Mechanical Forces and Their Effect on the Ribosome and Protein Translation Machinery
Abstract
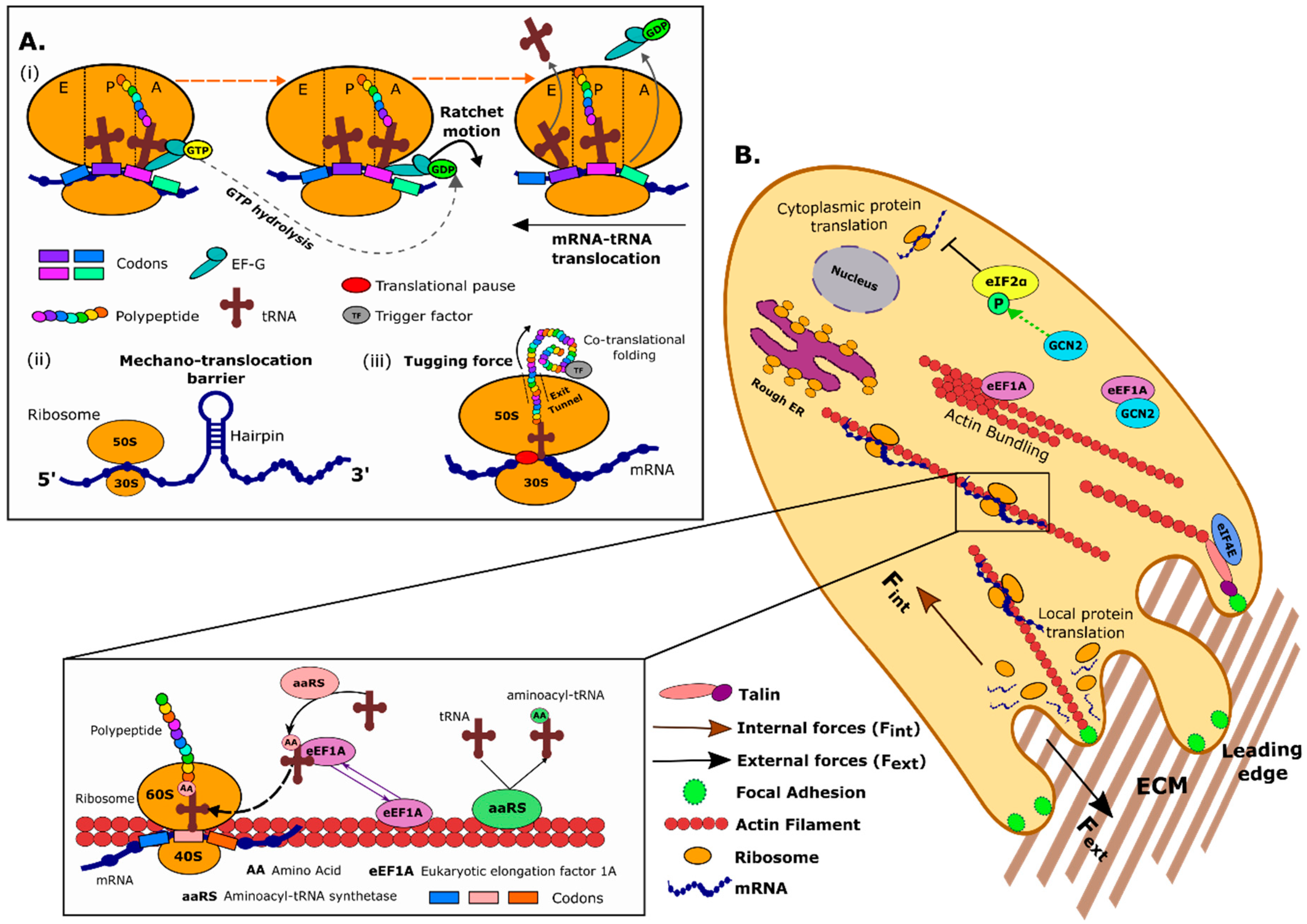
1. Protein Translation in Bacteria and Eukaryotes
2. Forces Generated at the Level of the Ribosomal Machinery
2.1. EF-G and the Ribosome Acts as a Molecular Ratchet System to Drive mRNA–tRNA Translocation
2.2. mRNA Secondary Structures as Mechanical Barriers to the Ribosome
2.3. Forces on the Nascent Polypeptide Generated during co-Translation Folding
3. Cell Mechanics and the Protein Translation Machinery
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Hershey, J.W.B.; Sonenberg, N.; Mathews, M.B. Principles of Translational Control: An Overview. Cold Spring Harb. Perspect. Boil. 2012, 4, a011528. [Google Scholar] [CrossRef] [PubMed]
- Noller, H.F. Evolution of Protein Synthesis from an RNA World. Cold Spring Harb. Perspect. Boil. 2010, 4, a003681. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Olsen, G.J.; Ibba, M.; Soll, D.; Bejuki, W.M. Aminoacyl-tRNA Synthetases, the Genetic Code, and the Evolutionary Process. Microbiol. Mol. Boil. Rev. 2000, 64, 202–236. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Ban, N. The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 A Resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef]
- Kaiser, C.M.; Tinoco, I. Probing the Mechanisms of Translation with Force. Chem. Rev. 2014, 114, 3266–3280. [Google Scholar] [CrossRef]
- Rheinberger, H.J.; Sternbach, H.; Nierhaus, K.H. Three tRNA binding sites on Escherichia coli ribosomes. Proc. Natl. Acad. Sci. USA 1981, 78, 5310–5314. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Polacek, N.; Vesper, O.; Staub, E.; Einfeldt, E.; Wilson, D.N.; Nierhaus, K.H. The Highly Conserved LepA Is a Ribosomal Elongation Factor that Back-Translocates the Ribosome. Cell 2006, 127, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Buskirk, A.R.; Green, R. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. B: Boil. Sci. 2017, 372, 20160183. [Google Scholar] [CrossRef] [PubMed]
- Scheper, G.C.; Van Der Knaap, M.S.; Proud, C. Translation matters: Protein synthesis defects in inherited disease. Nat. Rev. Genet. 2007, 8, 711–723. [Google Scholar] [CrossRef]
- Gandin, V.; Miluzio, A.; Barbieri, A.M.; Beugnet, A.; Kiyokawa, H.; Marchisio, P.C.; Biffo, S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 2008, 455, 684–688. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Shao, S.; Murray, J.; Hegde, R.S.; Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 2015, 524, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, N.; Gorman, A.M.; Gupta, S.; Samali, A. The eIF2α kinases: Their structures and functions. Cell. Mol. Life Sci. 2013, 70, 3493–3511. [Google Scholar] [CrossRef]
- A Hussmann, J.; Patchett, S.; Johnson, A.; Sawyer, S.; Press, W.H. Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. PLoS Genet. 2015, 11, e1005732. [Google Scholar] [CrossRef]
