Astrocyte-Derived Small Extracellular Vesicles Regulate Dendritic Complexity through miR-26a-5p Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Procedures
2.2. Plasmids
2.3. Primary Antibodies and Dilutions
2.4. Secondary Antibodies and Dilutions
2.5. Immunofluorescence (IF)
2.6. Western Blot
2.7. RNA Extraction
2.8. Reverse Transcription Quantitative PCR
2.9. In Utero Electroporation
2.10. Cell Cultures and Isolation of sEVs
2.11. Transwell Astrocyte-Neuron Co-Culture
2.12. Nanoparticle Tracking Analysis (NTA)
2.13. Sucrose Flotation Assay
2.14. Incubation with sEVs
2.15. Neuronal Magnetofection
2.16. Morphological Analysis
2.17. Bioinformatic Analysis
2.18. Statistical Analysis
3. Results
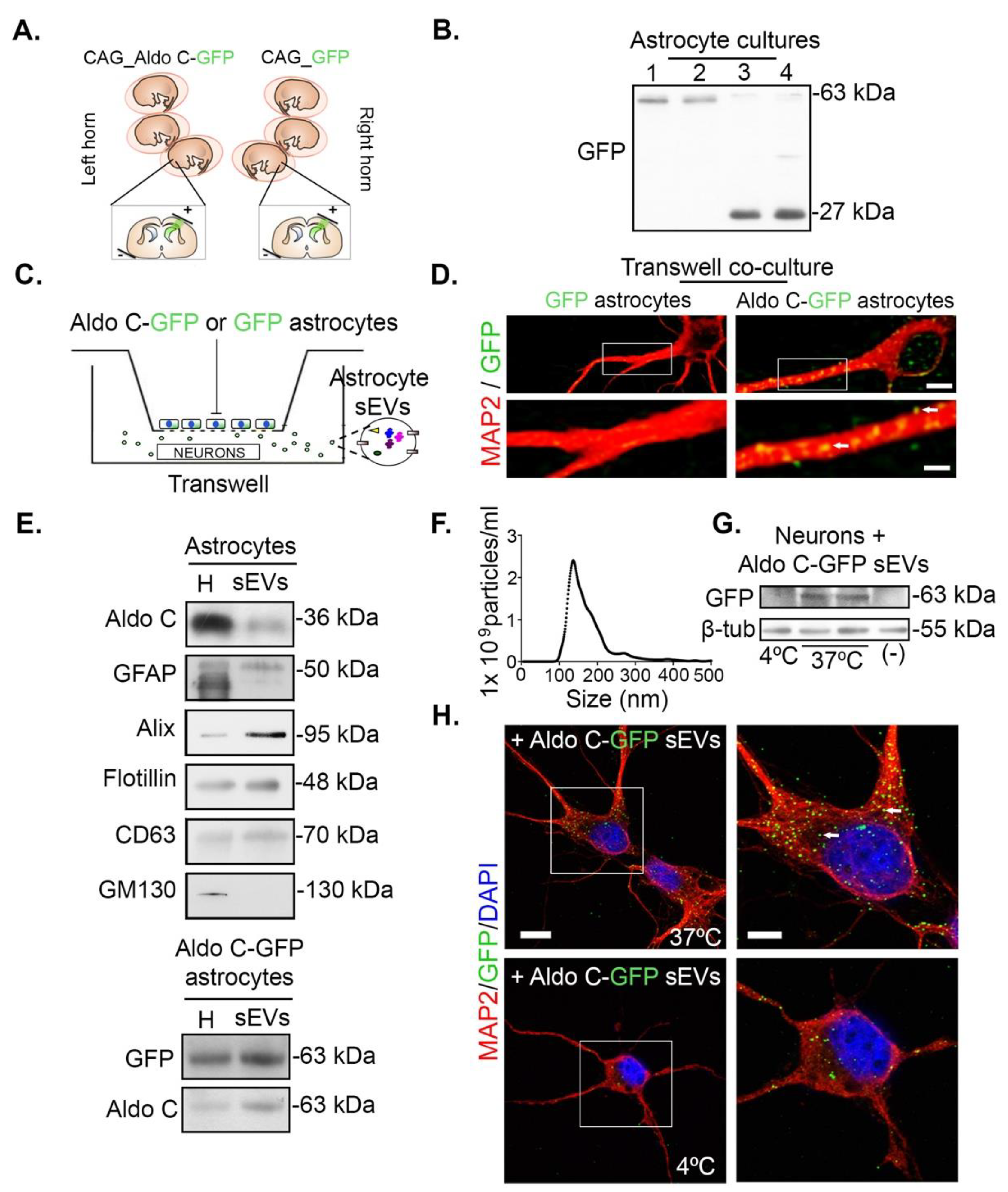
3.1. Astrocytes Transfer Aldolase C-GFP-Containing sEVs to Neurons
3.2. Astrocyte-Derived sEVs Decrease Dendritic Complexity in Neurons
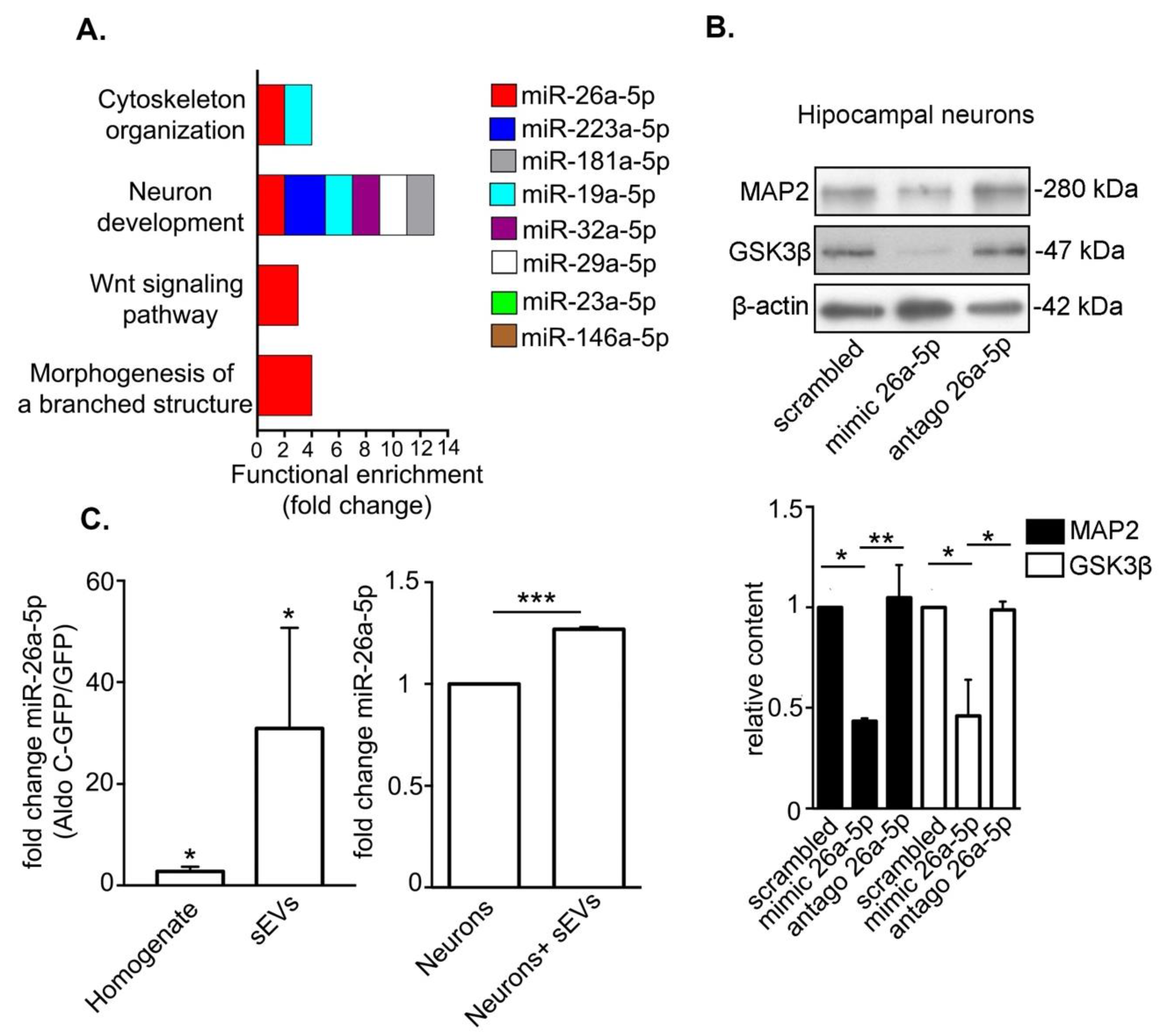
3.3. Astrocyte-Derived sEVs Carry miR-26-5p, Which Targets Gene Expression Associated to Neuronal Development and Morphology and Regulates Protein Expression in Neurons
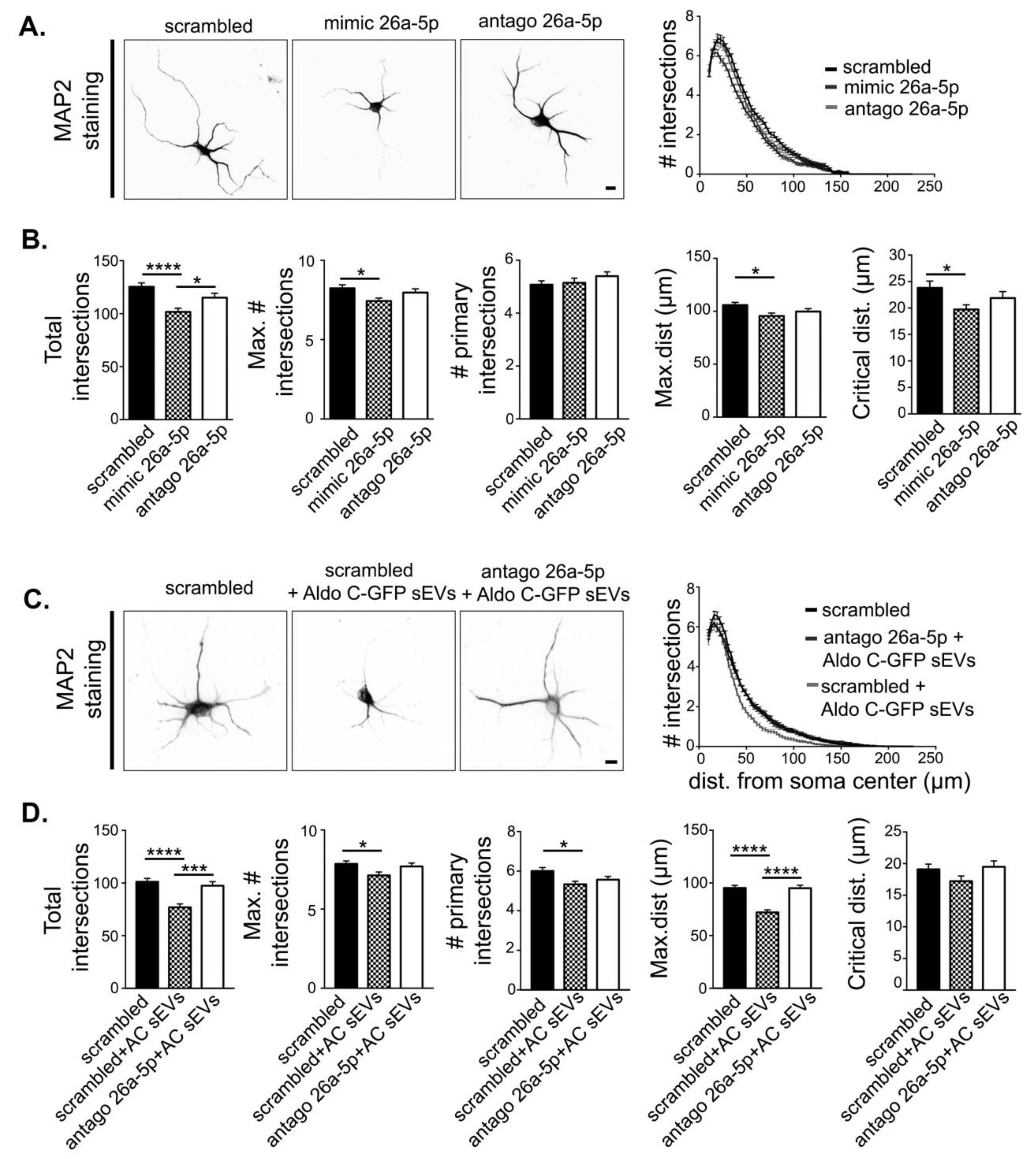
3.4. miR-26a-5p Mediates Aldo C-GFP sEVs-Induced Decrease of Dendritic Complexity
4. Discussion
4.1. Astrocyte-Derived sEVs Contain Regulated miRNAs: A Potential New Role for Aldo C
4.2. A Role for sEVs in Transferring Glial miRNAs to Neurons and Their Impact on Neuronal Morphology
4.3. Astrocyte-Derived sEVs in Neurological Conditions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shigyo, M.; Tohda, C. Extracellular vimentin is a novel axonal growth facilitator for functional recovery in spinal cord-injured mice. Sci. Rep. 2016, 6, 28293. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, P.; Reh, T. Regional differences in glial-derived factors that promote dendritic outgrowth from mouse cortical neurons in vitro. J. Neurosci. 1994, 14, 4639–4655. [Google Scholar] [CrossRef] [PubMed]
- Rousselet, A.; Autillo-Touati, A.; Araud, D.; Prochiantz, A. In vitro regulation of neuronal morphogenesis and polarity by astrocyte-derived factors. Dev. Boil. 1990, 137, 33–45. [Google Scholar] [CrossRef]
- Zhu, Y.-B.; Gao, W.; Zhang, Y.; Jia, F.; Zhang, H.-L.; Liu, Y.-Z.; Sun, X.-F.; Yin, Y.; Yin, D. Astrocyte-derived phosphatidic acid promotes dendritic branching. Sci. Rep. 2016, 6, 21096. [Google Scholar] [CrossRef] [PubMed]
- Mauch, D.H. CNS Synaptogenesis Promoted by Glia-Derived Cholesterol. Science 2001, 294, 1354–1357. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Stogsdill, J.; Pulimood, N.S.; Dingsdale, H.; Kim, Y.H.; Pilaz, L.-J.; Kim, I.H.; Manhães, A.C.; Rodrigues, W.S.; Pamukcu, A.; et al. Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1α and NL1 via Hevin. Cell 2016, 164, 183–196. [Google Scholar] [CrossRef]
