Molecular Rewiring of the Jasmonate Signaling Pathway to Control Auxin-Responsive Gene Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Cultural Conditions
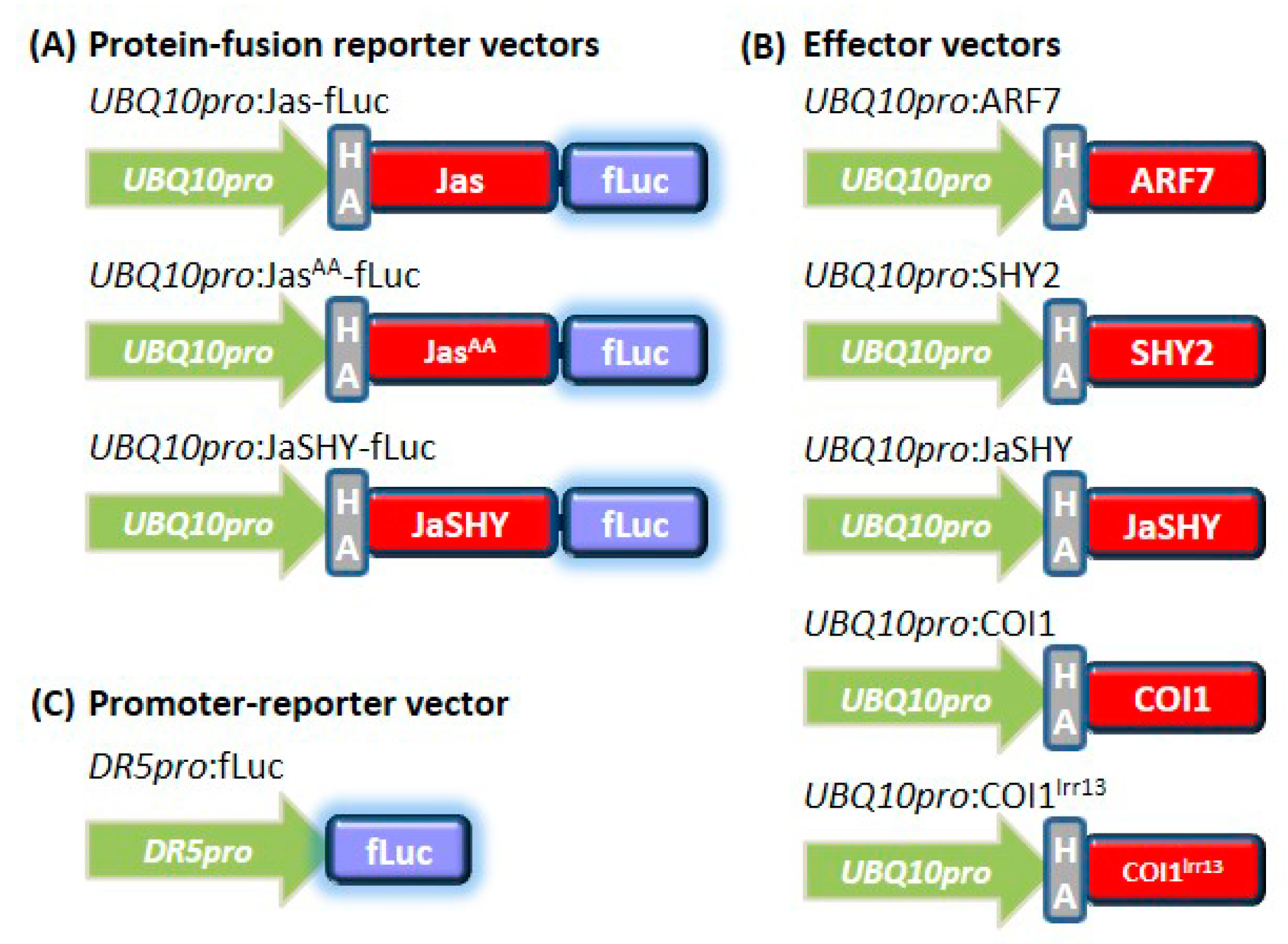
2.2. Molecular Cloning
2.3. Protoplast Transfection and Dual Luciferase Report Assay
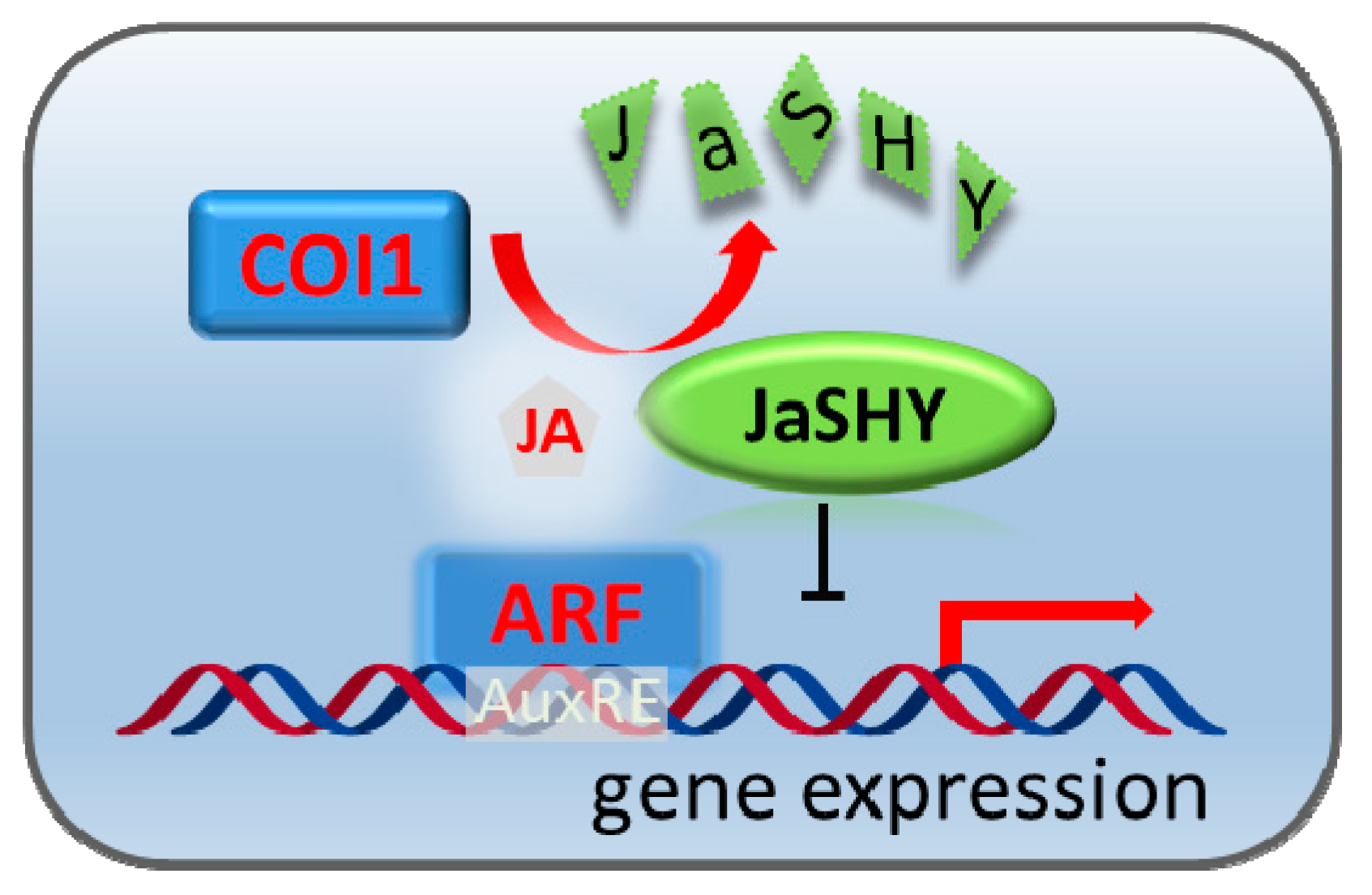
3. Results and Discussion
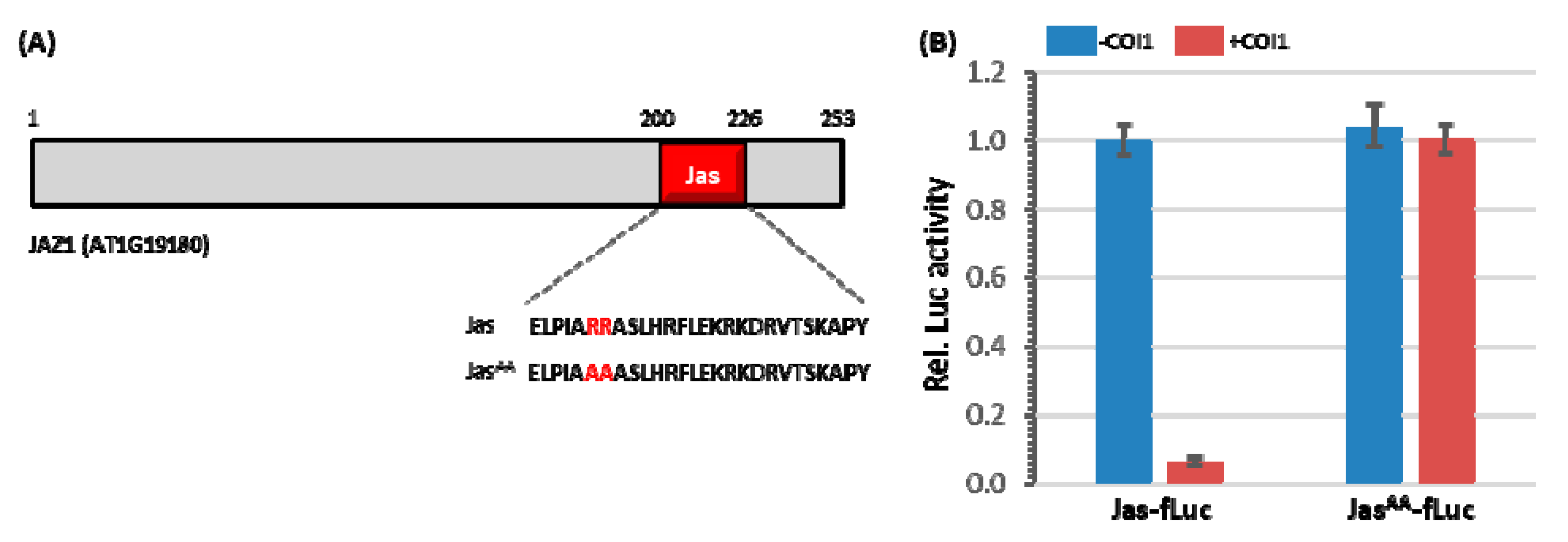
3.1. The Jas-Motif Is Sufficient to Target Luciferase Reporter for Degradation
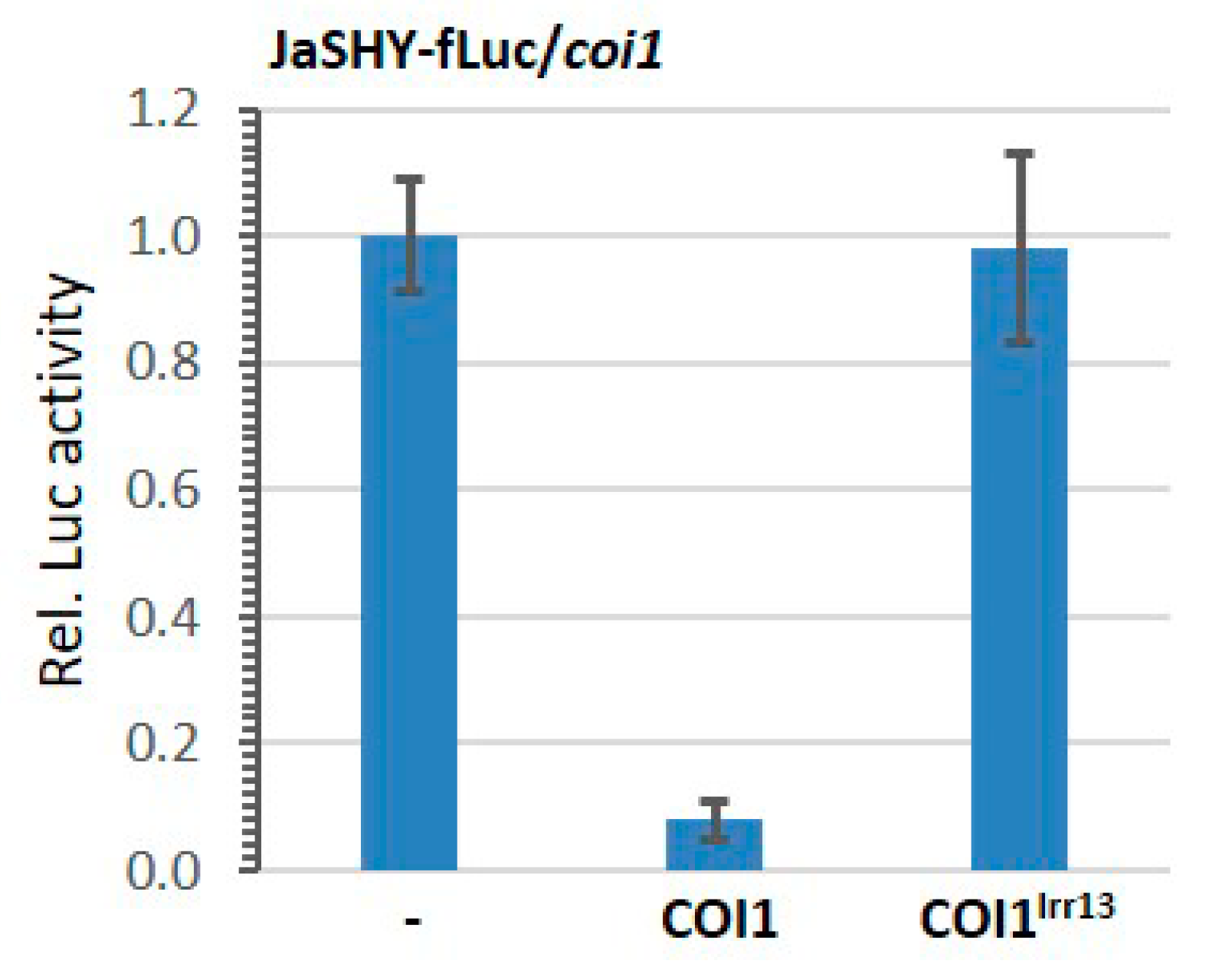
3.2. The Jas-Tagging Leads the Aux/IAA Protein SHY2 to Be Degraded by the JA Coreceptor COI1
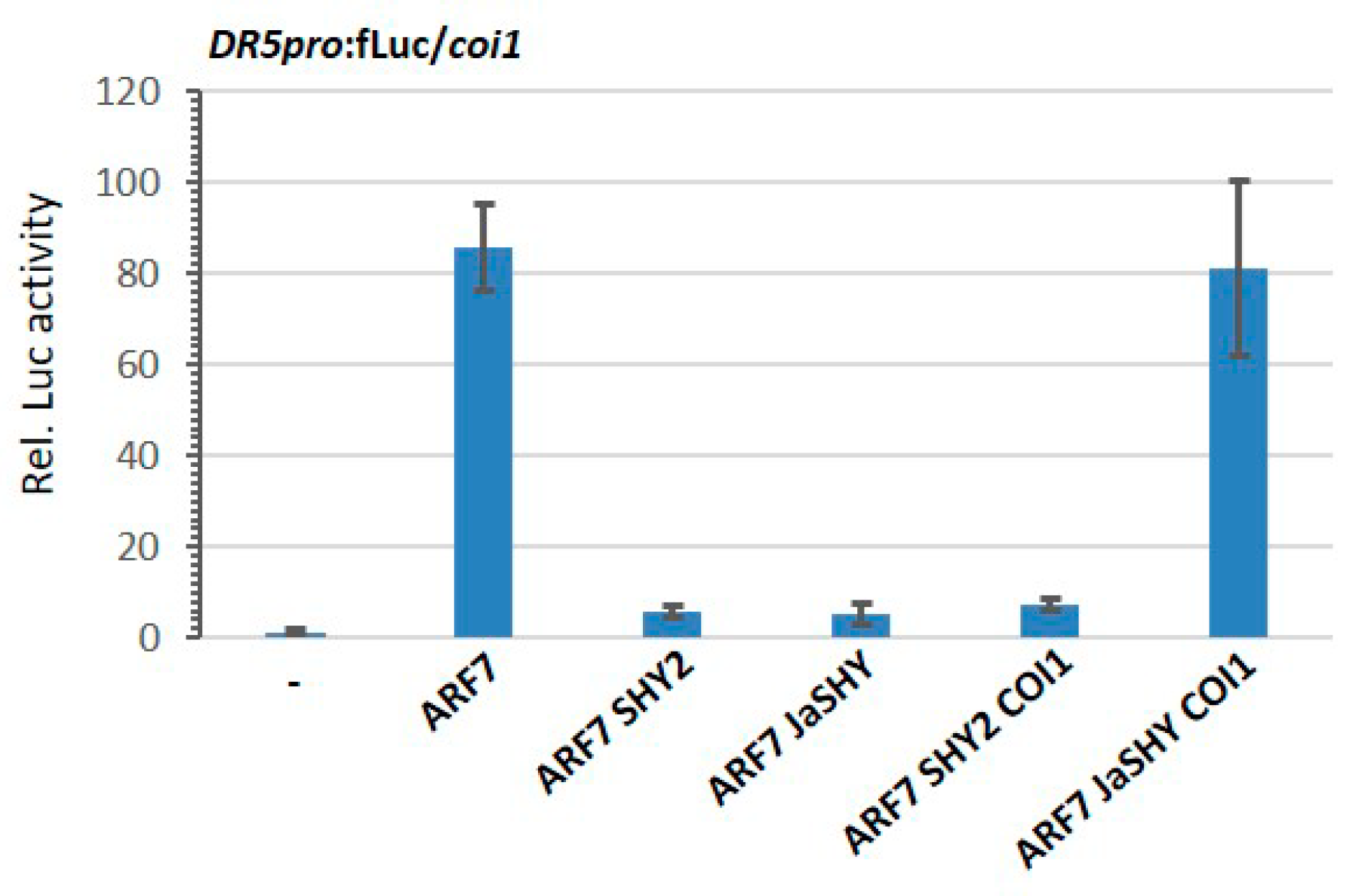
3.3. Degradation of the JaSHY Chimeric Repressor Liberates ARF7 Transcription Activity
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef]
- Cai, X.-T.; Xu, P.; Zhao, P.-X.; Liu, R.; Yu, L.-H.; Xiang, C.-B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, F.; Chen, L.; Pan, Y.; Sun, L.; Bao, N.; Zhang, T.; Cui, C.-X.; Qiu, Z.; Zhang, Y.; et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 2019, 5, 491–497. [Google Scholar] [CrossRef]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956.e14. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongelard, G.; Floková, K.; Pacurar, D.I.; Novák, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef]
- Anjum, S.A.; Xie, X.; Farooq, M.; Wang, L.; Xue, L.; Shahbaz, M.; Salhab, J. Effect of exogenous methyl jasmonate on growth, gas exchange and chlorophyll contents of soybean subjected to drought. Afr. J. Biotechnol. 2011, 10, 9640–9646. [Google Scholar]