- Schluenzen, F.; Tocilj, A.; Zarivach, R.; Harms, J.; Gluehmann, M.; Janell, D.; Bashan, A.; Bartels, H.; Agmon, I.; Franceschi, F.; et al. Structure of Functionally Activated Small Ribosomal Subunit at 3.3 Å Resolution. Cell 2000, 102, 615–623. [Google Scholar] [CrossRef]
- Wimberly, B.T.; Brodersen, D.; Jr, W.M.C.; Morgan-Warren, R.J.; Carter, A.P.; Vonrhein, C.; Hartsch, T.; Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature 2000, 407, 327–339. [Google Scholar] [CrossRef]
- Stark, H.; Rodnina, M.V.; Wieden, H.-J.; Zemlin, F.; Wintermeyer, W.; Van Heel, M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Genet. 2002, 9, 849–854. [Google Scholar] [CrossRef]
- Frank, J. The translation elongation cycle—capturing multiple states by cryo-electron microscopy. Philos. Trans. R. Soc. B: Boil. Sci. 2017, 372, 20160180. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Savelsbergh, A.; Katunin, V.I.; Wintermeyer, W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 1997, 385, 37–41. [Google Scholar] [CrossRef]
- Frank, J.; Agrawal, R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 2000, 406, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Ermolenko, D.N.; Majumdar, Z.K.; Hickerson, R.P.; Spiegel, P.C.; Clegg, R.M.; Noller, H.F. Observation of Intersubunit Movement of the Ribosome in Solution Using FRET. J. Mol. Boil. 2007, 370, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Cornish, P.V.; Ermolenko, D.N.; Noller, H.F.; Ha, T. Spontaneous Intersubunit Rotation in Single Ribosomes. Mol. Cell 2008, 30, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cui, X.; Beausang, J.F.; Zhang, H.; Farrell, I.; Cooperman, B.S.; Goldman, Y.E. Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. USA 2016, 113, 7515–7520. [Google Scholar] [CrossRef] [PubMed]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Dudko, O.K.; Hummer, A.G.; Szabo, A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA 2008, 105, 15755–15760. [Google Scholar] [CrossRef]
- Namy, O.; Moran, S.J.; Stuart, D.I.; Gilbert, R.J.C.; Brierley, I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 2006, 441, 244–247. [Google Scholar] [CrossRef]
- Liu, T.; Kaplan, A.; Alexander, L.; Yan, S.; Wen, J.-D.; Lancaster, L.; E Wickersham, C.; Fredrick, K.; Noller, H.; Tinoco, I.; et al. Direct measurement of the mechanical work during translocation by the ribosome. ELife 2014, 3, e03406. [Google Scholar] [CrossRef]
- Tinoco, I.; Collin, D.; Li, P. The effect of force on thermodynamics and kinetics: Unfolding single RNA molecules. Biochem. Soc. Trans. 2004, 32, 757–760. [Google Scholar] [CrossRef]
- Visscher, K. −1 Programmed Ribosomal Frameshifting as a Force-Dependent Process. Prog. in Mol. Biol. Transl. Sci. 2016, 139, 45–72. [Google Scholar]