- Guitart, K.; Loers, G.; Buck, F.; Bork, U.; Schachner, M.; Kleene, R. Improvement of neuronal cell survival by astrocyte-derived exosomes under hypoxic and ischemic conditions depends on prion protein. Glia 2016, 64, 896–910. [Google Scholar] [CrossRef]
- Chaudhuri, A.D.; Dastgheyb, R.M.; Yoo, S.W.; Trout, A.; Talbot, C.C., Jr.; Hao, H.; Witwer, K.W.; Haughey, N.J. TNFα and IL-1β modify the miRNA cargo of astrocyte shed extracellular vesicles to regulate neurotrophic signaling in neurons article. Cell Death Dis. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Rajendran, L.; Bali, J.; Barr, M.M.; Court, F.A.; Krämer-Albers, E.-M.; Picou, F.; Raposo, G.; Van Der Vos, K.E.; Van Niel, G.; Wang, J.; et al. Emerging roles of extracellular vesicles in the nervous system. J. Neurosci. 2014, 34, 15482–15489. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Boil. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, N.B.; Sullivan, M.L.G.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.; Zhang, Q.; Huang, W.-C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Pérez-Hernandez, D.; Vázquez, J.; Martín-Cófreces, N.B.; Martínez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef]
- Janas, T.; Janas, M.M.; Sapoń, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Gutierrez-Vazquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Boil. 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Sandoval, M.; Luarte, A.; Herrera-Molina, R.; Varas-Godoy, M.; Santibañez, M.; Rubio, F.J.; Smit, A.B.; Gundelfinger, E.D.; Li, K.-W.; Smalla, K.-H.; et al. The glycolytic enzyme aldolase C is up-regulated in rat forebrain microsomes and in the cerebrospinal fluid after repetitive fluoxetine treatment. Brain Res. 2013, 1520, 1–14. [Google Scholar] [CrossRef]
- Ampuero, E.; Luarte, A.; Santibañez, M.; Varas-Godoy, M.; Toledo, J.; Diaz-Veliz, G.; Cavada, G.; Rubio, F.J.; Wyneken, U. Two Chronic Stress Models Based on Movement Restriction in Rats Respond Selectively to Antidepressant Drugs: Aldolase C As a Potential Biomarker. Int. J. Neuropsychopharmacol. 2015, 18, 038. [Google Scholar] [CrossRef]
- Gómez-Molina, C.; Sandoval, M.; Henzi, R.; Ramírez, J.P.; Varas-Godoy, M.; Luarte, A.; Lafourcade, C.A.; Lopez-Verrilli, A.; Smalla, K.-H.; Kaehne, T.; et al. Small Extracellular Vesicles in Rat Serum Contain Astrocyte-Derived Protein Biomarkers of Repetitive Stress. Int. J. Neuropsychopharmacol. 2019, 22, 232–246. [Google Scholar] [CrossRef]
