Uncovering the Translational Regulatory Activity of the Tumor Suppressor BRCA1
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Case Selection
2.3. Tissue Microarray (TMA) and Immunohistochemistry (IHC)
2.4. Cell Culture
2.5. Transfection with Plasmids
2.6. RNA Interference (RNAi)
2.7. Immunoblotting
2.8. Isolation of Polysomes and Total Cytoplasmic RNA
2.9. RNA-Binding Protein Immunoprecipitation (RIP)
2.10. Microarray Analysis
2.11. Quantitative RT-PCR
2.11.1. RIP Analysis
2.11.2. Polysome Profile Analysis
2.12. Quantification of A-to-I Editing
2.13. Statistical Analysis
3. Results
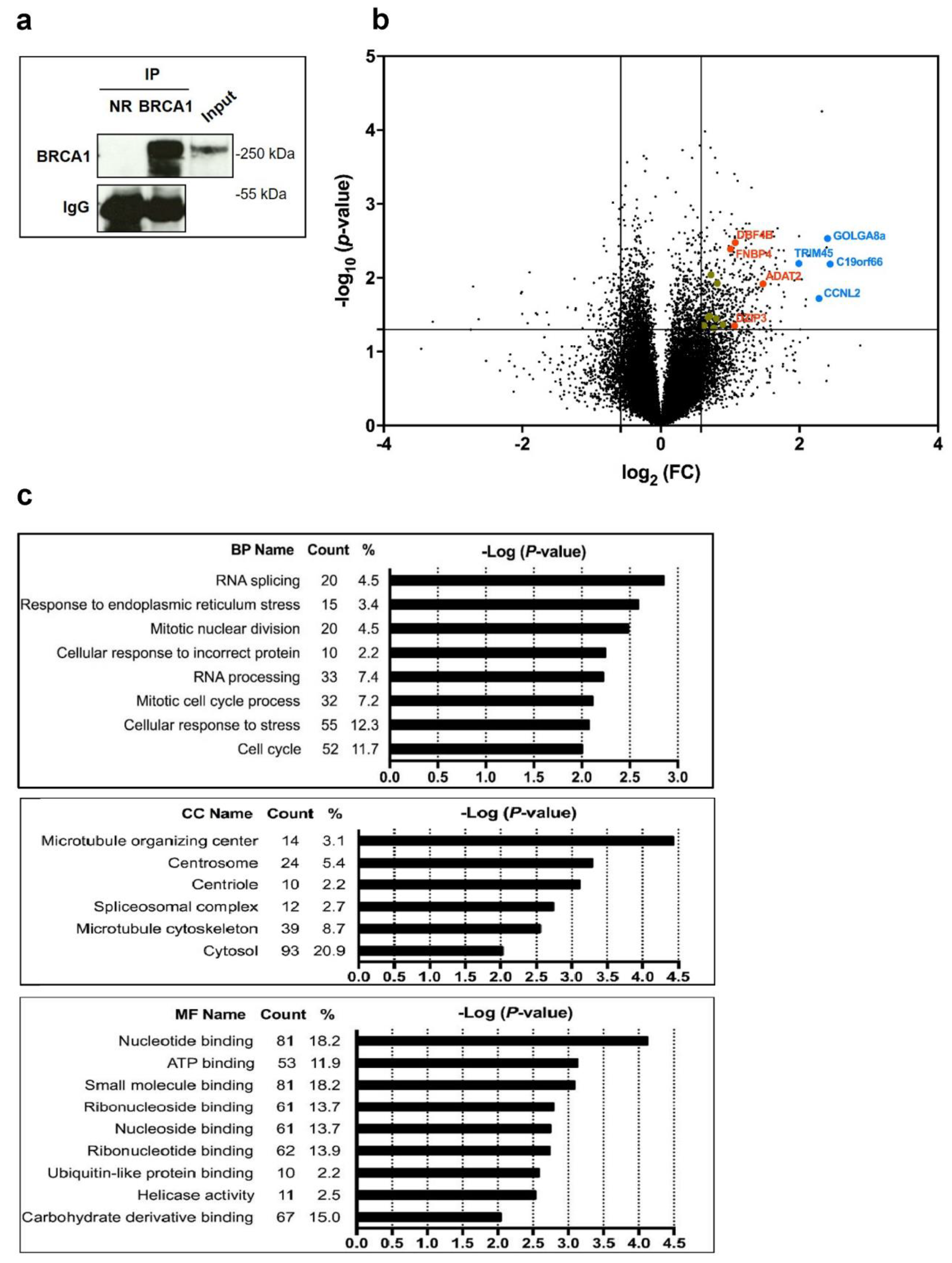
3.1. BRCA1 Associates with mRNAs
3.2. BRCA1 Controls Translation of a Subset of BRCA1-Associated mRNAs
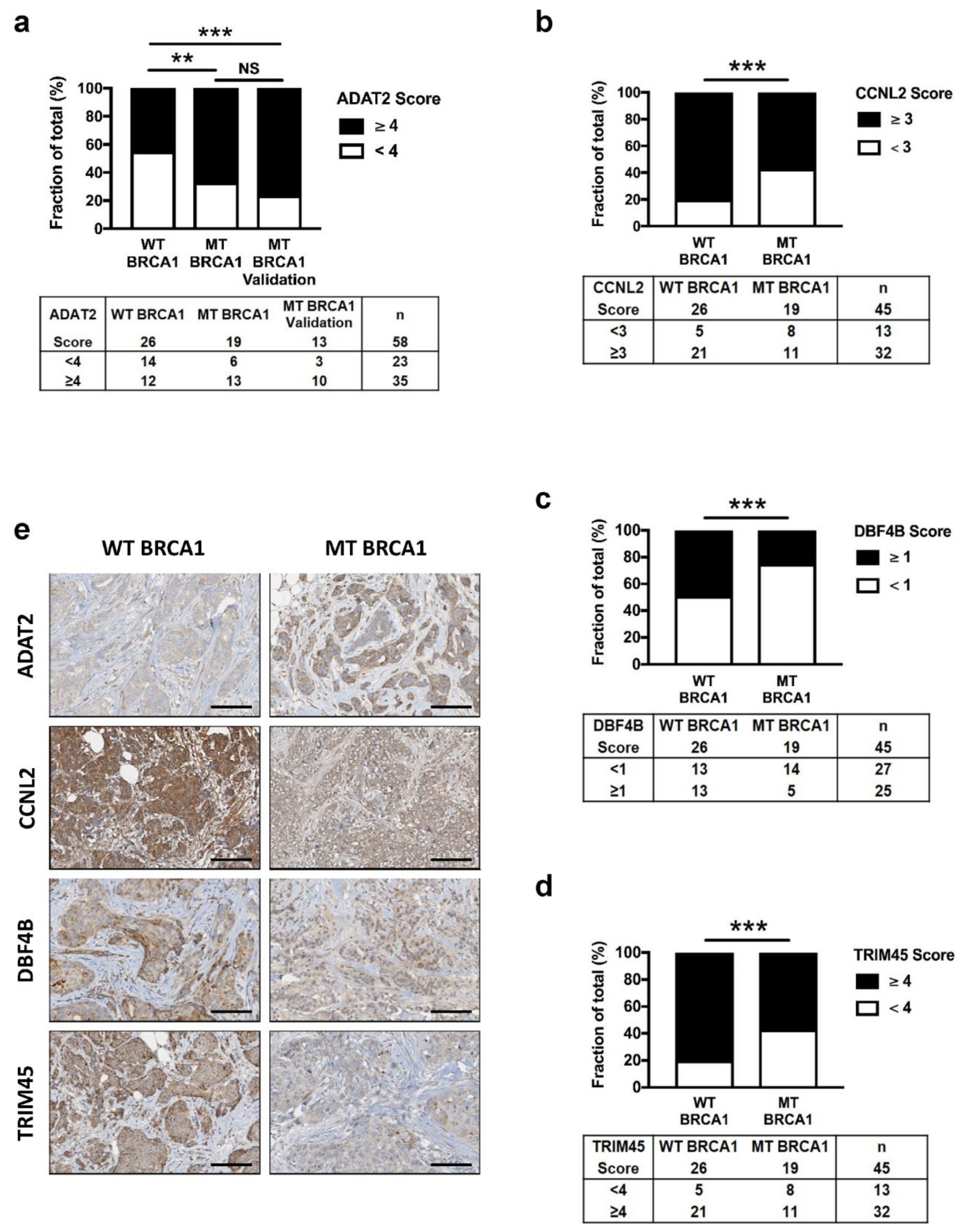
3.3. BRCA1 Controls ADAT2, CCNL2, DBF4B, and TRIM45 Proteins in Mammary Epithelial Cell Lines
3.4. Altered Expression of ADAT2, CCNL2, DBF4B, and TRIM45 Proteins in BRCA1 Deficient Human Breast Cancers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Malone, K.E.; Daling, J.R.; Doody, D.R.; Hsu, L.; Bernstein, L.; Coates, R.J.; Marchbanks, P.A.; Simon, M.S.; McDonald, J.A.; Norman, S.A.; et al. Prevalence and predictors of BRCA1 and BRCA2 mutations in a population-based study of breast cancer in white and black American women ages 35 to 64 years. Cancer Res. 2006, 66, 8297–8308. [Google Scholar] [CrossRef] [PubMed]
- Kurian, A.W.; Gong, G.D.; John, E.M.; Miron, A.; Felberg, A.; Phipps, A.I.; West, D.W.; Whittemore, A.S. Performance of prediction models for BRCA mutation carriage in three racial/ethnic groups: Findings from the Northern California Breast Cancer Family Registry. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1084–1091. [Google Scholar] [CrossRef]
- Armstrong, N.; Ryder, S.; Forbes, C.; Ross, J.; Quek, R.G. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin. Epidemiol. 2019, 11, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.A.; Mooij, T.M.; Roos-Blom, M.J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef] [PubMed]
- Engel, C.; Fischer, C.; Zachariae, S.; Bucksch, K.; Rhiem, K.; Giesecke, J.; Herold, N.; Wappenschmidt, B.; Hubbel, V.; Maringa, M.; et al. Breast cancer risk in BRCA1/2 mutation carriers and noncarriers under prospective intensified surveillance. Int. J. Cancer 2019. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef]
- Staff, S.; Isola, J.; Tanner, M. Haplo-insufficiency of BRCA1 in sporadic breast cancer. Cancer Res. 2003, 63, 4978–4983. [Google Scholar]
