1. Introduction
Recently, clustered regularly interspaced short palindromic repeats (CRISPR)/caspase-9 (Cas9) (CRISPR/Cas9) gene editing technology has been widely employed for the rapid generation of genetically modified (GM) animals, due to its simplicity, versatility, and efficiency [
1,
2]. This technology has enabled the production of mice with knockout, knock-in (KI), or conditional alleles, or those with single point mutations, within a few weeks. For the generation of GM animals, many researchers employed the microinjection of CRISPR components into zygotes using a micromanipulator system or in vitro electroporation (EP) of zygotes in a solution containing the CRISPR components using an electroporator [
3,
4,
5,
6,
7,
8,
9,
10,
11,
12,
13,
14,
15]. These processes require ex vivo handling of zygotes and embryo transfer of the genome editing-treated zygotes to the reproductive tract of a recipient female for further development, all of which are costly, time-consuming, and labor-intensive.
We developed a new method, called genome editing via oviductal nucleic acid delivery (GONAD), which was subsequently renamed “improved GONAD (
i-GONAD)”, for the production of genome-edited mice [
16,
17,
18], rats [
19,
20], and hamsters [
21]. This technology is based on the injection of a solution (1–1.5 μL) containing genome editing reagents into the lumen of an oviduct via the oviductal wall of pregnant female animals at the late zygote to two-cell stage following in vivo EP of the entire oviduct using tweezer-type electrodes under a dissecting microscope [
22,
23]. In other words, in the case of GONAD/
i-GONAD, genome editing occurs in preimplantation embryos floating in the oviductal lumen in situ. The genome-edited embryos will develop into fetuses after implantation, and the fetuses will be delivered naturally as genome-edited newborn animals. Thus, this technology does not require ex vivo handling of embryos, which is strictly required for the above-mentioned zygote microinjection and in vitro EP. In this context, GONAD/
i-GONAD appears to be more convenient and simpler than the methods that are based on the ex vivo handling of embryos.
According to Ohtsuka et al. [
17] and Gurumurthy et al. [
22], GONAD/
i-GONAD is easier to use on hybrid mice such as B6C3F1 (a hybrid between C57BL/6 and C3H/He) and randomly bred Institute of Cancer Research (ICR) mice than on the widely used inbred strain C57BL/6 (B6). Specifically, the electric conditions used for hybrid mice and ICR were often found to be deleterious to the development of B6 embryos. Ohtsuka et al. [
17] demonstrated successful acquisition of genome-edited B6 offspring when
i-GONAD was carried out using the CUY21EditII electroporator provided by BEX Co., Ltd. (Tokyo, Japan), which has the capacity to generate a constant current. According to Gurumurthy et al. [
22], the optimal electric conditions for B6 involve in vivo EP being carried out under a constant current of 100 mA. To our knowledge, two electroporators, NEPA21 (NEPA GENE Co. Ltd., Chiba, Japan) and CUY21EditII, are those that have been most frequently used for successful GONAD/
i-GONAD [
16,
17,
18,
19,
20,
21]. CUY21EditII can provide a constant current, whereas NEPA21 cannot. In this study, we attempted to explore the electric conditions (resistance and voltage) that provide a current of 100 mA when the NEPA21 electroporator is used.
Another problem in using B6 mice for
i-GONAD is the difficulty in consistently obtaining pregnant B6 females because estrous females often fail to be found. The administration of gonadotrophins has been used for inducing superovulation in many mouse strains to obtain a number of early embryos [
24]. This approach has an additional advantage in that it is capable of synchronizing the estrous cycle of females; thus, it is unnecessary to check the estrous cycle through smear testing or to inspect the state of the vagina visually. Sato et al. [
25] reported that intraperitoneal (IP) administration of low-dose (0.02–0.2 international units (IU)) pregnant mare’s serum gonadotrophin (PMSG) following IP administration of 5 IU human chorionic gonadotrophin (hCG) 48 h apart is effective for natural ovulation induction before
i-GONAD. Unfortunately, this regime has only been used successfully on B6C3F1 hybrid mice. In this study, we examined whether the administration of a single IP injection of low-dose PMSG is effective for synchronizing the estrous cycle in B6 females.