- Chen, F.; LoTurco, J.J. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods 2012, 207, 172–180. [Google Scholar] [CrossRef]
- Hitoshi, N.; Ken-Ichi, Y.; Jun-Ichi, M. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108, 193–199. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 5, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rosen, G.D.; Bai, J.; Wang, Y.; Fiondella, C.G.; Threlkeld, S.W.; LoTurco, J.J.; Galaburda, A.M. Disruption of neuronal migration by RNAi of Dyx1c1 results in neocortical and hippocampal malformations. Cereb. Cortex 2007, 17, 2562–2572. [Google Scholar] [CrossRef] [PubMed]
- Kaech, S.; Banker, G. Culturing hippocampal neurons. Nat. Protoc. 2006, 1, 2406–2415. [Google Scholar] [CrossRef]
- Ramírez, G.; Toro, R.; Döbeli, H.; Von Bernhardi, R. Protection of rat primary hippocampal cultures from Aβ cytotoxicity by pro-inflammatory molecules is mediated by astrocytes. Neurobiol. Dis. 2005, 19, 243–254. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Boil. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Pribiag, H.; Peng, H.; Shah, W.A.; Stellwagen, D.; Carbonetto, S. Dystroglycan mediates homeostatic synaptic plasticity at GABAergic synapses. Proc. Natl. Acad. Sci. USA 2014, 111, 6810–6815. [Google Scholar] [CrossRef]
- Sholl, D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953, 87, 387–406. [Google Scholar]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA–target interactions. Nucleic Acids Res. 2008, 37, D105–D110. [Google Scholar] [CrossRef]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. AmiGO: Online access to ontology and annotation data. Bioinformatic 2008, 25, 288–289. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Jovičić, A.; Gitler, A.D. Distinct repertoires of microRNAs present in mouse astrocytes compared to astrocyte-secreted exosomes. PLoS ONE 2017, 12, e0171418. [Google Scholar] [CrossRef]
- Jovičić, A.; Roshan, R.; Moisoi, N.; Pradervand, S.; Moser, R.; Pillai, B.; Luthi-Carter, R. Comprehensive Expression Analyses of Neural Cell-Type-Specific miRNAs Identify New Determinants of the Specification and Maintenance of Neuronal Phenotypes. J. Neurosci. 2013, 33, 5127–5137. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, L.; Gräfe, A.; Seiler, A.; Schumacher, S.; Nitsch, R.; Wulczyn, F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005, 21, 1469–1477. [Google Scholar] [CrossRef]
- Pan, Z.; Shan, Q.; Gu, P.; Wang, X.M.; Tai, L.W.; Sun, M.; Luo, X.; Sun, L.; Cheung, C.-W. miRNA-23a/CXCR4 regulates neuropathic pain via directly targeting TXNIP/NLRP3 inflammasome axis. J. Neuroinflammation 2018, 15, 29. [Google Scholar] [CrossRef]
