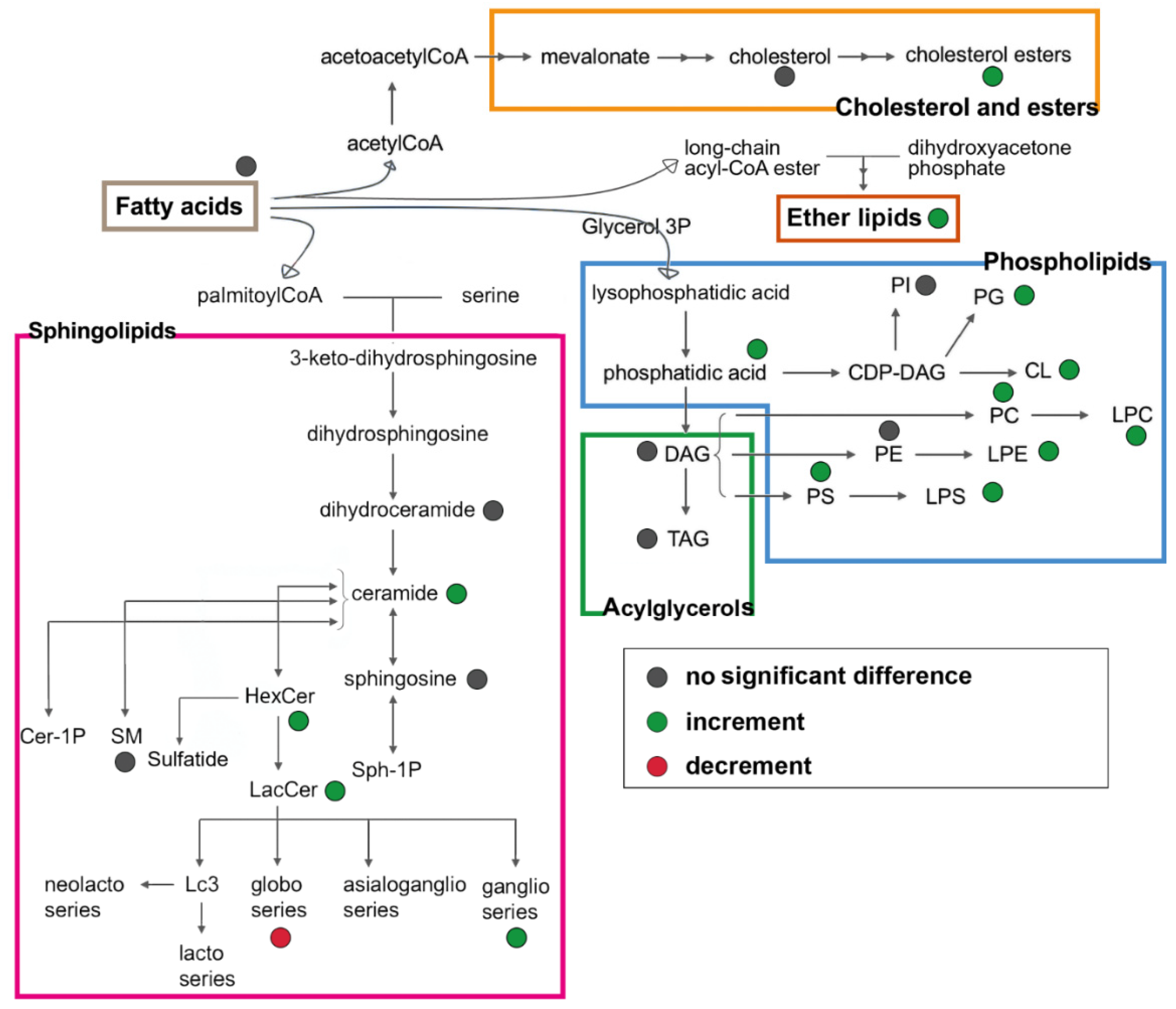
3.4. Untargeted Lipidomics of Cystic Fibrosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| H | healthy phenotype |
| CF | cystic fibrosis |
| Cer | ceramides |
| DHCer | dihydroceramides |
| HexCer | glucosylceramides |
| LacCer | lactosylceramides |
| GM3 | gangliosides |
| Gb3 | globotriaosylceramide |
| SM | sphingomyelins |
| Chol | free cholesterol |
| CE | cholesterol esters |
| LPE | lysophosphatidyletanolamines |
| PC | phosphatidylcholines |
| PG | phosphatidylglicerols |
| PI | phosphatidylinositoles |
| LPI | lysophosphatidylinositoles |
| PG | phosphatidylglicerols |
| LPG | lysophosphatidylglicerols |
| FA | free fatty acids |
| ACar | acylcarnitines |
| CL | cardiolipins |
| etherPL | ether-linked phospholipids |
| SFA | saturated fatty acids |
| MUFA | monounsaturated fatty acids |
| PUFA | polyunsaturated fatty acids |
References
- Tang, Q.Q. Lipid metabolism and diseases. Sci. Bull. 2016, 61, 1471–1472. [Google Scholar] [CrossRef]
- Yan, F.; Wen, Z.; Wang, R.; Luo, W.; Du, Y.; Wang, W.; Chen, X. Identification of the lipid biomarkers from plasma in idiopathic pulmonary fibrosis by Lipidomics. BMC Pulm. Med. 2017, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Wenk, M.R. Lipidomics: New tools and applications. Cell 2010, 143, 888–895. [Google Scholar] [CrossRef]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Brietzke, E. Recent advances in lipidomics: Analytical and clinical perspectives. Prostaglandins Other. Lipid. Mediat. 2017, 128, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.R.; Jeucken, A.; Wassenaar, T.A.; Van De Lest, C.H.A.; Brouwers, J.F.; Helms, J.B. LION/web: A web-based ontology enrichment tool for lipidomic data analysis. Gigascience 2019, 8, giz061. [Google Scholar] [CrossRef] [PubMed]
- Elborn, J.S. Cystic fibrosis. Lancet 2016, 388, 2519–2531. [Google Scholar] [CrossRef]
- De Boeck, K.; Amaral, M.D. Progress in therapies for cystic fibrosis. Lancet Respir. Med. 2016, 4, 662–674. [Google Scholar] [CrossRef]
- Risé, P.; Volpi, S.; Colombo, C.; Padoan, R.F.; D’Orazio, C.; Ghezzi, S.; Melotti, P.; Bennato, V.; Agostoni, C.; Assael, B.M.; et al. Whole blood fatty acid analysis with micromethod in cystic fibrosis and pulmonary disease. J. Cyst. Fibros. 2010, 9, 228–233. [Google Scholar] [CrossRef]
- Freedman, S.D.; Blanco, P.G.; Shea, J.C.; Alvarez, J.G. Analysis of lipid abnormalities in CF mice. Methods Mol. Med. 2002, 70, 517–524. [Google Scholar]