2. Materials and Methods
2.1. Animals
C57BL/6JJmsSlc (B6), BALB/cCrSIc (BALB/c), and Slc:ICR (ICR) mice were purchased from Japan SLC, Inc. (Shizuoka, Japan). Adult mice at the age ranges of 8–10 weeks (female) and 10–12 weeks (male) were used. All mice were maintained under temperature-controlled conditions (24 ± 2 °C) with a 12L/12D light cycle (lights on at 7:00 a.m.). A solid diet and water were provided ad libitum. The experiments described were performed in accordance with the guidelines of Hamamatsu University School of Medicine Committee on Recombinant DNA Security (No. 29–17). Additionally, the experiments were approved by Hamamatsu University School of Medicine Animal Care and Use Committee (No. 2017097). The experiments involving in vivo transfection of mouse preimplantation embryos by i-GONAD were accompanied by surgery (exposure of ovary/oviducts/uterus) and operation/manipulation (DNA injection via the oviductal wall and in vivo EP). All efforts were made to minimize the number of animals used and their suffering.
2.2. Mating Protocol
In the natural mating group, each female mouse was coupled with two males in a cage in the evening. In the PMSG treatment group, female mice received an IP injection of 1.2 IU PMSG/10 g in the evening (5:00–6:00 p.m.). Then, 47–49 h later, each treated female mouse was mated with two males in a cage. The morning after mating, females were checked for the presence of a copulation plug to confirm that mating had occurred. The plugged females were subjected to i-GONAD or remained untreated until birth to evaluate the following pregnancy indices: plug rate (number of plugged females/number of mated females × 100%), pregnancy rate (number of pregnant females/number of plugged females × 100%), and size of first litters (number of pups/number of females delivering live pups).
2.3. Measurement of Current
We used two types of square-pulse generator: NEPA21 (NEPA GENE Co. Ltd. Chiba, Japan) and GEB15 (BEX Co. Ltd., Tokyo, Japan). NEPA21 generates a poring pulse (Pp) and a transfer pulse (Tp). The electroporation parameters were as follows: 3 Pp (40 V, 5 ms wavelength, 50 ms pulse interval, 10% decay (±pulse orientation)) and 3 Tp (10 V, 50 ms wavelength, 50 ms pulse interval, 40% decay (±pulse orientation)). For GEB15, the electroporation parameters were as follows: driving pulse voltage (Pd V): 40 V, Pd on: 5.00 ms, Pd off: 50 ms, pulse cycles: 3, and decay: 10%. Notably, the voltage was set to 40 V (Pp for NEPA21 and PdV for GEB15).
Non-pregnant adult B6 females were euthanized with a mixture of three anesthetic agents (medetomidine, midazolam, and butorphanol) and incised at the dorsal skin for exposure of the ovary/oviduct/uterus. The oviduct regions were then covered with a piece of wet paper (KimWipe; Jujo-Kimberly Co. Ltd., Tokyo, Japan) soaked in Dulbecco’s modified phosphate-buffered saline (DPBS) and then sandwiched in tweezer-type electrodes, CUY652-3 (NEPA GENE Co. Ltd.). In this case, the interval between the two electrodes varied from 0 to 2 mm (
Figure S1). First, the resistance value was measured by pressing the Ω button set in each electroporator when the oviduct was sandwiched with the electrodes. Next, immediately after measuring the resistance, the start button was pressed to check the current.