- Gioia, U.; Di Carlo, V.; Caramanica, P.; Toselli, C.; Cinquino, A.; Marchioni, M.; Laneve, P.; Biagioni, S.; Bozzoni, I.; Cacci, E.; et al. Mir-23a and mir-125b regulate neural stem/progenitor cell proliferation by targeting Musashi1. RNA Boil. 2014, 11, 1105–1112. [Google Scholar] [CrossRef][Green Version]
- Shin, J.H.; Park, Y.M.; Kim, D.H.; Moon, G.J.; Bang, O.Y.; Ohn, T.; Kim, H.H. Ischemic brain extract increases SDF-1 expression in astrocytes through the CXCR2/miR-223/miR-27b pathway. Biochim. Biophys. Acta Gene Regul. Mech. 2014, 1839, 826–836. [Google Scholar] [CrossRef]
- Howng, S.-Y.B.; Huang, Y.; Ptáček, L.; Fu, Y.-H. Understanding the Role of Dicer in Astrocyte Development. PLoS ONE 2015, 10, e0126667. [Google Scholar] [CrossRef][Green Version]
- Hutchison, E.R.; Kawamoto, E.; Taub, D.D.; Lal, A.; Abdelmohsen, K.; Zhang, Y.; Wood, W.H.; Lehrmann, E.; Camandola, S.; Becker, K.G.; et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 2013, 61, 1018–1028. [Google Scholar] [CrossRef]
- Stary, C.M.; Sun, X.; Ouyang, Y.; Li, L.; Giffard, R.G. miR-29a differentially regulates cell survival in astrocytes from cornu ammonis 1 and dentate gyrus by targeting VDAC1. Mitochondrion 2016, 30, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.T.S.; Fuh, S.-C.; Karamchandani, J.R.; Woulfe, J.M.J.; Munoz, D.G.; Ellezam, B.; Blain, M.; Ho, M.-K.; Bedell, B.J.; Antel, J.P.; et al. Astrocytes in the Pathogenesis of Multiple Sclerosis: An In Situ MicroRNA Study. J. Neuropathol. Exp. Neurol. 2019, 78, 1130–1146. [Google Scholar] [CrossRef] [PubMed]
- Kye, M.J.; Liu, T.; Levy, S.; Xu, N.L.; Groves, B.B.; Bonneau, R.; Lao, K.; Kosik, K.S. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA 2007, 13, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.-J.; Liu, C.-M.; Zhang, B.-Y.; Wang, X.; Zhang, M.; Zhang, S.-R.; Hall, P.; Hu, Y.-W.; Zhou, F.-Q. MicroRNA-26a supports mammalian axon regeneration in vivo by suppressing GSK3β expression. Cell Death Dis. 2015, 6, e1865. [Google Scholar] [CrossRef] [PubMed]
- Cáceres, A.; Mautino, J.; Kosik, K.S. Suppression of MAP2 in cultured cerebeller macroneurons inhibits minor neurite formation. Neuron 1992, 9, 607–618. [Google Scholar] [CrossRef]
- Llorens-Martín, M.; Fuster-Matanzo, A.; Teixeira, C.; Jurado-Arjona, J.; Ulloa, F.; DeFelipe, J.; Rabano, A.; Hernández, F.; Soriano, E.; Ávila, J. GSK-3β overexpression causes reversible alterations on postsynaptic densities and dendritic morphology of hippocampal granule neurons in vivo. Mol. Psychiatry 2013, 18, 451–460. [Google Scholar] [CrossRef]
