Functions of Vertebrate Ferlins
Abstract
:1. Introduction
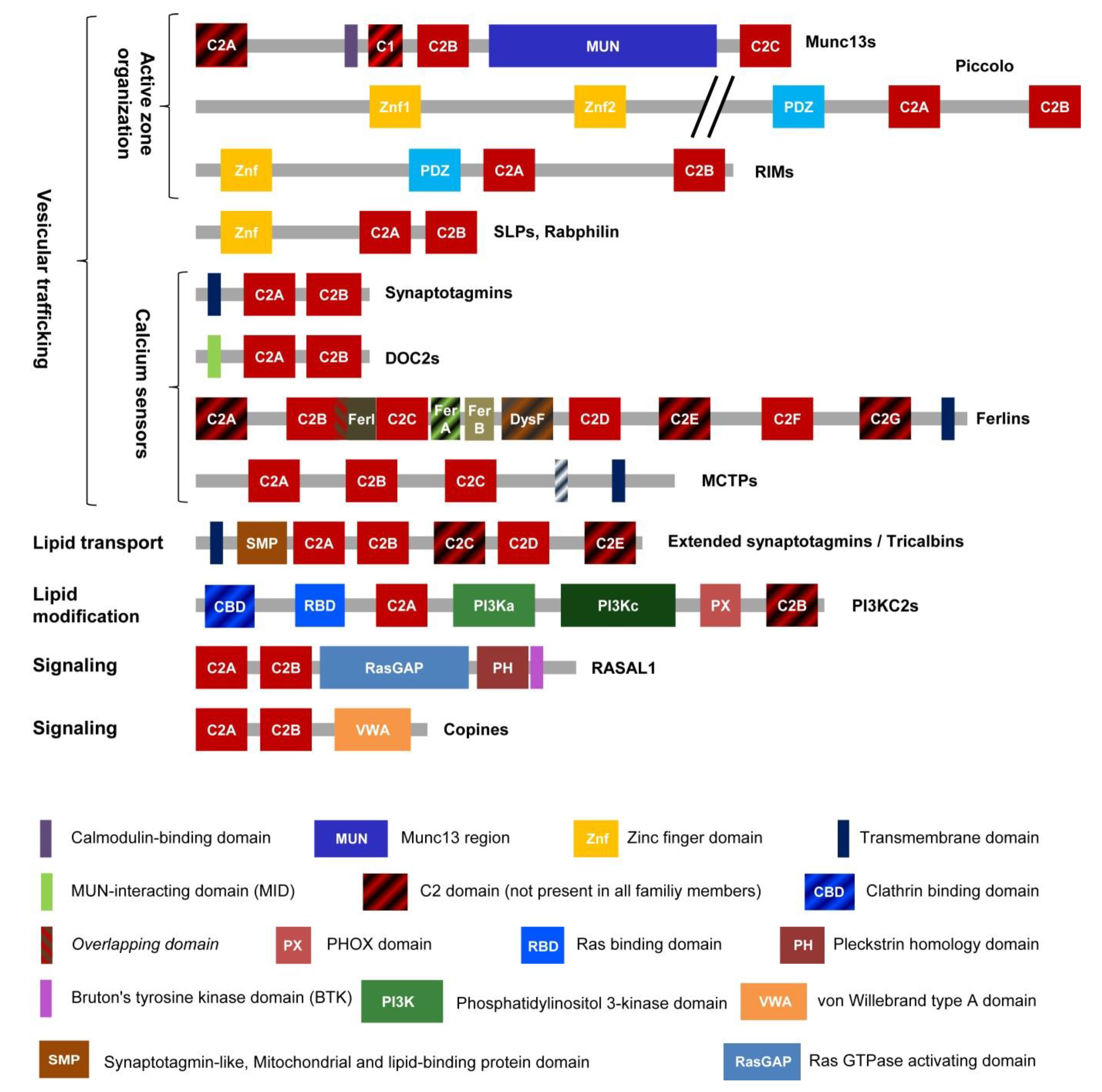
2. Proteins with Multiple C2 Domains (MC2D)
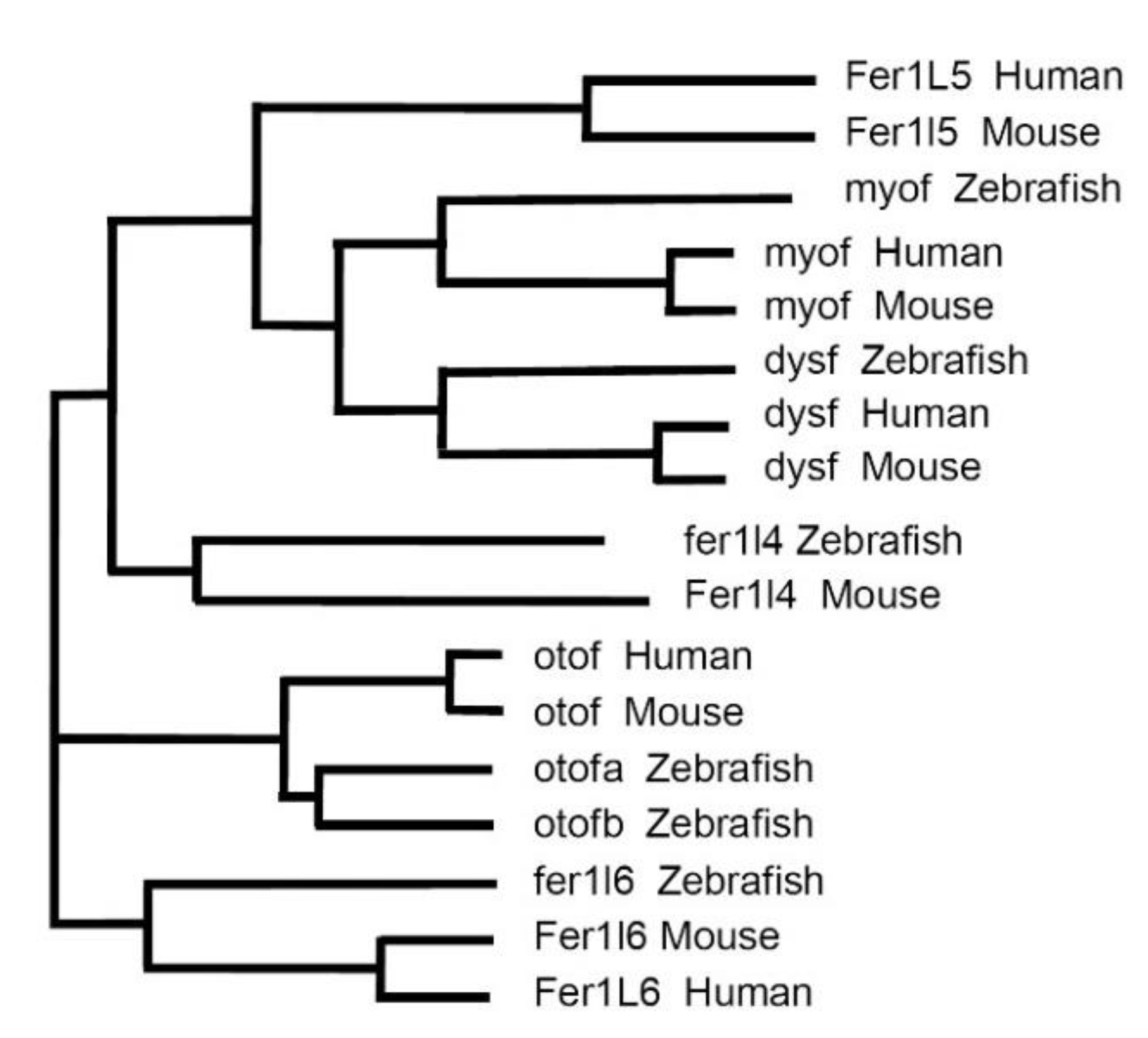
3. Vertebrate Ferlins: Family Members and Domain Organization
4. Ferlin Domains: Properties and Function
5. Tissue Distribution of the Ferlins
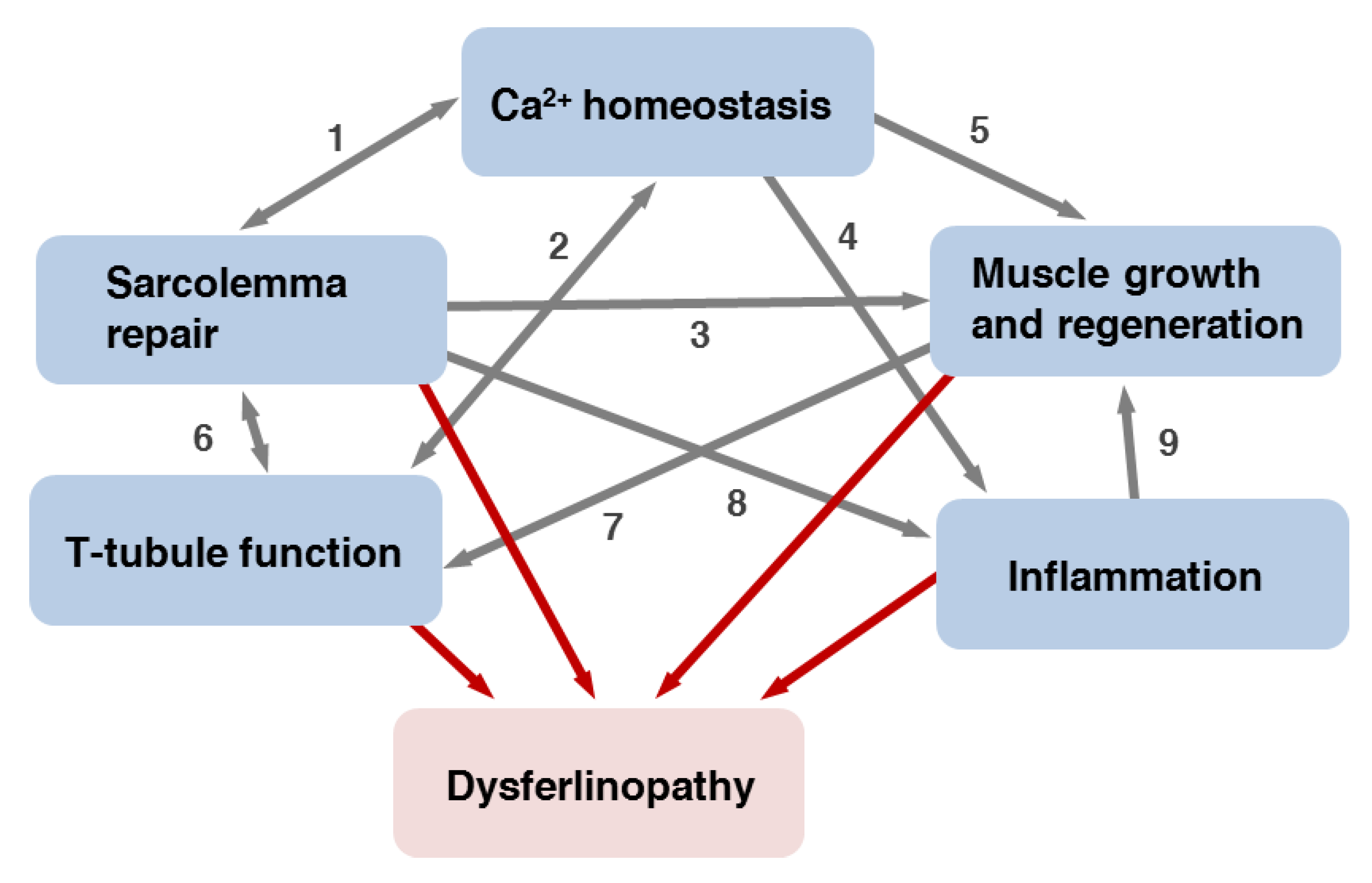
6. Functions of Dysferlin in Muscle
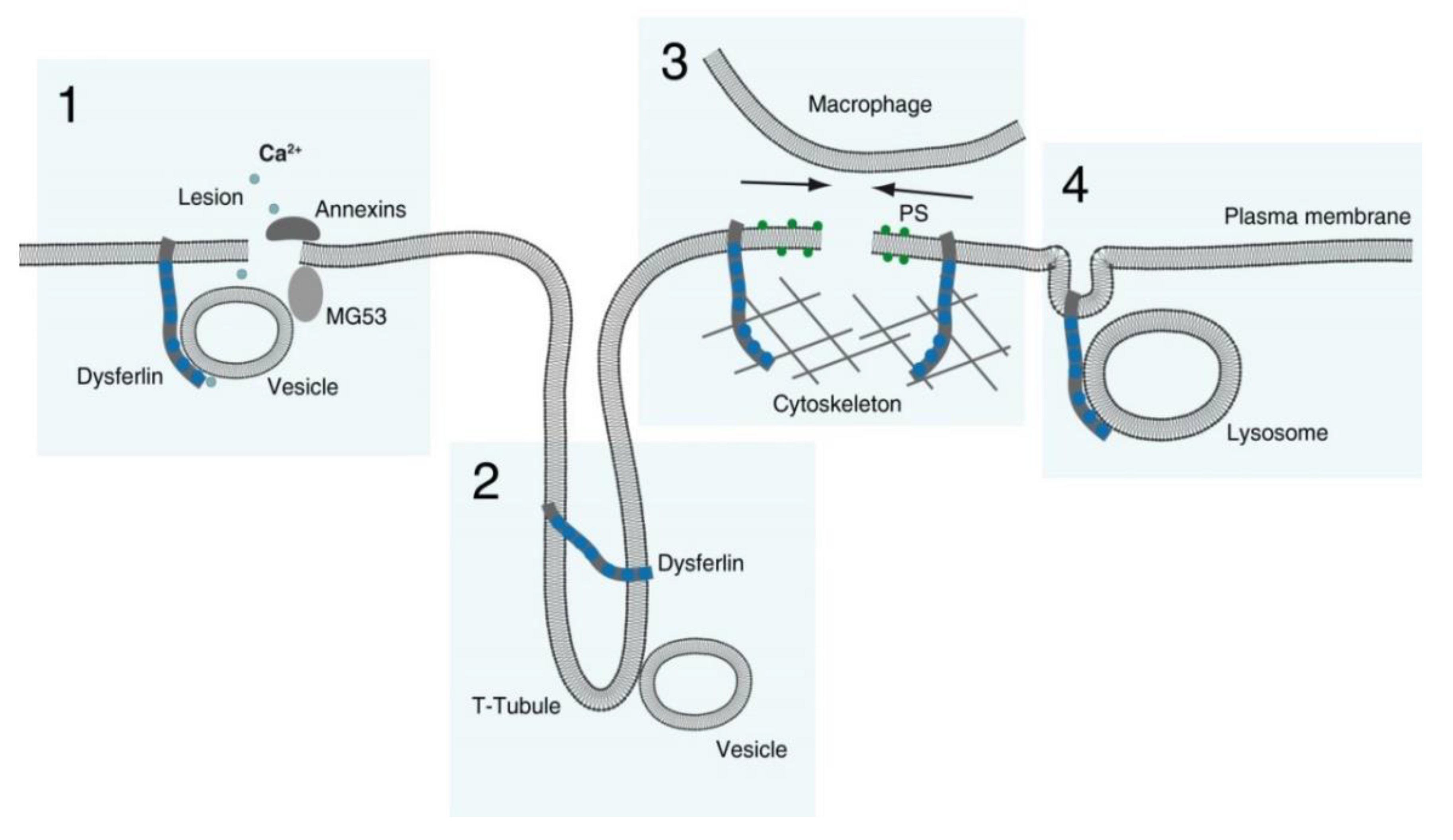
6.1. Dysferlin Functions in Sarcolemma Repair
- Membranous repair patch or plug formation;
- T-tubule stabilization (see below) with T-tubule as a possible membrane reservoir;
- PS-sorting; recruitment of macrophages and contraction of the membrane wound, and;