2.4. Preparation of Genome Editing Reagents Used for i-GONAD
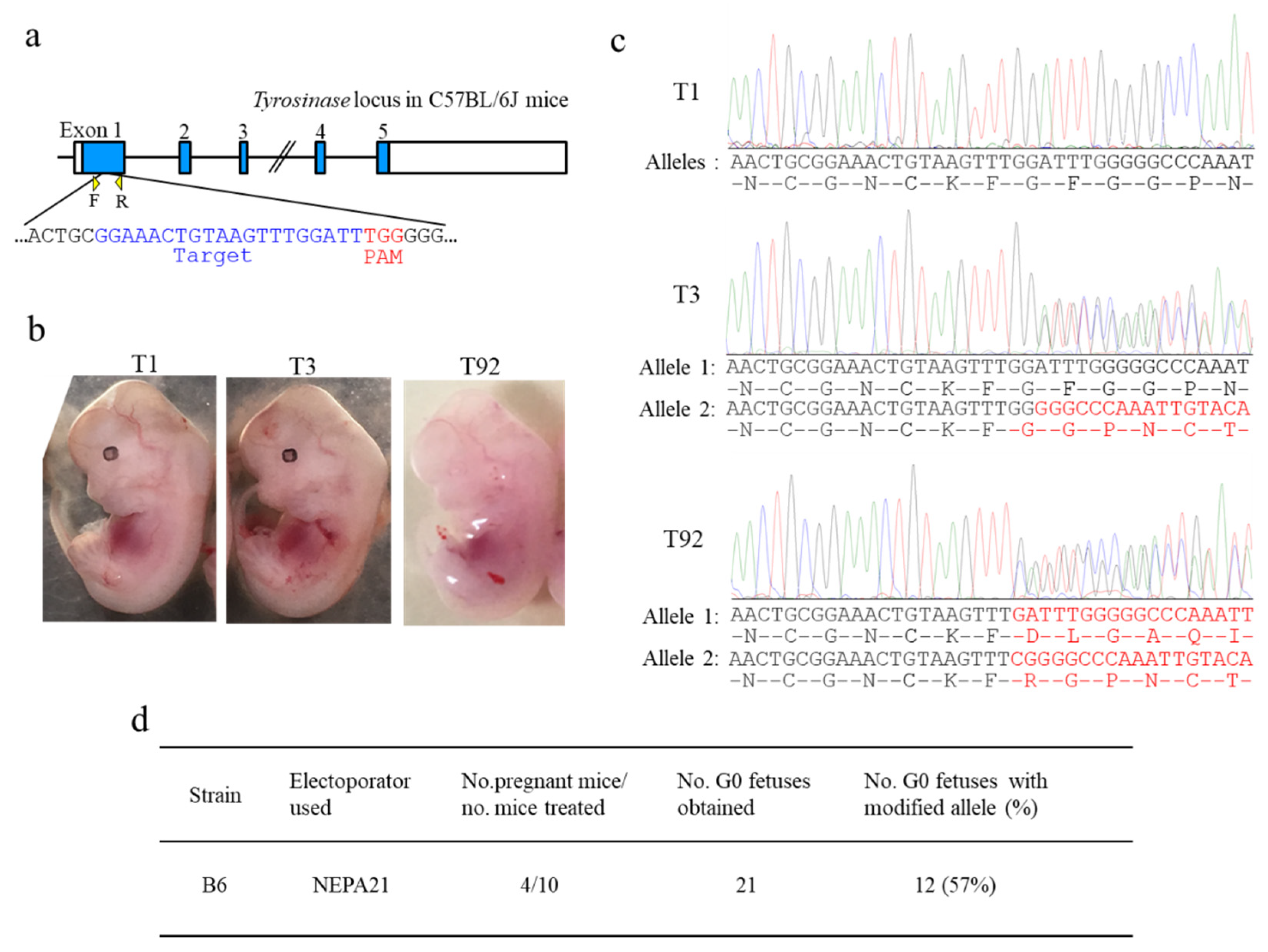
For the CRISPR/Cas9-mediated induction of insertions and deletions (indels) in B6, Alt-R CRISPR-Cas9 CRISPR RNAs (crRNAs) were designed to recognize the target site (exon 1 of wild mouse tyrosinase gene (Tyr) that matches a 20 bp DNA sequence (Tyr-wild crRNA: GGAAACTGTAAGTTTGGATT) just upstream of the protospacer adjacent motif (PAM). crRNAs were synthesized by Integrated DNA Technologies, Inc. (IDT) (Coralville, IA, USA). Alt-R CRISPR-Cas9 trans-activating small RNA (tracrRNA) was also purchased from IDT. Each reagent was dissolved in Opti-MEM medium (#31985062; Thermo Fisher Scientific Inc., Waltham, MA, USA) at a final concentration of 100 μM, and stored at −80 °C until use. crRNAs and tracrRNA were annealed by mixing equimolar amounts of each and incubating at room temperature for about 10 min to allow the formation of crRNA/tracrRNA duplexes. The annealed crRNA/tracrRNA duplexes (6 μL) were mixed with 1 μL of Cas9 protein (10 μg/μL; purchased from IDT) to form ribonucleoprotein (RNP) complexes in 2 μL of Opti-MEM medium in a 0.5-mL PCR tube. Thus, the solution used for i-GONAD comprised crRNA/tracrRNA duplexes (final concentration of 30 μM), Cas9 protein (1 μg/μL), and 0.02% Fast Green FCF (#15939-54; Nacalai Tesque Inc., Kyoto, Japan) dissolved in Opti-MEM medium.