- Ollero, M.; Astarita, G.; Guerrera, I.C.; Sermet-Gaudelus, I.; Trudel, S.; Piomelli, D.; Edelman, A. Plasma lipidomics reveals potential prognostic signatures within a cohort of cystic fibrosis patients. J. Lipid Res. 2011, 52, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Gelzo, M.; Sica, C.; Elce, A.; Dello Russo, A.; Iacotucci, P.; Carnovale, V.; Raia, V.; Salvatore, D.; Corso, G.; Castaldo, G. Reduced absorption and enhanced synthesis of cholesterol in patients with cystic fibrosis: A preliminary study of plasma sterols. Clin. Chem. Lab. Med. 2016, 54, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, V.; Milla, C.; Parks, E.J.; Schwarzenberg, S.J.; Moran, A. Abnormal lipid concentrations in cystic fibrosis. Am. J. Clin. Nutr. 2002, 75, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- White, N.M.; Jiang, D.; Burgess, J.D.; Bederman, I.R.; Previs, S.F.; Kelley, T.J. Altered cholesterol homeostasis in cultured and in vivo models of cystic fibrosis. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2007, 292, 476–486. [Google Scholar] [CrossRef]
- Teichgräber, V.; Ulrich, M.; Endlich, N.; Riethmüller, J.; Wilker, B.; De Oliveira-Munding, C.C.; Van Heeckeren, A.M.; Barr, M.L.; Von Kürthy, G.; Schmid, K.W.; et al. Ceramide accumulation mediates inflammation, cell death and infection susceptibility in cystic fibrosis. Nat. Med. 2008, 14, 382–391. [Google Scholar] [CrossRef]
- Ernst, W.L.; Shome, K.; Wu, C.C.; Gong, X.; Frizzell, R.A.; Aridor, M. VAMP-associated proteins (VAP) as receptors that couple cystic fibrosis transmembrane conductance regulator (CFTR) proteostasis with lipid homeostasis. J. Biol. Chem. 2016, 291, 5206–5220. [Google Scholar] [CrossRef]
- Fang, D.; West, R.H.; Manson, M.E.; Ruddy, J.; Jiang, D.; Previs, S.F.; Sonawane, N.D.; Burgess, J.D.; Kelley, T.J. Increased plasma membrane cholesterol in cystic fibrosis cells correlates with CFTR genotype and depends on de novo cholesterol synthesis. Respir. Res. 2010, 11, 61. [Google Scholar] [CrossRef]
- Mingione, A.; Dei Cas, M.; Bonezzi, F.; Caretti, A.; Piccoli, M.; Anastasia, L.; Ghidoni, R.; Paroni, R.; Signorelli, P. Inhibition of Sphingolipid Synthesis as a Phenotype-Modifying Therapy in Cystic Fibrosis. Cell. Physiol. Biochem. 2020, 50323257, 110–125. [Google Scholar]
- Caretti, A.; Torelli, R.; Perdoni, F.; Falleni, M.; Tosi, D.; Zulueta, A.; Casas, J.; Sanguinetti, M.; Ghidoni, R.; Borghi, E.; et al. Inhibition of ceramide de novo synthesis by myriocin produces the double effect of reducing pathological inflammation and exerting antifungal activity against A. fumigatus airways infection. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1089–1097. [Google Scholar] [CrossRef]
- Della Corte, A.; Chitarrini, G.; Di Gangi, I.M.; Masuero, D.; Soini, E.; Mattivi, F.; Vrhovsek, U. A rapid LC-MS/MS method for quantitative profiling of fatty acids, sterols, glycerolipids, glycerophospholipids and sphingolipids in grapes. Talanta 2015, 140, 52–61. [Google Scholar] [CrossRef]
- Cajka, T.; Smilowitz, J.T.; Fiehn, O. Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal. Chem. 2017, 89, 12360–12368. [Google Scholar] [CrossRef] [PubMed]