- Wang, S.; Cesca, F.; Loers, G.; Schweizer, M.; Buck, F.; Benfenati, F.; Schachner, M.; Kleene, R. Synapsin I Is an Oligomannose-Carrying Glycoprotein, Acts As an Oligomannose-Binding Lectin, and Promotes Neurite Outgrowth and Neuronal Survival When Released via Glia-Derived Exosomes. J. Neurosci. 2011, 31, 7275–7290. [Google Scholar] [CrossRef]
- Wang, G.; Dinkins, M.; He, Q.; Zhu, G.; Poirier, C.; Campbell, A.; Mayer-Proschel, M.; Bieberich, E. Astrocytes Secrete Exosomes Enriched with Proapoptotic Ceramide and Prostate Apoptosis Response 4 (PAR-4). J. Boil. Chem. 2012, 287, 21384–21395. [Google Scholar] [CrossRef]
- Ramirez, M.I.; De Amorim, M.G.; Gadelha, C.; Milic, I.; Welsh, J.A.; Freitas, V.M.; Nawaz, M.; Akbar, N.; Couch, Y.; Makin, L.; et al. Technical challenges of working with extracellular vesicles. Nanoscale 2018, 10, 881–906. [Google Scholar] [CrossRef]
- Ragni, E.; Viganò, M.; Rebulla, P.; Giordano, R.; Lazzari, L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: How to choose the most reliable housekeeping genes. J. Cell. Mol. Med. 2013, 17, 168–180. [Google Scholar] [CrossRef]
- Gouin, K.; Peck, K.; Antes, T.; Johnson, J.L.; Li, C.; Vaturi, S.D.; Middleton, R.; De Couto, G.; Walravens, A.-S.; Rodriguez-Borlado, L.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Glycolysis Is an Energy-Conversion Pathway in Many Organisms-Biochemistry-NCBI Bookshelf; WH Freeman: New York, NY, USA, 2002. [Google Scholar]
- Lew, C.R.; Tolan, D.R. Targeting of Several Glycolytic Enzymes Using RNA Interference Reveals Aldolase Affects Cancer Cell Proliferation through a Non-glycolytic Mechanism. J. Boil. Chem. 2012, 287, 42554–42563. [Google Scholar] [CrossRef] [PubMed]
- Barros, L.F. Metabolic signaling by lactate in the brain. Trends Neurosci. 2013, 36, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Kusakabe, T.; Motoki, K.; Hori, K. Human Aldolase C: Characterization of the Recombinant Enzyme Expressed in Escherichia coli1. J. Biochem. 1994, 115, 1172–1177. [Google Scholar] [CrossRef] [PubMed]
- Kao, A.W.; Noda, Y.; Johnson, J.H.; Pessin, J.E.; Saltiel, A.R. Aldolase mediates the association of F-actin with the insulin-responsive glucose transporter GLUT4. J. Boil. Chem. 1999, 274, 17742–17747. [Google Scholar] [CrossRef] [PubMed]
- Volker, K.; Knull, H.R. A Glycolytic Enzyme Binding Domain on Tubulin. Arch. Biochem. Biophys. 1997, 338, 237–243. [Google Scholar] [CrossRef]
- Merkulova, M.; Hurtado-Lorenzo, A.; Hosokawa, H.; Zhuang, Z.; Brown, D.; Ausiello, D.A.; Marshansky, V. Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am. J. Physiol. Physiol. 2011, 300, C1442–C1455. [Google Scholar] [CrossRef]