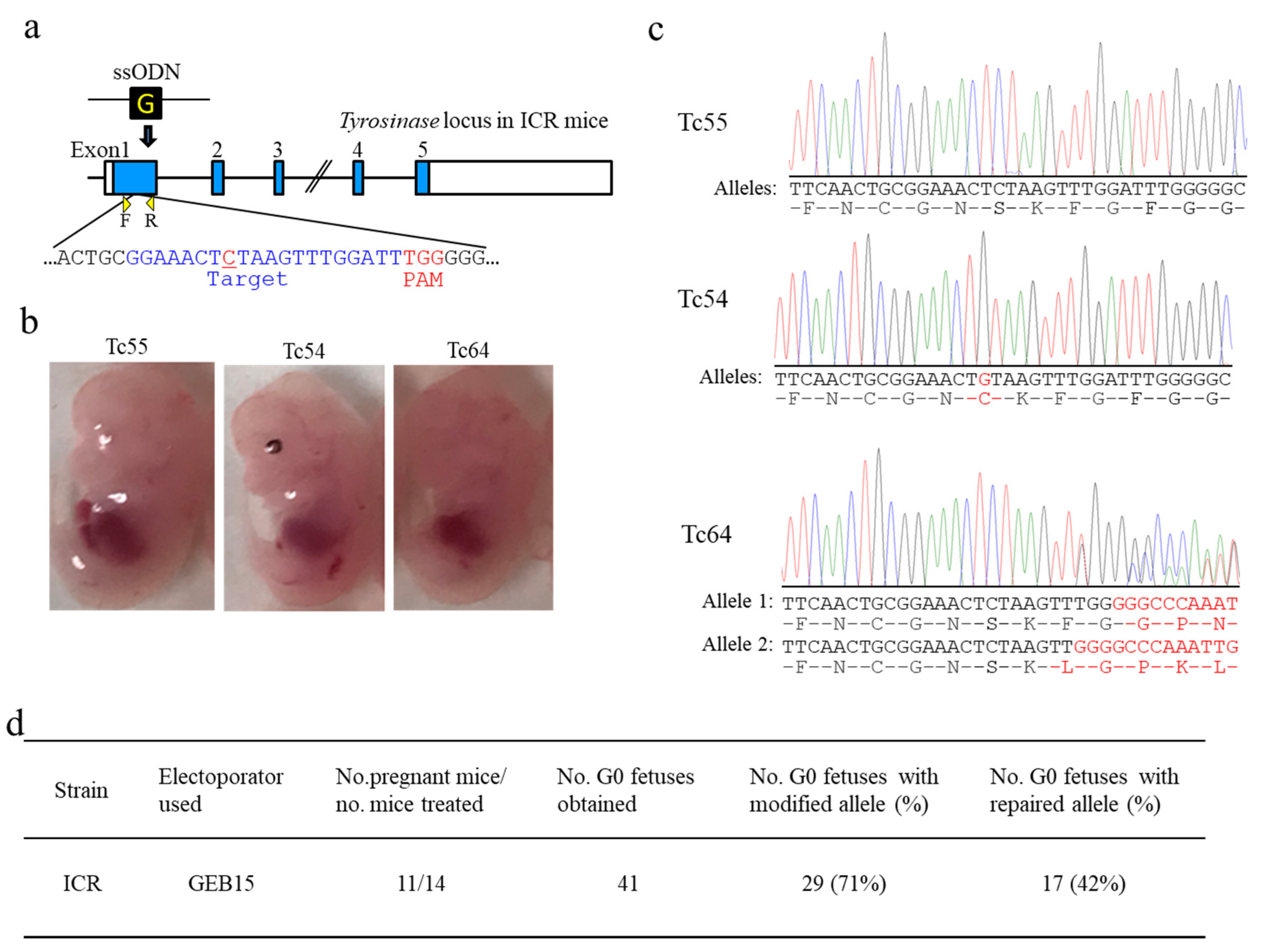
For CRISPR/Cas9-mediated KI using a single-stranded oligodeoxynucleotide (ssODN), the Tyr-ssODN (5′-TCT GTG TTT TAT AAT AGG ACC TGC CAG TGC TCA GGC AAC TTC ATG GGT TTC AAC TGC GGA AAC TGT AAG TTT GGA TTT GGG GGC CCA AAT TGT ACA GAG AAG CGA GTC TTG ATT AGA AGA AAC ATT TTT G-3′) with 64 bp (left arm) and 65 bp (right arm) homologous arms at both ends was custom-synthesized (Macrogen, Seoul, South Korea). The lyophilized ssODN was re-suspended in Opti-MEM to a concentration of 10 µg/µL. Alt-R CRISPR-Cas9 crRNAs were designed to recognize the target site (exon 1 of albino mouse
Tyr (Figure 3a and Figure 4a)) that matched a 20 bp DNA sequence (Tyr-albino crRNA: GGAAACTCTAAGTTTGGATT) just upstream of the PAM [
17]. Annealed crRNA/tracrRNA duplexes (3 μL each) were mixed with Cas9 protein (1 μL) and ssODN (2 μL) in Opti-MEM to a total of 10 μL in a 0.5 mL PCR tube, so that the final concentrations of components were 30 µM crRNA/tracrRNA duplexes, 1 μg/μL Cas9 protein, 2 µg/µL ssODN, and 0.02% Fast Green FCF.