- Huan, T.; Forsberg, E.M.; Rinehart, D.; Johnson, C.H.; Ivanisevic, J.; Benton, H.P.; Fang, M.; Aisporna, A.; Hilmers, B.; Poole, F.L.; et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods 2017, 14, 461–462. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Qian, W.; Shen, C.; Lin, L.; Xie, T.; Peng, L.; Xu, J.; Yang, R.; Ji, J.; Zhao, X. High-resolution lipidomics reveals dysregulation of lipid metabolism in respiratory syncytial virus pneumonia mice. RSC Adv. 2018, 8, 29368–29377. [Google Scholar] [CrossRef]
- Hu, C.; Zhou, Y.; Feng, J.; Zhou, S.; Li, C.; Zhao, S.; Shen, Y.; Hong, L.; Xuan, Q.; Liu, X.; et al. Untargeted Lipidomics Reveals Specific Lipid Abnormalities in Nonfunctioning Human Pituitary Adenomas. J. Proteome Res. 2019, 19, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinforma. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, 486–494. [Google Scholar] [CrossRef]
- Rahman, M.A.; Akond, M.; Babar, M.A.; Beecher, C.; Erickson, J.; Thomason, K.; De Jong, F.A.; Mason, R.E. LC-HRMS Based Non-Targeted Metabolomic Profiling of Wheat (Triticum aestivum; L.) under Post-Anthesis Drought Stress. Am. J. Plant Sci. 2017, 8, 3024–3061. [Google Scholar] [CrossRef]
- Jiang, W.; Gao, L.; Li, P.; Kan, H.; Qu, J.; Men, L.; Liu, Z.; Liu, Z. Metabonomics study of the therapeutic mechanism of fenugreek galactomannan on diabetic hyperglycemia in rats, by ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1044, 8–16. [Google Scholar] [CrossRef]
- Platania, C.B.M.; Dei Cas, M.; Cianciolo, S.; Fidilio, A.; Lazzara, F.; Paroni, R.; Pignatello, R.; Strettoi, E.; Ghidoni, R.; Drago, F.; et al. Novel ophthalmic formulation of myriocin: implications in retinitis pigmentosa. Drug Deliv. 2019, 26, 237–243. [Google Scholar] [CrossRef]
- La Corte, E.; Cas, M.D.; Raggi, A.; Patan, M.; Broggi, M.; Schiavolin, S.; Calatozzolo, C.; Pollo, B.; Pipolo, C.; Bruzzone, M.G.; et al. Long and Very-Long-Chain Ceramides Correlate with A More Aggressive Behavior in Skull Base Chordoma Patients. Int. J. Mol. Sci. 2019, 20, 4480. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Cheng, H.; Yang, K.; Gross, R.W.; Han, X. Alkaline methanolysis of lipid extracts extends shotgun lipidomics analyses to the low-abundance regime of cellular sphingolipids. Anal. Biochem. 2007, 371, 135–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Merrill, A.H.; Sullards, M.C.; Allegood, J.C.; Kelly, S.; Wang, E. Sphingolipidomics: High-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 2005, 36, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ser, Z.; Locasale, J.W. Development and quantitative evaluation of a high-resolution metabolomics technology. Anal. Chem. 2014, 86, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Rombouts, C.; De Spiegeleer, M.; Van Meulebroek, L.; De Vos, W.H.; Vanhaecke, L. Validated comprehensive metabolomics and lipidomics analysis of colon tissue and cell lines. Anal. Chim. Acta 2019, 1066, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Han, T.-L.; Yang, Y.; Zhang, H.; Law, K.P. Analytical challenges of untargeted GC-MS-based metabolomics and the critical issues in selecting the data processing strategy. F1000Research 2017, 6, 967. [Google Scholar] [CrossRef]