- Atchley, D.P.; Albarracin, C.T.; Lopez, A.; Valero, V.; Amos, C.I.; Gonzalez-Angulo, A.M.; Hortobagyi, G.N.; Arun, B.K. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J. Clin. Oncol. 2008, 26, 4282–4288. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef]
- Venkitaraman, A.R. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 2014, 343, 1470–1475. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.P.; Livingston, D.M. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2012, 2, 679–684. [Google Scholar] [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Bade, S.; Le Guillou, M.; Burke, K.; Reed, R.; Bowman-Colin, C.; Su, Y.; Ting, D.T.; Polyak, K.; Richardson, A.L.; et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat. Commun. 2014, 5, 5496. [Google Scholar] [CrossRef]
- Zhang, X.; Chiang, H.C.; Wang, Y.; Zhang, C.; Smith, S.; Zhao, X.; Nair, S.J.; Michalek, J.; Jatoi, I.; Lautner, M.; et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 2017, 8, 15908. [Google Scholar] [CrossRef]
- Savage, K.I.; Harkin, D.P. BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. Febs J. 2015, 282, 630–646. [Google Scholar] [CrossRef] [PubMed]
- Dizin, E.; Gressier, C.; Magnard, C.; Ray, H.; Decimo, D.; Ohlmann, T.; Dalla Venezia, N. BRCA1 interacts with poly(A)-binding protein: Implication of BRCA1 in translation regulation. J. Biol. Chem. 2006, 281, 24236–24246. [Google Scholar] [CrossRef] [PubMed]
- Dacheux, E.; Vincent, A.; Nazaret, N.; Combet, C.; Wierinckx, A.; Mazoyer, S.; Diaz, J.J.; Lachuer, J.; Venezia, N.D. BRCA1-Dependent Translational Regulation in Breast Cancer Cells. PLoS ONE 2013, 8, e67313. [Google Scholar] [CrossRef] [PubMed]
- Eisinger, F.; Bressac, B.; Castaigne, D.; Cottu, P.H.; Lansac, J.; Lefranc, J.P.; Lesur, A.; Nogues, C.; Pierret, J.; Puy-Pernias, S.; et al. Identification and management of hereditary predisposition to cancer of the breast and the ovary (update 2004). Bull. Du Cancer 2004, 91, 219–237. [Google Scholar]
- Vincent, A.; Berthel, E.; Dacheux, E.; Magnard, C.; Venezia, N.L. BRCA1 affects protein phosphatase 6 signalling through its interaction with ANKRD28. Biochem. J. 2016, 473, 949–960. [Google Scholar] [CrossRef]
- Keene, J.D.; Komisarow, J.M.; Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006, 1, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Pineyro, D.; Rodriguez-Escriba, M.; Camacho, N.; Reina, O.; Saint-Leger, A.; Filonava, L.; Batlle, E.; Ribas de Pouplana, L. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015, 43, 5145–5157. [Google Scholar] [CrossRef] [PubMed]