- Anjum, S.A.; Tanveer, M.; Hussain, S.; Tung, S.A.; Samad, R.A.; Wang, L.; Khan, I.; ur Rehman, N.; Shah, A.N.; Shahzad, B. Exogenously applied methyl jasmonate improves the drought tolerance in wheat imposed at early and late developmental stages. Acta Physiol. Plant 2016, 38, 25. [Google Scholar] [CrossRef]
- Kang, D.-J.; Seo, Y.-J.; Lee, J.-D.; Ishii, R.; Kim, K.U.; Shin, D.H.; Park, S.K.; Jang, S.W.; Lee, I.-J. asmonic Acid Differentially Affects Growth, Ion Uptake and Abscisic Acid Concentration in Salt-tolerant and Salt-sensitive Rice Cultivars. J. Agron. Crop Sci. 2005, 191, 273–282. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, J.; Withers, J.; Xin, X.-F.; Banerjee, R.; Fariduddin, Q.; Nakamura, Y.; Nomura, K.; Howe, G.A.; Boland, W.; et al. Host target modification as a strategy to counter pathogen hijacking of the jasmonate hormone receptor. Proc. Natl. Acad. Sci. USA 2015, 112, 14354–14359. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonates: News on Occurrence, Biosynthesis, Metabolism and Action of an Ancient Group of Signaling Compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in Jasmonate Signaling for Multistress Resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Duan, G.; Li, C.; Liu, L.; Han, G.; Zhang, Y.; Wang, C. The Crosstalks Between Jasmonic Acid and Other Plant Hormone Signaling Highlight the Involvement of Jasmonic Acid as a Core Component in Plant Response to Biotic and Abiotic Stresses. Front. Plant Sci. 2019, 10, 1349. [Google Scholar] [CrossRef]