- Van Der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef]
- Drotleff, B.; Lämmerhofer, M. Guidelines for Selection of Internal Standard-based Normalization Strategies in Untargeted Lipidomic Profiling by LC-HR-MS/MS. Anal. Chem. 2019, 91, 9836–9843. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fan, S.; Wancewicz, B.; Cajka, T.; Sa, M.; Showalter, M.R.; Baillie, R.; Tenenbaum, J.D.; Louie, G.; Kaddurah-Daouk, R.; et al. Generation and quality control of lipidomics data for the alzheimer’s disease neuroimaging initiative cohort. Sci. Data 2018, 5, 1–13. [Google Scholar] [CrossRef]
- Chiu, C.-Y.; Yeh, K.-W.; Lin, G.; Chiang, M.-H.; Yang, S.-C.; Chao, W.-J.; Yao, T.-C.; Tsai, M.-H.; Hua, M.-C.; Liao, S.-L.; et al. Metabolomics Reveals Dynamic Metabolic Changes Associated with Age in Early Childhood. PLoS ONE 2016, 11, e0149823. [Google Scholar] [CrossRef]
- Ollero, M. Methods for the study of lipid metabolites in cystic fibrosis. J. Cyst. Fibros. 2004, 3, 97–98. [Google Scholar] [CrossRef]
- Del Ciampo, I.R.L.; Sawamura, R.; Machado Fernandes, M.I. Cystic fibrosis: From protein-energy malnutrition to obesity with dyslipidemia. Iran. J. Pediatr. 2013, 23, 605–606. [Google Scholar] [PubMed]
- Ziobro, R.; Henry, B.; Edwards, M.J.; Lentsch, A.B.; Gulbins, E. Ceramide mediates lung fibrosis in cystic fibrosis. Biochem. Biophys. Res. Commun. 2013, 434, 705–709. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. The ceramide-centric universe of lipid-mediated cell regulation: Stress encounters of the lipid kind. J. Biol. Chem. 2002, 277, 25847–25850. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef]
- Caretti, A.; Bragonzi, A.; Facchini, M.; De Fino, I.; Riva, C.; Gasco, P.; Musicanti, C.; Casas, J.; Fabriàs, G.; Ghidoni, R.; et al. Anti-inflammatory action of lipid nanocarrier-delivered myriocin: Therapeutic potential in cystic fibrosis. Biochim. Biophys. Acta-Gen. Subj. 2014, 1840, 586–594. [Google Scholar] [CrossRef]
- Ramu, Y.; Xu, Y.; Lu, Z. Inhibition of CFTR Cl- channel function caused by enzymatic hydrolysis of sphingomyelin. Proc. Natl. Acad. Sci. USA 2007, 104, 6448–6453. [Google Scholar] [CrossRef]
- Grassmé, H.; Henry, B.; Ziobro, R.; Becker, K.A.; Riethmüller, J.; Gardner, A.; Seitz, A.P.; Steinmann, J.; Lang, S.; Ward, C.; et al. β1-Integrin Accumulates in Cystic Fibrosis Luminal Airway Epithelial Membranes and Decreases Sphingosine, Promoting Bacterial Infections. Cell Host Microbe 2017, 21, 707–718.e8. [Google Scholar] [CrossRef]