- Wulff, T.F.; Arguello, R.J.; Molina Jordan, M.; Roura Frigole, H.; Hauquier, G.; Filonava, L.; Camacho, N.; Gatti, E.; Pierre, P.; Ribas de Pouplana, L.; et al. Detection of a Subset of Posttranscriptional Transfer RNA Modifications in Vivo with a Restriction Fragment Length Polymorphism-Based Method. Biochemistry 2017, 56, 4029–4038. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Amano, A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J. Cell Biol. 2012, 197, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.I.; Gorski, J.J.; Barros, E.M.; Irwin, G.W.; Manti, L.; Powell, A.J.; Pellagatti, A.; Lukashchuk, N.; McCance, D.J.; McCluggage, W.G.; et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell 2014, 54, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Khurana, S.; Kubben, N.; Abdelmohsen, K.; Oberdoerffer, P.; Gorospe, M.; Misteli, T. A BRCA1-interacting lncRNA regulates homologous recombination. Embo Rep. 2015, 16, 1520–1534. [Google Scholar] [CrossRef]
- Baltz, A.G.; Munschauer, M.; Schwanhausser, B.; Vasile, A.; Murakawa, Y.; Schueler, M.; Youngs, N.; Penfold-Brown, D.; Drew, K.; Milek, M.; et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 2012, 46, 674–690. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Yang, L.; Li, N.; Wang, C.; Yu, Y.; Yuan, L.; Zhang, M.; Cao, X. Cyclin L2, a novel RNA polymerase II-associated cyclin, is involved in pre-mRNA splicing and induces apoptosis of human hepatocellular carcinoma cells. J. Biol. Chem. 2004, 279, 11639–11648. [Google Scholar] [CrossRef] [PubMed]
- Loyer, P.; Trembley, J.H.; Grenet, J.A.; Busson, A.; Corlu, A.; Zhao, W.; Kocak, M.; Kidd, V.J.; Lahti, J.M. Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: Influence of cyclin L isoforms on splice site selection. J. Biol. Chem. 2008, 283, 7721–7732. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Huang, D.Z.; Deng, T.; Zhou, L.K.; Wang, X.; Bai, M.; Ba, Y. Overexpression of cyclin L2 inhibits growth and enhances chemosensitivity in human gastric cancer cells. Asian Pac. J. Cancer Prev. 2012, 13, 1425–1430. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, H.; Tao, Y.; Chen, M.; Yu, J.; Li, W.J.; Tao, L.; Li, Y.; Li, F. Upregulation of MicroRNA-214 Contributes to the Development of Vascular Remodeling in Hypoxia-induced Pulmonary Hypertension Via Targeting CCNL2. Sci. Rep. 2016, 6, 24661. [Google Scholar] [CrossRef] [PubMed]