- Caspi, M.; Perry, G.; Skalka, N.; Meisel, S.; Firsow, A.; Amit, M.; Rosin-Arbesfeld, R. Aldolase positively regulates of the canonical Wnt signaling pathway. Mol. Cancer 2014, 13, 164. [Google Scholar] [CrossRef]
- Codocedo, J.F.; Inestrosa, N.C. Wnt-5a-regulated miR-101b controls COX2 expression in hippocampal neurons. Boil. Res. 2016, 49, 9. [Google Scholar] [CrossRef]
- Smith, S.; Kimyon, R.S.; Watters, J.J. Cell-Type-Specific Jumonji Histone Demethylase Gene Expression in the Healthy Rat CNS: Detection by a Novel Flow Cytometry Method. ASN Neuro 2014, 6. [Google Scholar] [CrossRef]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nature 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Wayman, G.A.; Davare, M.; Ando, H.; Fortin, D.; Varlamova, O.; Cheng, H.-Y.M.; Marks, D.; Obrietan, K.; Soderling, T.R.; Goodman, R.H.; et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. USA 2008, 105, 9093–9098. [Google Scholar] [CrossRef] [PubMed]
- Fiore, R.; Khudayberdiev, S.; Christensen, M.; Siegel, G.; Flavell, S.; Kim, T.-K.; Greenberg, M.E.; Schratt, G.M. Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009, 28, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Magill, S.T.; Cambronne, X.; Luikart, B.W.; Lioy, D.T.; Leighton, B.H.; Westbrook, G.L.; Mandel, G.; Goodman, R.H. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. USA 2010, 107, 20382–20387. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, Z.; Yang, F.; Gao, Y.; Song, J.; Wan, Y. microRNA-181a is involved in insulin-like growth factor-1-mediated regulation of the transcription factor CREB1. J. Neurochem. 2013, 126, 771–780. [Google Scholar] [CrossRef]
- Van Spronsen, M.; Van Battum, E.Y.; Kuijpers, M.; Vangoor, V.R.; Rietman, M.L.; Pothof, J.; Gumy, L.F.; Van Ijcken, W.F.; Akhmanova, A.; Pasterkamp, J.; et al. Developmental and Activity-Dependent miRNA Expression Profiling in Primary Hippocampal Neuron Cultures. PLoS ONE 2013, 8, e74907. [Google Scholar] [CrossRef]
- Jiang, H.; Guo, W.; Liang, X.; Rao, Y. Both the Establishment and the Maintenance of Neuronal Polarity Require Active Mechanisms. Cell 2005, 120, 123–135. [Google Scholar] [CrossRef]
- Zhao, S.; Ye, X.; Xiao, L.; Lian, X.; Feng, Y.; Li, F.; Li, L. MiR-26a inhibits prostate cancer progression by repression of Wnt5a. Tumor Boil. 2014, 35, 9725–9733. [Google Scholar] [CrossRef]
- Horigane, S.-I.; Ageta-Ishihara, N.; Kamijo, S.; Fujii, H.; Okamura, M.; Kinoshita, M.; Takemoto-Kimura, S.; Bito, H. Facilitation of axon outgrowth via a Wnt5a-CaMKK-CaMKIα pathway during neuronal polarization. Mol. Brain 2016, 9, 8. [Google Scholar] [CrossRef]