- Hamai, H.; Keyserman, F.; Quittell, L.M.; Worgall, T.S. Defective CFTR increases synthesis and mass of sphingolipids that modulate membrane composition and lipid signaling. J. Lipid Res. 2009, 50, 1101–1108. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Astudillo, L.; Therville, N.; Colacios, C.; Ségui, B.; Andrieu-Abadie, N.; Levade, T. Glucosylceramidases and malignancies in mammals. Biochimie 2016, 125, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Nagata, M.; Izumi, Y.; Ishikawa, E.; Kiyotake, R.; Doi, R.; Iwai, S.; Omahdi, Z.; Yamaji, T.; Miyamoto, T.; Bamba, T.; et al. Intracellular metabolite β-glucosylceramide is an endogenous Mincle ligand possessing immunostimulatory activity. Proc. Natl. Acad. Sci. USA 2017, 114, e3285–e3294. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulou, M.; Gordillo, R.; Koliaki, C.; Gancheva, S.; Jelenik, T.; De Filippo, E.; Herder, C.; Markgraf, D.; Jankowiak, F.; Esposito, I.; et al. Specific hepatic sphingolipids relate to insulin resistance, oxidative stress, and inflammation in nonalcoholic steato hepatitis. Diabetes Care 2018, 41, 1235–1243. [Google Scholar] [CrossRef] [PubMed]
- Barth, B.M.; Shanmugavelandy, S.S.; Tacelosky, D.M.; Kester, M.; Morad, S.A.F.; Cabot, M.C. Gaucher’s disease and cancer: A sphingolipid perspective. Crit. Rev. Oncog. 2013, 18, 221–234. [Google Scholar] [CrossRef]
- Chiurchiù, V.; Leuti, A.; Maccarrone, M. Bioactive lipids and chronic inflammation: Managing the fire within. Front. Immunol. 2018, 9, 38. [Google Scholar] [CrossRef]
- Seidl, E.; Kiermeier, H.; Liebisch, G.; Ballmann, M.; Hesse, S.; Paul-Buck, K.; Ratjen, F.; Rietschel, E.; Griese, M. Lavage lipidomics signatures in children with cystic fibrosis and protracted bacterial bronchitis. J. Cyst. Fibros. 2019, 18, 790–795. [Google Scholar] [CrossRef]
- Tselepis, A.D.; Chapman, M.J. Inflammation, bioactive lipids and atherosclerosis: Potential roles of a lipoprotein-associated phospholipase A2, platelet activating factor-acetylhydrolase. Atheroscler. Suppl. 2002, 3, 57–68. [Google Scholar] [CrossRef]
- Gentzsch, M.; Choudhury, A.; Chang, X.B.; Pagano, R.E.; Riordan, J.R. Misassembled mutant ΔF508 CFTR in the distal secretory pathway alters cellular lipid trafficking. J. Cell Sci. 2007, 120, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.C.; Yoon, A.J.; Faull, K.F.; Desharnais, R.; Zemanick, E.T.; Porter, E. Cholesteryl esters are elevated in the lipid fraction of bronchoalveolar lavage fluid collected from pediatric cystic fibrosis patients. PLoS ONE 2015, 10, e0125326. [Google Scholar] [CrossRef]
- Ma, D.; Yoon, A.; Bartlett, J.; Faull, K.F.; McCray, P.B., Jr.; Zemanick, E.T.; Porter, E. Antimicrobial cholesteryl esters in cystic fibrosis airway secretions. Pediatr. Pulmonol. 2013, 10, e0125326. [Google Scholar]