- Kren, B.T.; Unger, G.M.; Abedin, M.J.; Vogel, R.I.; Henzler, C.M.; Ahmed, K.; Trembley, J.H. Preclinical evaluation of cyclin dependent kinase 11 and casein kinase 2 survival kinases as RNA interference targets for triple negative breast cancer therapy. Breast Cancer Res. 2015, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa-Sugata, N.; Ishii, A.; Taniyama, C.; Matsui, E.; Arai, K.; Masai, H. A second human Dbf4/ASK-related protein, Drf1/ASKL1, is required for efficient progression of S and M phases. J. Biol. Chem. 2005, 280, 13062–13070. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.S.; Walter, J.C. Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 2005, 19, 2295–2300. [Google Scholar] [CrossRef]
- Collart, C.; Smith, J.C.; Zegerman, P. Chk1 Inhibition of the Replication Factor Drf1 Guarantees Cell-Cycle Elongation at the Xenopus laevis Mid-blastula Transition. Dev. Cell 2017, 42, 82–96.e83. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Qi, X.; Yuan, W.; Ai, J.; Zhu, C.; Cao, L.; Yang, H.; Liu, F.; Wu, X.; et al. TRIM45, a novel human RBCC/TRIM protein, inhibits transcriptional activities of ElK-1 and AP-1. Biochem. Biophys. Res. Commun. 2004, 323, 9–16. [Google Scholar] [CrossRef]
- Shibata, M.; Sato, T.; Nukiwa, R.; Ariga, T.; Hatakeyama, S. TRIM45 negatively regulates NF-kappaB-mediated transcription and suppresses cell proliferation. Biochem. Biophys. Res. Commun. 2012, 423, 104–109. [Google Scholar] [CrossRef]
- Huang, C.C.; Tu, S.H.; Lien, H.H.; Jeng, J.Y.; Huang, C.S.; Huang, C.J.; Lai, L.C.; Chuang, E.Y. Concurrent gene signatures for han chinese breast cancers. PLoS ONE 2013, 8, e76421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, C.; Cui, J.; Ou, J.; Han, J.; Qin, Y.; Zhi, F.; Wang, R.F. TRIM45 functions as a tumor suppressor in the brain via its E3 ligase activity by stabilizing p53 through K63-linked ubiquitination. Cell Death Dis. 2017, 8, e2831. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Keller, W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 1999, 286, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- Novoa, E.M.; Pavon-Eternod, M.; Pan, T.; Ribas de Pouplana, L. A role for tRNA modifications in genome structure and codon usage. Cell 2012, 149, 202–213. [Google Scholar] [CrossRef]
- Torres, A.G.; Pineyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I editing on tRNAs: Biochemical, biological and evolutionary implications. Febs Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef]
- Rafels-Ybern, A.; Torres, A.G.; Grau-Bove, X.; Ruiz-Trillo, I.; Ribas de Pouplana, L. Codon adaptation to tRNAs with Inosine modification at position 34 is widespread among Eukaryotes and present in two Bacterial phyla. Rna Biol. 2017, 1–8. [Google Scholar] [CrossRef]