- Jang, G.; Yoon, Y.; Choi, Y.D. Crosstalk with Jasmonic Acid Integrates Multiple Responses in Plant Development. Int. J. Mol. Sci. 2020, 21, 305. [Google Scholar] [CrossRef]
- Gfeller, A.; Liechti, R.; Farmer, E.E. Arabidopsis jasmonate signaling pathway. Sci Signal 2010, 3, cm4. [Google Scholar] [CrossRef]
- Browse, J.; Wallis, J.G. Arabidopsis Flowers Unlocked the Mechanism of Jasmonate Signaling. Plants 2019, 8, 285. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Wasternack, C.; Feussner, I. The Oxylipin Pathways: Biochemistry and Function. Annu. Rev. Plant Biol. 2018, 69, 363–386. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Xie, D.X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Nagels Durand, A.; Pauwels, L.; Goossens, A. The Ubiquitin System and Jasmonate Signaling. Plants 2016, 5, 6. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Chico, J.M.; Bossche, R.V.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Liu, Z.; Karmarkar, V. Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 2008, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Zhu, J.-K.; Yang, Z. Epigenetic Modifications and Plant Hormone Action. Mol. Plant 2016, 9, 57–70. [Google Scholar] [CrossRef]
- Zuin, A.; Isasa, M.; Crosas, B. Ubiquitin signaling: Extreme conservation as a source of diversity. Cells 2014, 3, 690–701. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hellmann, H. Plant E3 ligases: Flexible enzymes in a sessile world. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef]