- Lafourcade, C.; Ramírez, J.P.; Luarte, A.; Fernández, A.; Wyneken, U.; Fernández, A. MIRNAS in Astrocyte-Derived Exosomes as Possible Mediators of Neuronal Plasticity. J. Exp. Neurosci. 2016, 10, JEN.S39916. [Google Scholar] [CrossRef]
- Harada, A.; Teng, J.; Takei, Y.; Oguchi, K.; Hirokawa, N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J. Cell Boil. 2002, 158, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Chamak, B.; Fellous, A.; Glowinski, J.; Prochiantz, A. MAP2 expression and neuritic outgrowth and branching are coregulated through region-specific neuro-astroglial interactions. J. Neurosci. 1987, 7, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Caputo, V.; Sinibaldi, L.; Fiorentino, A.; Parisi, C.; Catalanotto, C.; Pasini, A.; Cogoni, C.; Pizzuti, A. Brain Derived Neurotrophic Factor (BDNF) Expression Is Regulated by MicroRNAs miR-26a and miR-26b Allele-Specific Binding. PLoS ONE 2011, 6, e28656. [Google Scholar] [CrossRef] [PubMed]
- Moya-Alvarado, G.; Gonzalez, A.; Stuardo, N.; Bronfman, F.C. Brain-Derived Neurotrophic Factor (BDNF) Regulates Rab5-Positive Early Endosomes in Hippocampal Neurons to Induce Dendritic Branching. Front. Cell. Neurosci. 2018, 12, 493. [Google Scholar] [CrossRef]
- Xu, B.; Zang, K.; Ruff, N.L.; Zhang, Y.A.; McConnell, S.K.; Stryker, M.P.; Reichardt, L.F. Cortical degeneration in the absence of neurotrophin signaling: Dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron 2000, 26, 233–245. [Google Scholar] [CrossRef]
- Beasley, C.L.; Pennington, K.; Behan, A.; Wait, R.; Dunn, M.J.; Cotter, D.R. Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: Evidence for disease-associated changes. Proteom. 2006, 6, 3414–3425. [Google Scholar] [CrossRef]
- Johnston-Wilson, N.L.; Sims, C.D.; Hofmann, J.P.; Anderson, L.; Shore, A.D.; Torrey, E.F.; Yolken, R.H. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depresssive disorder. Mol. Psychiatry 2000, 5, 142–149. [Google Scholar] [CrossRef]
- Ditzen, C.; Tang, N.; Jastorff, A.M.; Teplytska, L.; Yassouridis, A.; Maccarrone, G.; Uhr, M.; Bronisch, T.; Miller, C.A.; Holsboer, F.; et al. Cerebrospinal Fluid Biomarkers for Major Depression Confirm Relevance of Associated Pathophysiology. Neuropsychopharmacology 2011, 37, 1013–1025. [Google Scholar] [CrossRef][Green Version]
- Chadderton, P.; Schaefer, A.T.; Williams, S.; Margrie, T.W. Sensory-evoked synaptic integration in cerebellar and cerebral cortical neurons. Nat. Rev. Neurosci. 2014, 15, 71–83. [Google Scholar] [CrossRef]
- Irwin, S.A.; Galvez, R.; Greenough, W.T. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb. Cortex 2000, 10, 1038–1044. [Google Scholar] [CrossRef]