- Qu, X.; Wen, J.-D.; Lancaster, L.; Noller, H.F.; Bustamante, C.; Tinoco, I. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 2011, 475, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Desai, V.P.; Frank, F.; Lee, A.; Righini, M.; Lancaster, L.; Noller, H.F.; Tinoco, I.; Bustamante, C. Co-temporal Force and Fluorescence Measurements Reveal a Ribosomal Gear Shift Mechanism of Translation Regulation by Structured mRNAs. Mol. Cell 2019, 75, 1007–1019.e5. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D.H.; Kaiser, C.M.; Milin, A.; Righini, M.; Tinoco, I.; Bustamante, C. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science 2015, 348, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, O.B.; Müller-Lucks, A.; Kramer, G.; Bukau, B.; Von Heijne, G. Trigger Factor Reduces the Force Exerted on the Nascent Chain by a Cotranslationally Folding Protein. J. Mol. Boil. 2016, 428, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Fritch, B.; Kosolapov, A.; Hudson, P.; Nissley, D.A.; Woodcock, H.; Deutsch, C.; O’Brien, E.P. Origins of the Mechanochemical Coupling of Peptide Bond Formation to Protein Synthesis. J. Am. Chem. Soc. 2018, 140, 5077–5087. [Google Scholar] [CrossRef]
- Leininger, S.E.; Trovato, F.; Nissley, D.A.; O’Brien, E.P. Domain topology, stability, and translation speed determine mechanical force generation on the ribosome. Proc. Natl. Acad. Sci. USA 2019, 116, 5523–5532. [Google Scholar] [CrossRef]
- Marino, J.; Von Heijne, G.; Beckmann, R. Small protein domains fold inside the ribosome exit tunnel. FEBS Lett. 2016, 590, 655–660. [Google Scholar] [CrossRef]
- Niesen, M.J.; Müller-Lucks, A.; Hedman, R.; Von Heijne, G.; Miller, T.F. Forces on Nascent Polypeptides during Membrane Insertion and Translocation via the Sec Translocon. Biophys. J. 2018, 115, 1885–1894. [Google Scholar] [CrossRef]
- Komar, A.A. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009, 34, 16–24. [Google Scholar] [CrossRef]
- Nilsson, O.B.; Hedman, R.; Marino, J.; Wickles, S.; Bischoff, L.; Johansson, M.; Müller-Lucks, A.; Trovato, F.; Puglisi, J.D.; O’Brien, E.P.; et al. Cotranslational Protein Folding inside the Ribosome Exit Tunnel. Cell Rep. 2015, 12, 1533–1540. [Google Scholar] [CrossRef]
- Ito, K.; Chiba, S. Arrest Peptides:Cis-Acting Modulators of Translation. Annu. Rev. Biochem. 2013, 82, 171–202. [Google Scholar] [CrossRef] [PubMed]
- Farias-Rico, J.A.; Selin, F.R.; Myronidi, I.; Frühauf, M.; Von Heijne, G. Effects of protein size, thermodynamic stability, and net charge on cotranslational folding on the ribosome. Proc. Natl. Acad. Sci. USA 2018, 115, E9280–E9287. [Google Scholar] [CrossRef] [PubMed]
- Knight, A.M.; Culviner, P.H.; Kurt-Yilmaz, N.; Zou, T.; Ozkan, S.B.; Cavagnero, S. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem. Biol. 2013, 8, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, C.M.; Goldman, D.H.; Chodera, J.D.; Tinoco, I., Jr.; Bustamante, C. The ribosome modulates nascent protein folding. Science 2011, 334, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Merz, F.; Rutkowska, A.; Zachmann-Brand, B.; Deuerling, E.; Bukau, B. Trigger Factor Forms a Protective Shield for Nascent Polypeptides at the Ribosome. J. Boil. Chem. 2006, 281, 6539–6545. [Google Scholar] [CrossRef]