- Pérez, A.C.; Goossens, A. Jasmonate signalling: A copycat of auxin signalling? Plant Cell Environ. 2013, 36, 2071–2084. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef]
- Winkler, M.; Niemeyer, M.; Hellmuth, A.; Janitza, P.; Christ, G.; Samodelov, S.L.; Wilde, V.; Majovsky, P.; Trujillo, M.; Zurbriggen, M.D.; et al. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 2017, 8, 15706. [Google Scholar] [CrossRef]
- Li, N.; Uhrig, J.F.; Thurow, C.; Huang, L.-J.; Gatz, C. Reconstitution of the Jasmonate Signaling Pathway in Plant Protoplasts. Cells 2019, 8, 1532. [Google Scholar] [CrossRef]
- Sheen, J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001, 127, 1466–1475. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Mosblech, A.; Thurow, C.; Gatz, C.; Feussner, I.; Heilmann, I. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 2011, 65, 949–957. [Google Scholar] [CrossRef]
- Köster, J.; Thurow, C.; Kruse, K.; Meier, A.; Iven, T.; Feussner, I.; Gatz, C. Xenobiotic- and jasmonic acid-inducible signal transduction pathways have become interdependent at the Arabidopsis CYP81D11 promoter. Plant Physiol. 2012, 159, 391–402. [Google Scholar] [CrossRef]
- Uhrig, J.F.; Huang, L.-J.; Barghahn, S.; Willmer, M.; Thurow, C.; Gatz, C. CC-type glutaredoxins recruit the transcriptional co-repressor TOPLESS to TGA-dependent target promoters in Arabidopsis thaliana. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; De Meyer, B.; Hilson, P. Modular cloning in plant cells. Trends Plant Sci. 2005, 10, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [PubMed]