- Murmu, M.S.; Salomon, S.; Biala, Y.; Weinstock, M.; Braun, K.; Bock, J. Changes of spine density and dendritic complexity in the prefrontal cortex in offspring of mothers exposed to stress during pregnancy. Eur. J. Neurosci. 2006, 24, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, M. Prenatal stressors in rodents: Effects on behavior. Neurobiol. Stress 2016, 6, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, V.; Firestein, B.L. The dendritic tree and brain disorders. Mol. Cell. Neurosci. 2012, 50, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiatry 2019, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- McEwen, B.S.; Bowles, N.P.; Gray, J.D.; Hill, M.N.; Hunter, R.G.; Karatsoreos, I.N.; Nasca, C. Mechanisms of stress in the brain. Nat. Neurosci. 2015, 18, 1315. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, L.; Darbinian, N.; Goetzl, E.J. Novel window on early human neurodevelopment via fetal exosomes in maternal blood. Ann. Clin. Transl. Neurol. 2016, 3, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Agliardi, C.; Clerici, M. Blood extracellular vesicles (EVs) of central nervous system origin: A window into the brain. Neural Regen. Res. 2019, 15, 55–56. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luarte, A.; Henzi, R.; Fernández, A.; Gaete, D.; Cisternas, P.; Pizarro, M.; Batiz, L.F.; Villalobos, I.; Masalleras, M.; Vergara, R.; et al. Astrocyte-Derived Small Extracellular Vesicles Regulate Dendritic Complexity through miR-26a-5p Activity. Cells 2020, 9, 930. https://doi.org/10.3390/cells9040930
Luarte A, Henzi R, Fernández A, Gaete D, Cisternas P, Pizarro M, Batiz LF, Villalobos I, Masalleras M, Vergara R, et al. Astrocyte-Derived Small Extracellular Vesicles Regulate Dendritic Complexity through miR-26a-5p Activity. Cells. 2020; 9(4):930. https://doi.org/10.3390/cells9040930
Chicago/Turabian StyleLuarte, Alejandro, Roberto Henzi, Anllely Fernández, Diego Gaete, Pablo Cisternas, Matias Pizarro, Luis Federico Batiz, Isabel Villalobos, Matias Masalleras, Rodrigo Vergara, and et al. 2020. "Astrocyte-Derived Small Extracellular Vesicles Regulate Dendritic Complexity through miR-26a-5p Activity" Cells 9, no. 4: 930. https://doi.org/10.3390/cells9040930
APA StyleLuarte, A., Henzi, R., Fernández, A., Gaete, D., Cisternas, P., Pizarro, M., Batiz, L. F., Villalobos, I., Masalleras, M., Vergara, R., Varas-Godoy, M., Abarzua-Catalan, L., Herrera-Molina, R., Lafourcade, C., & Wyneken, U. (2020). Astrocyte-Derived Small Extracellular Vesicles Regulate Dendritic Complexity through miR-26a-5p Activity. Cells, 9(4), 930. https://doi.org/10.3390/cells9040930