- Pavon-Eternod, M.; Gomes, S.; Geslain, R.; Dai, Q.; Rosner, M.R.; Pan, T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009, 37, 7268–7280. [Google Scholar] [CrossRef]
- Goodarzi, H.; Nguyen, H.C.B.; Zhang, S.; Dill, B.D.; Molina, H.; Tavazoie, S.F. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 2016, 165, 1416–1427. [Google Scholar] [CrossRef]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Goncalves, A.; Lee, K.H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Popova, T.; Manie, E.; Rieunier, G.; Caux-Moncoutier, V.; Tirapo, C.; Dubois, T.; Delattre, O.; Sigal-Zafrani, B.; Bollet, M.; Longy, M.; et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012, 72, 5454–5462. [Google Scholar] [CrossRef] [PubMed]
- Katz, S.J.; Ward, K.C.; Hamilton, A.S.; McLeod, M.C.; Wallner, L.P.; Morrow, M.; Jagsi, R.; Hawley, S.T.; Kurian, A.W. Gaps in Receipt of Clinically Indicated Genetic Counseling After Diagnosis of Breast Cancer. J. Clin. Oncol. 2018, 36, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Fernandez, J.A.; McArt, D.G.; Boyle, D.P.; Li, G.; Loughrey, M.B.; Irwin, G.W.; Harkin, D.P.; James, J.A.; McQuaid, S.; et al. Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer. Lab. Invest. 2018, 98, 15–26. [Google Scholar] [CrossRef]
- Vorrius, T.R.; Snyder, K.; Pica-Mendez, A.; Tan, C.; Laterza, O.; Toniatti, C.; Carpenter, C.; Lee, H.; Tanaka, W.; Zhang, Z.Q. Immunohistochemical Detection of BRCA-1 and BRCA-2 Expression in Human Breast and Ovarian Tumors. J. Histotechnol. 2009, 32, 202–203. [Google Scholar] [CrossRef]
- Perez-Valles, A.; Martorell-Cebollada, M.; Nogueira-Vazquez, E.; Garcia-Garcia, J.A.; Fuster-Diana, E. The usefulness of antibodies to the BRCA1 protein in detecting the mutated BRCA1 gene. An immunohistochemical study. J. Clin. Pathol. 2001, 54, 476–480. [Google Scholar] [CrossRef]
- Milner, R.; Wombwell, H.; Eckersley, S.; Barnes, D.; Warwicker, J.; Van Dorp, E.; Rowlinson, R.; Dearden, S.; Hughes, G.; Harbron, C.; et al. Validation of the BRCA1 antibody MS110 and the utility of BRCA1 as a patient selection biomarker in immunohistochemical analysis of breast and ovarian tumours. Virchows Arch. 2013, 462, 269–279. [Google Scholar] [CrossRef]
- Mangia, A.; Chiriatti, A.; Tommasi, S.; Menolascina, F.; Petroni, S.; Zito, F.A.; Simone, G.; Schittulli, F.; Paradiso, A. BRCA1 expression and molecular alterations in familial breast cancer. Histol. Histopathol. 2009, 24, 69–76. [Google Scholar] [CrossRef]