- Li, N.; Muthreich, M.; Huang, L.-J.; Thurow, C.; Sun, T.; Zhang, Y.; Gatz, C. TGACG-BINDING FACTORs (TGAs) and TGA-interacting CC-type glutaredoxins modulate hyponastic growth in Arabidopsis thaliana. New Phytol. 2019, 221, 1906–1918. [Google Scholar] [CrossRef] [PubMed]
- Larrieu, A.; Champion, A.; Legrand, J.; Lavenus, J.; Mast, D.; Brunoud, G.; Oh, J.; Guyomarc’h, S.; Pizot, M.; Farmer, E.E.; et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat. Commun. 2015, 6, 6043. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signalling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef]
- Zhang, L.; Ward, J.D.; Cheng, Z.; Dernburg, A.F. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 2015, 142, 4374–4384. [Google Scholar] [CrossRef]
- Natsume, T.; Kiyomitsu, T.; Saga, Y.; Kanemaki, M.T. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 2016, 15, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Sathyan, K.M.; McKenna, B.D.; Anderson, W.D.; Duarte, F.M.; Core, L.; Guertin, M.J. An improved auxin-inducible degron system preserves native protein levels and enables rapid and specific protein depletion. Genes Dev. 2019, 33, 1441–1455. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.C.; Soh, M.C.; Kang, B.J.; Furuya, M.; Nam, H.G. Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J. 1996, 9, 441–456. [Google Scholar] [CrossRef]
- Tian, Q.; Reed, J.W. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 1999, 126, 711–721. [Google Scholar]