2.5. i-GONAD Procedure
We performed
i-GONAD at 0.7 days post-coitum (dpc) (4:00 p.m.). Surgical procedures were performed on adult females anesthetized with a mixture of three anesthetic agents (medetomidine (0.75 mg/kg; Nippon Zenyaku Kogyo Co. Ltd., Fukushima, Japan), midazolam (4 mg/kg; Sandoz K.K., Tokyo, Japan), and butorphanol (5 mg/kg; Meiji Seika Pharma Co., Ltd., Tokyo, Japan)) under a dissecting microscope, on the basis of a slightly modified versions of previously reported procedures [
17,
22,
23]. The ovary/oviduct/uterus were exposed after making an incision (≈1 cm in length) in the dorsal skin and a subsequent incision (≈0.5 cm in length) in the muscle layer. The exposed ovary, oviduct, as well as part of the uterus, were placed on the back skin of mice, and adipose tissue was anchored with an Aorta-Klemme to prevent return of the exposed tissues. Approximately 1.0 μL of solution was injected into the oviduct lumen upstream of the ampulla using a micropipette (prepared using an electric puller (#PN-3; NARISHIGE Co. Ltd., Tokyo, Japan)) and an attached mouthpiece. Immediately after the injection, the oviduct regions were covered with a piece of wet KimWipe paper, soaked in DPBS, and grasped in tweezer-type electrodes (#CUY652-3; NEPA GENE Co. Ltd.). Electroporation was performed using a square-wave pulse generator: NEPA21 (NEPA GENE Co. Ltd.) or GEB15 (BEX Co. Ltd.). The electroporation parameters were as follows, based on previous papers [
22,
23]: 3 Pp (50 V/5 ms wavelength/50 ms duration/10% decay rate/± polarity) and 3 Tp (10 V/50 ms wavelength/50 ms duration/40% decay rate/± polarity) for NEPA21 and Pd; V: 40 V or 60 V, Pd on: 5.00 ms, Pd off: 50 ms, Pd N: 3/10% decay for GEB15. After EP, oviducts were returned to their original position, the incisions made in the internal dorsal muscle were sutured, and the dorsal skin was closed using a surgical stapler. The animals were then given an intradermal detoxicant, atipamezole (3.75 mg/kg; ANTISEDAN, Nippon Zenyaku Kogyo Co. Ltd., Fukushima, Japan), monitored for anesthesia recovery, and housed for further analysis.
2.6. Analysis of CRISPR/Cas9-Induced Indels
Pregnant female mice at 12.5 or 13.5 dpc were subjected to euthanasia. The fetuses were dissected out in DPBS. Pigmentation of the eyes and morphological abnormalities were assessed under a dissecting microscope and photographed. Tail biopsies were then taken for the isolation of genomic DNA from the fetuses. Genomic DNA was isolated from the tail biopsies by incubation in 100 μL of 50 mM NaOH at 95 °C for 10 min. Then, 10 μL of 1 M Tris-HCl (pH 8.0) was added to the aliquot and mixed. This crude DNA extract was used as a template for PCR.