- Kaiser, C.M.; Chang, H.-C.; Agashe, V.R.; Lakshmipathy, S.K.; Etchells, S.A.; Hayer-Hartl, M.; Hartl, F.U.; Barral, J.M. Real-time observation of trigger factor function on translating ribosomes. Nature 2006, 444, 455–460. [Google Scholar] [CrossRef]
- Haldar, S.; Tapia-Rojo, R.; Eckels, E.C.; Valle-Orero, J.; Fernández, J.M. Trigger factor chaperone acts as a mechanical foldase. Nat. Commun. 2017, 8, 668. [Google Scholar] [CrossRef]
- Jahn, M.; Buchner, J.; Hugel, T.; Rief, M. Folding and assembly of the large molecular machine Hsp90 studied in single-molecule experiments. Proc. Natl. Acad. Sci. USA 2016, 113, 1232–1237. [Google Scholar] [CrossRef]
- Liu, K.; Rehfus, J.E.; Mattson, E.; Kaiser, C.M. The ribosome destabilizes native and non-native structures in a nascent multidomain protein. Protein Sci. 2017, 26, 1439–1451. [Google Scholar] [CrossRef]
- Scholl, Z.N.; Yang, W.; Marszalek, P.E. Competing Pathways and Multiple Folding Nuclei in a Large Multidomain Protein, Luciferase. Biophys. J. 2017, 112, 1829–1840. [Google Scholar] [CrossRef]
- Kim, S.; Coulombe, P.A. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat. Rev. Mol. Cell Boil. 2010, 11, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.; Sattlegger, E.; Castilho, B.A. Perturbations in actin dynamics reconfigure protein complexes that modulate GCN2 activity and promote an eIF2 response. J. Cell Sci. 2016, 129, 4521–4533. [Google Scholar] [CrossRef] [PubMed]
- Stapulionis, R.; Kolli, S.; Deutscher, M.P. Efficient mammalian protein synthesis requires an intact F-actin system. J. Boil. Chem. 1997, 272, 24980–24986. [Google Scholar] [CrossRef] [PubMed]
- Gross, S.; Kinzy, T. Improper Organization of the Actin Cytoskeleton Affects Protein Synthesis at Initiation. Mol. Cell. Boil. 2006, 27, 1974–1989. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.C.; Ephrussi, A. mRNA Localization: Gene Expression in the Spatial Dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Wolosewick, J.J.; Porter, K.R. Stereo high-voltage electron microscopy of whole cells of the human diploid line, WI-38. Am. J. Anat. 1976, 147, 303–323. [Google Scholar] [CrossRef]
- Lenk, R.; Ransom, L.; Kaufmann, Y.; Penman, S. A cytoskeletal structure with associated polyribosomes obtained from HeLa cells. Cell 1977, 10, 67–78. [Google Scholar] [CrossRef]
- Fulton, A.B.; Wan, K.M.; Penman, S. The spatial distribution of polyribosomes in 3T3 cells and the associated assembly of proteins into the skeletal framework. Cell 1980, 20, 849–857. [Google Scholar] [CrossRef]
- Medalia, O.; Weber, I.; Frangakis, A.; Nicastro, D.; Gerisch, G.; Baumeister, W. Macromolecular Architecture in Eukaryotic Cells Visualized by Cryoelectron Tomography. Science 2002, 298, 1209–1213. [Google Scholar] [CrossRef]
- Hamill, D.; Davis, J.; Drawbridge, J.; A Suprenant, K. Polyribosome targeting to microtubules: Enrichment of specific mRNAs in a reconstituted microtubule preparation from sea urchin embryos. J. Cell Boil. 1994, 127, 973–984. [Google Scholar] [CrossRef]