- Tian, Q.; Uhlir, N.J.; Reed, J.W. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 2002, 14, 301–319. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Benkova, E.; Jäger, K.E.; Schlereth, A.; Hamann, T.; Kientz, M.; Wilmoth, J.C.; Reed, J.W.; Jürgens, G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005, 24, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Kasahara, H.; Mimura, T.; Kamiya, Y.; Fukaki, H. Multiple AUX/IAA-ARF modules regulate lateral root formation: The role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos. Trans. R. Soc. Lond. 2012, 367, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Orosa-Puente, B.; Leftley, N.; von Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M.; et al. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hentrich, M.; Pollmann, S. Auxin-oxylipin crosstalk: Relationship of antagonists. J. Integr. Plant Biol. 2011, 53, 429–445. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
| Primer Symbol | Primer Sequence (5′-3′) |
|---|---|
| JAS-gw-d1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGAACTTCCTATTGCTAGAAG |
| JASnostop-gw-r1 | GGGGACCACTTTGTACAAGAAAGCTGGGTGAGTATGGTGCCTTTGACGTAAC |
| JASA-gw-d1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGAACTTCCTATTGCTGCAGCAGC |
| SHY2-gw-d1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGATGAGTTTGTTAACCTC |
| SHY2nostop-gw-r1 | GGGGACCACTTTGTACAAGAAAGCTGGGTGATACACCACAGCCTAAACC |
| SHY2-gw-r1 | GGGGACCACTTTGTACAAGAAAGCTGGGTCATACACCACAGCCTAAACC |