PCR amplification of a sequence corresponding to
Tyr was performed in a volume of 20 µL containing 10 µL of 2× PCR buffer for KOD FX (TOYOBO, Osaka, Japan), 0.4 mM deoxyribonucleotides (dNTPs) mix, 1 µL of crude lysate, 0.25 µM primer pairs (Tyr–F: TCT CTG ATG GCC ATT TTC CTC/Tyr–R: AAC ATG GGT GTT GAC CCA TT) [
17], and 0.1 U KOD FX (TOYOBO, Osaka, Japan) under cycling conditions of denaturation at 94 °C for 3 min; amplification with 33 cycles of 95 °C for 20 s, 57 °C for 30 s, and 68 °C for 1 min; and extension at 68 °C for 5 min. Amplification products (5 μL) were separated by 2% agarose gel electrophoresis.
Direct sequencing was performed using the PCR products and the primer Tyr–F with a BigDye Terminator v3.1 Cycle Sequencing kit (Thermo Fisher Scientific Inc.), and then analyzed on an automated ABI PRISM 3100 DNA sequencer (Thermo Fisher Scientific Inc.).
4. Discussion
i-GONAD is now recognized as an efficient and convenient method for producing genome-edited mice such as ICR and hybrid mice. It strictly requires an electroporator, a square pulse-generating apparatus. However, it has been difficult to apply the technology to inbred strains, as exemplified by the B6 strain, which has been widely used as experimental mice [
17]. To overcome this issue, Gurumurthy et al. [
22] focused on the use of a constant current upon
i-GONAD, and found that
i-GONAD performed using an electroporator capable of generating a constant current of 100 mA was suitable for acquiring genome-edited B6 mice. Notably, this was in contrast with the case of ICR mice, which usually required 100–200 mA for their genome editing upon
i-GONAD [
22]. Unfortunately, the electroporators most widely used appear to be those capable of generating a constant voltage. To adapt these apparatuses for
i-GONAD using B6 mice, we here explored electric conditions enabling the generation of a current of 100 mA through changing the resistance (which can be achieved by changing the distance between the two electrodes when an intact oviduct is sandwiched by those electrodes). When we used two electroporators driven under a constant voltage, NEPA21 and GEB15, they both exhibited current values on the basis of Ohm’s law, although in some cases a high current (which deviated from Ohm’s law) was observed (see
Figure 1). We thus examined the resistance (R or Ω) in the case with a voltage fixed at 40 V, as the current can be easily determined through the following formula: R (Ω) = −40 V/0.1 A. Our results showed that, under resistance of 400 Ω, a current of ≈100 mA is generated. When
i-GONAD was performed using NEPA21 under the conditions in which the resistance varied from 350 to 400 Ω, genome-edited B6 and BALB/c mice were successfully obtained (see
Table 1,
Tables S2 and S4;
Figure 2 and
Figure 3). In contrast, under more stringent conditions (40 V/100–200 Ω/ ≈300 mA), which are known to be efficient for successfully producing genome-edited mice such as ICR and other hybrid mice [
17,
22], there were no fetuses from the B6 and BALB/c strains (see
Table 1,
Tables S1 and S3). From these experiments,
i-GONAD employing a 100 mA current was found to be important for these inbred strains.
Notably, it still remains a problem that the pregnancy rate of the
i-GONAD-treated B6 females appears to be low. There are several reports describing the fact that the pregnancy rate of plug-positive B6 females ranged from 35% to 85%, which clearly varied among the laboratories tested [
26,
27]. To examine whether our success in the
i-GONAD-based production of genome-edited fetuses was a result of appropriate in vivo EP or was simply related to the pregnancy rate of B6 mice themselves, we checked the pregnancy rate of intact B6 females as well as their litter size. When B6 mice were subjected to
i-GONAD, the rate of successful genome editing in fetuses was 40% and the average litter size was 5.3 (see
Table 1). On the other hand, the pregnancy rate of intact B6 females was 33% and their average litter size was 4.6 (see
Table 2), suggesting no appreciable difference between these two groups. This in turn means that the electric conditions employed here appear to be optimal for the
i-GONAD-based production of genome-edited B6 mice. In the case of BALB/c mice, the pregnancy rate of intact females was 58% and their average litter size was 8.4 (see
Table 2); however, the pregnancy rate of the
i-GONAD-treated females was 21% and their average litter size was 5.3 (see
Table 1). This means that the electric conditions employed here appear to still be suboptimal for generating genome-edited BALB/c mice using
i-GONAD. In this context, further exploration is needed with regards to the optimal conditions enabling increases of the pregnancy rate and litter size of the
i-GONAD-treated BALB/c females.