- Thornell, L.E.; Eriksson, A. Filament systems in the Purkinje fibers of the heart. Am. J. Physiol. Circ. Physiol. 1981, 241, H291–H305. [Google Scholar] [CrossRef] [PubMed]
- Boyde, A.; Bailey, E. Observations on the marginal ruffles of an established fibroblast-like cell line. Cell Tissue Res. 1977, 179, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Chicurel, M.E.; Singer, R.H.; Meyer, C.J.; Ingber, N.E. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature 1998, 392, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Grant, W.M.; Persky, D.; Latham, V.M.; Singer, R.H.; Condeelis, J. Interactions of Elongation Factor 1α with F-Actin and β-Actin mRNA: Implications for Anchoring mRNA in Cell Protrusions. Mol. Boil. Cell 2002, 13, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Woods, A.J.; Roberts, M.S.; Choudhary, J.S.; Barry, S.T.; Mazaki, Y.; Sabe, H.; Morley, S.J.; Critchley, D.R.; Norman, J. Paxillin Associates with Poly(A)-binding Protein 1 at the Dense Endoplasmic Reticulum and the Leading Edge of Migrating Cells. J. Boil. Chem. 2001, 277, 6428–6437. [Google Scholar] [CrossRef] [PubMed]
- Mingle, L.A.; Okuhama, N.N.; Shi, J.; Singer, R.H.; Condeelis, J.; Liu, G. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 2005, 118, 2425–2433. [Google Scholar] [CrossRef]
- Babic, I.; Sharma, S.; Black, U.L. A Role for Polypyrimidine Tract Binding Protein in the Establishment of Focal Adhesions. Mol. Cell. Boil. 2009, 29, 5564–5577. [Google Scholar] [CrossRef]
- Willett, M.; Pollard, H.J.; Vlasak, M.; Morley, S.J. Localization of ribosomes and translation initiation factors to talin/β3-integrin-enriched adhesion complexes in spreading and migrating mammalian cells. Boil. Cell 2010, 102, 265–276. [Google Scholar] [CrossRef]
- Willett, M.; Brocard, M.; David, A.; Morley, S.J. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem. J. 2011, 438, 217–227. [Google Scholar] [CrossRef][Green Version]
- Yang, F.; Demma, M.; Warren, V.; Dharmawardhane, S.; Condeelis, J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1a. Nature 1990, 347, 494–496. [Google Scholar] [CrossRef]
- Demma, M.; Warren, V.; Hock, R.; Dharmawardhane, S.; Condeelis, J. Isolation of an abundant 50,000-dalton actin filament bundling protein from Dictyostelium amoebae. J. Boil. Chem. 1990, 265, 2286–2291. [Google Scholar]
- Liu, G.; Tang, J.; Edmonds, B.T.; Murray, J.; Levin, S.; Condeelis, J. F-actin sequesters elongation factor 1alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J. Cell Boil. 1996, 135, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Owen, C.H.; DeRosier, D.J.; Condeelis, J. Actin crosslinking protein EF-1 a of Dictyostelium discoideum has a unique bonding rule that allows square-packed bundles. J. Struct. Boil. 1992, 109, 248–254. [Google Scholar] [CrossRef]
- Fujimura, K.; Choi, S.; Wyse, M.; Strnadel, J.; Wright, T.; Klemke, R.L. Eukaryotic Translation Initiation Factor 5A (EIF5A) Regulates Pancreatic Cancer Metastasis by Modulating RhoA and Rho-associated Kinase (ROCK) Protein Expression Levels. J. Boil. Chem. 2015, 290, 29907–29919. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. The Scanning Mechanism of Eukaryotic Translation Initiation. Annu. Rev. Biochem. 2014, 83, 779–812. [Google Scholar] [CrossRef]