| JASHY-d1 | TACGTCAAAGGCACCATACATGGATGAGTTTGTTAACCTC |
| JASHY-r1 | TGAGGTTAACAAACTCATCCATGTATGGTGCCTTTGACGT |
| DR5-gw-d1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGCCTGCAGGTCGACGGTAT |
| DR5-gw-r1 | GGGGACCACTTTGTACAAGAAAGCTGGGTTTGTAATTGTAATTGTAAATAGT |
| ARF7-gw-d1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGAAAGCTCCTTCATCAAATGGAG |
| ARF7-gw-r1 | GGGGACCACTTTGTACAAGAAAGCTGGGTCACCGGTTAAACGAAGTGGCTGAG |
| SeqL1 | TCGCGTTAACGCTAGCATGGATCTC |
| SeqL2 | GTAACATCAGAGATTTTGAGACAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Cao, L.; Miu, W.; Cao, R.; Peng, M.; Wan, W.; Huang, L.-J. Molecular Rewiring of the Jasmonate Signaling Pathway to Control Auxin-Responsive Gene Expression. Cells 2020, 9, 641. https://doi.org/10.3390/cells9030641
Li N, Cao L, Miu W, Cao R, Peng M, Wan W, Huang L-J. Molecular Rewiring of the Jasmonate Signaling Pathway to Control Auxin-Responsive Gene Expression. Cells. 2020; 9(3):641. https://doi.org/10.3390/cells9030641
Chicago/Turabian StyleLi, Ning, Linggai Cao, Wenzhuo Miu, Ruibin Cao, Mingbo Peng, Wenkai Wan, and Li-Jun Huang. 2020. "Molecular Rewiring of the Jasmonate Signaling Pathway to Control Auxin-Responsive Gene Expression" Cells 9, no. 3: 641. https://doi.org/10.3390/cells9030641
APA StyleLi, N., Cao, L., Miu, W., Cao, R., Peng, M., Wan, W., & Huang, L.-J. (2020). Molecular Rewiring of the Jasmonate Signaling Pathway to Control Auxin-Responsive Gene Expression. Cells, 9(3), 641. https://doi.org/10.3390/cells9030641