According to previous reports [
16,
17,
22], the available electroporators used for
i-GONAD were BTX T-820 (Harvard Apparatus, Inc., Holliston, MA, USA), NEPA21, and CUY21EditII. The GEB15 apparatus is a cheap edition of CUY21EditII and employs a constant voltage. Unlike NEPA21, GEB15 generates a driving pulse (corresponding to Tp) alone. Thus, GEB15 has been considered as a machine specifically for use for in vitro EP. We considered the fact that GEB15 is also useful for in vivo EP like NEPA21 if the resistance is appropriately adjusted. When plugged ICR females were subjected to
i-GONAD using GEB15, we successfully obtained genome-edited fetuses with relatively high efficiency (71%) (see
Table 1 and
Table S5;
Figure 4). Furthermore, GEB15 was found to be applicable to the production of genome-edited B6 individuals when the current was set to ≈100 mA. More surprisingly, the genome editing efficiency achieved by using GEB15 was comparable to that achieved by using NEPA21 (see
Table 1). It is thus conceivable that any type of electroporator currently or previously commercially available can be fitted to
i-GONAD if the voltage and resistance are appropriately adjusted.
It is generally difficult to perform planned breeding in B6 mice. In contrast with the case of albino mice such as ICR, which are easy to recognize as being suitable for mating through visual inspection of the vagina, it is difficult to use this approach on B6 females. Recently, Sato et al. [
25] demonstrated the usefulness of low-dose (0.02–0.2 IU) administration of PMSG, a gonadotrophic hormone widely used for inducing superovulation in experimental mice, to simultaneously prepare a number of plugged females. Unfortunately, this approach was only confined to hybrid mice such as B6C3F1, and employed hCG, a gonadotrophic hormone, to stimulate ovulation. We here examined whether this approach is applicable to B6 females to examine pregnancy indices such as the rate of plugged females, pregnancy rate of plugged females, and first litter size. In this case, we modified the protocol [
25] and employed a single administration of low-dose (1.2 IU) PMSG alone. When B6 or BALB/c females were mated with males 2 days after the administration of PMSG, the rates of plugged females in the PMSG-treated group were increased 2- or 1.5-fold (for B6 and BALB/c, respectively) compared with those in the PMSG-untreated group. This means that about 50% of females became successfully plugged after PMSG treatment. The pregnancy rate of plugged females slightly increased in both strains after PMSG treatment (see
Table 2). The first litter size increased in B6 females but decreased slightly in BALB/c females after PMSG treatment (see
Table 2). These findings support the usefulness of low-dose PMSG as a convenient way of achieving planned breeding. When this approach was combined with
i-GONAD, the pregnancy rates of B6 and BALB/c females increased 1.6- and 1.7-fold, respectively (see
Table 1). Thus, it is concluded that the administration of low-dose PMSG is beneficial for the
i-GONAD-based production of genome-edited mice.
5. Conclusions
In this study, we explored optimal electric conditions that allow the
i-GONAD-based production of the genome-edited B6 strain, as well as other inbred strains such as BALB/c when a universal electroporator (driven under a constant voltage) such as NEAP21 and GEB15 is employed. We found that zygotes from those inbred strains could be successfully genome-edited without overt developmental damage when
i-GONAD was performed under the electric conditions of 40 V/350–400 Ω/100 mA. Furthermore, we found that low-dose PMSG was effective for simultaneously acquiring a number of plugged females. Consequently, the pregnancy rates of B6 and BALB/c mice after
i-GONAD treatment increased 1.6- and 1.7-fold, respectively, compared with those of PMSG-untreated females (see
Table 1). This pretreatment before mating is thus beneficial to increase the production rate of genome-edited mice.
Notably, we succeeded in producing genome-edited rats (including Sprague-Dawley (SD), Lewis, Brown Norway (BN), and SDBNF1 strains) using
i-GONAD [
20]. Unfortunately, the BN strain was insensitive to this treatment. Given that the BN strain is recognized as one of the standard strains for the Rat Genome Sequencing Consortium (RGSC), the development of a method to generate gene-engineered BN rats is highly desirable [
28]. Our previous
i-GONAD method relied on a constant voltage. If each electric parameter (Ω and mA) is tested under a fixed voltage of 40 V, it may be possible to establish genome-edited BN rats through
i-GONAD. Our present approach based on checking each electric parameter in pursuit of the optimal
i-GONAD can also be applied to other inbred strains, as well as wild-derived mice, for which
i-GONAD has not been considered applicable.