- Castilho, B.A.; Shanmugam, R.; Silva, R.; Ramesh, R.; Himme, B.M.; Sattlegger, E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1843, 1948–1968. [Google Scholar] [CrossRef]
- Perez, W.B.; Kinzy, T. Translation Elongation Factor 1A Mutants with Altered Actin Bundling Activity Show Reduced Aminoacyl-tRNA Binding and Alter Initiation via eIF2α Phosphorylation. J. Boil. Chem. 2014, 289, 20928–20938. [Google Scholar] [CrossRef]
- Singer, R.H.; Langevin, G.L.; Lawrence, J.B. Ultrastructural visualization of cytoskeletal mRNAs and their associated proteins using double-label in situ hybridization. J. Cell Boil. 1989, 108, 2343–2353. [Google Scholar] [CrossRef]
- Steward, O.; Levy, W. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J. Neurosci. 1982, 2, 284–291. [Google Scholar] [CrossRef]
- Tiedge, H.; Brosius, J. Translational Machinery in Dendrites of Hippocampal Neurons in Culture. J. Neurosci. 1996, 16, 7171–7181. [Google Scholar] [CrossRef]
- Hafner, A.-S.; Donlin-Asp, P.G.; Leitch, B.; Herzog, E.; Schuman, E.M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 2019, 364, eaau3644. [Google Scholar] [CrossRef] [PubMed]
- Bramham, C.R. Local protein synthesis, actin dynamics, and LTP consolidation. Curr. Opin. Neurobiol. 2008, 18, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Katz, Z.; Wells, A.L.; Park, H.Y.; Wu, B.; Shenoy, S.; Singer, R.H. β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012, 26, 1885–1890. [Google Scholar] [CrossRef] [PubMed]
- Katz, Z.; English, B.P.; Lionnet, T.; Yoon, Y.J.; Monnier, N.; Ovryn, B.; Bathe, M.; Singer, R.H. Mapping translation ’hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. ELife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Willett, M.; Brocard, M.; Pollard, H.J.; Morley, S.J. mRNA encoding WAVE–Arp2/3-associated proteins is co-localized with foci of active protein synthesis at the leading edge of MRC5 fibroblasts during cell migration. Biochem. J. 2013, 452, 45–55. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simpson, L.J.; Tzima, E.; Reader, J.S. Mechanical Forces and Their Effect on the Ribosome and Protein Translation Machinery. Cells 2020, 9, 650. https://doi.org/10.3390/cells9030650
Simpson LJ, Tzima E, Reader JS. Mechanical Forces and Their Effect on the Ribosome and Protein Translation Machinery. Cells. 2020; 9(3):650. https://doi.org/10.3390/cells9030650
Chicago/Turabian StyleSimpson, Lisa J., Ellie Tzima, and John S. Reader. 2020. "Mechanical Forces and Their Effect on the Ribosome and Protein Translation Machinery" Cells 9, no. 3: 650. https://doi.org/10.3390/cells9030650
APA StyleSimpson, L. J., Tzima, E., & Reader, J. S. (2020). Mechanical Forces and Their Effect on the Ribosome and Protein Translation Machinery. Cells, 9(3), 650. https://doi.org/10.3390/cells9030650