- Johnston, R.; D’Costa, Z.; Ray, S.; Gorski, J.; Harkin, D.P.; Mullan, P.; Panov, K.I. The identification of a novel role for BRCA1 in regulating RNA polymerase I transcription. Oncotarget 2016, 7, 68097–68110. [Google Scholar] [CrossRef]
- Veras, I.; Rosen, E.M.; Schramm, L. Inhibition of RNA polymerase III transcription by BRCA1. J. Mol. Biol. 2009, 387, 523–531. [Google Scholar] [CrossRef]
- Marcel, V.; Catez, F.; Diaz, J.J. p53, a translational regulator: Contribution to its tumour-suppressor activity. Oncogene 2015, 34, 5513–5523. [Google Scholar] [CrossRef] [PubMed]
- Marcel, V.; Nguyen Van Long, F.; Diaz, J.J. 40 Years of Research Put p53 in Translation. Cancers 2018, 10, 152. [Google Scholar] [CrossRef] [PubMed]



| Exploratory Cohort | Validation Cohort | ||
|---|---|---|---|
| Characteristics | WT BRCA1 | MT BRCA1 | MT BRCA1 |
| n = 26 | n = 19 | n = 13 | |
| Tumor Type (%) | |||
| Invasive ductal carcinoma | 26 (100) | 19 (100) | 13 (100) |
| SBR Grade (%) | |||
| 1 | 0 (0) | 0 (0) | 0 (0) |
| 2 | 8 (31) | 0 (0) | 2 (15) |
| 3 | 18 (69) | 19 (100) | 11 (85) |
| Subtype (%) | |||
| HR negative HER2 negative | 17 (65) | 13 (68) | 10 (77) |
| HR positive HER2 negative | 9 (35) | 6 (32) | 3 (23) |
| Age (year) | |||
| Median (min-max) | 43 (28–65) | 38 (23–61) | 43 (36–65) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berthel, E.; Vincent, A.; Eberst, L.; Torres, A.G.; Dacheux, E.; Rey, C.; Marcel, V.; Paraqindes, H.; Lachuer, J.; Catez, F.; et al. Uncovering the Translational Regulatory Activity of the Tumor Suppressor BRCA1. Cells 2020, 9, 941. https://doi.org/10.3390/cells9040941
Berthel E, Vincent A, Eberst L, Torres AG, Dacheux E, Rey C, Marcel V, Paraqindes H, Lachuer J, Catez F, et al. Uncovering the Translational Regulatory Activity of the Tumor Suppressor BRCA1. Cells. 2020; 9(4):941. https://doi.org/10.3390/cells9040941
Chicago/Turabian StyleBerthel, Elise, Anne Vincent, Lauriane Eberst, Adrian Gabriel Torres, Estelle Dacheux, Catherine Rey, Virginie Marcel, Hermes Paraqindes, Joël Lachuer, Frédéric Catez, and et al. 2020. "Uncovering the Translational Regulatory Activity of the Tumor Suppressor BRCA1" Cells 9, no. 4: 941. https://doi.org/10.3390/cells9040941
APA StyleBerthel, E., Vincent, A., Eberst, L., Torres, A. G., Dacheux, E., Rey, C., Marcel, V., Paraqindes, H., Lachuer, J., Catez, F., de Pouplana, L. R., Treilleux, I., Diaz, J.-J., & Dalla Venezia, N. (2020). Uncovering the Translational Regulatory Activity of the Tumor Suppressor BRCA1. Cells, 9(4), 941. https://doi.org/10.3390/cells9040941