- Ghidoni, R.; Caretti, A.; Signorelli, P. Role of sphingolipids in the pathobiology of lung inflammation. Mediators Inflamm. 2015, 2015, 487508. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.; Naren, A.P. Pharmacological Correction of Cystic Fibrosis: Molecular Mechanisms at the Plasma Membrane to Augment Mutant CFTR Function. Curr. Drug Targets 2016, 17, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Matos, A.M.; Pinto, F.R.; Barros, P.; Amaral, M.D.; Pepperkok, R.; Matos, P. Inhibition of calpain 1 restores plasma membrane stability to pharmacologically rescued Phe508del-CFTR variant. J. Biol. Chem. 2019, 294, 13396–13410. [Google Scholar] [CrossRef] [PubMed]
- Wallner, S.; Orso, E.; Grandl, M.; Konovalova, T.; Liebisch, G.; Schmitz, G. Phosphatidylcholine and phosphatidylethanolamine plasmalogens in lipid loaded human macrophages. PLoS ONE 2018, 13, 1–21. [Google Scholar] [CrossRef]
- Dean, J.M.; Lodhi, I.J. Structural and functional roles of ether lipids. Protein Cell 2018, 9, 196–206. [Google Scholar] [CrossRef]
- Drzymała-Czyż, S.; Krzyżanowska, P.; Koletzko, B.; Nowak, J.; Miśkiewicz-Chotnicka, A.; Moczko, J.A.; Lisowska, A.; Walkowiak, J. Determinants of serum glycerophospholipid fatty acids in cystic fibrosis. Int. J. Mol. Sci. 2017, 18, 185. [Google Scholar] [CrossRef]
- Zhou, J.J.; Linsdell, P. Molecular mechanism of arachidonic acid inhibition of the CFTR chloride channel. Eur. J. Pharmacol. 2007, 563, 88–91. [Google Scholar] [CrossRef]
- Li, Y.; Wang, W.; Parker, W.; Clancy, J.P. Adenosine regulation of cystic fibrosis transmembrane conductance regulator through prostenoids in airway epithelia. Am. J. Respir. Cell Mol. Biol. 2006, 34, 600–608. [Google Scholar] [CrossRef]
- Seegmiller, A.C. Abnormal unsaturated fatty acid metabolism in cystic fibrosis: Biochemical mechanisms and clinical implications. Int. J. Mol. Sci. 2014, 15, 16083–16099. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dei Cas, M.; Zulueta, A.; Mingione, A.; Caretti, A.; Ghidoni, R.; Signorelli, P.; Paroni, R. An Innovative Lipidomic Workflow to Investigate the Lipid Profile in a Cystic Fibrosis Cell Line. Cells 2020, 9, 1197. https://doi.org/10.3390/cells9051197
Dei Cas M, Zulueta A, Mingione A, Caretti A, Ghidoni R, Signorelli P, Paroni R. An Innovative Lipidomic Workflow to Investigate the Lipid Profile in a Cystic Fibrosis Cell Line. Cells. 2020; 9(5):1197. https://doi.org/10.3390/cells9051197
Chicago/Turabian StyleDei Cas, Michele, Aida Zulueta, Alessandra Mingione, Anna Caretti, Riccardo Ghidoni, Paola Signorelli, and Rita Paroni. 2020. "An Innovative Lipidomic Workflow to Investigate the Lipid Profile in a Cystic Fibrosis Cell Line" Cells 9, no. 5: 1197. https://doi.org/10.3390/cells9051197
APA StyleDei Cas, M., Zulueta, A., Mingione, A., Caretti, A., Ghidoni, R., Signorelli, P., & Paroni, R. (2020). An Innovative Lipidomic Workflow to Investigate the Lipid Profile in a Cystic Fibrosis Cell Line. Cells, 9(5), 1197. https://doi.org/10.3390/cells9051197