- Lysosome exocytosis.
6.2. Dysferlin Functions in Triad Biology
6.3. Dysferlin in the Differentiation, Growth and Regeneration of Skeletal Muscle
7. Functions of Myoferlin and Fer1L5
8. Function of Fer1L6
9. Functions of Muscle Ferlins in Non-Muscle Cells
10. Functions of Otoferlin
11. Fer1L4—A Non-Muscle Ferlin Long Non-Coding RNA
12. Mechanism of Action of Ferlins in Membrane Fusion
13. Ferlins in Human Diseases: Dysferlinopathies and Their Pathomechanisms
13.1. Defective Repair of Myofiber Sarcolemma and, Possibly, T-Tubules
13.2. Changes in Muscle Fibers Ca2+ Homeostasis
13.3. Impaired Muscle Growth and Regeneration
13.4. Inflammatory Processes
14. Ferlins in Human Diseases: Otoferlin and Deafness DFNB9
15. Ferlins in Human Diseases: Cancer
16. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Glossary
| Ca2+-sensor for vesicle exocytosis | a protein capable to trigger exocytosis upon binding of calcium ions (Ca2+), but unable to fuse membranes in the absence of proteins—fusogenes. |
| Synaptic vesicle (SV) tethering | primary recruitment of SV to the active zone. |
| SV docking | tighter attachment of SV to the membrane than tethering. |
| SV priming | the sequence of events rendering a SV competent for exocytosis. |
| Inner ear | innermost part of the vertebrate ear, responsible for hearing (cochlea) and sense of balance (vestibular system: semicircular canals, utricle and saccule). |
| Hair cells | cells characterized by a hair bundle and ability to sense and amplify mechanical stimuli (the outer hair cells (OHCs) of the mammalian cochlea) or transducing the latter into chemical signals (e.g., the inner hair cells (IHCs) of the mammalian cochlea). Hair cells are found in the inner ear and in the lateral line of vertebrates. |
| Lateral line | a system of sense organs found in aquatic vertebrates, detecting movement and pressure gradients of surrounding water. |
| Rhabdomyosarcoma | rare, aggressive and malignant pediatric skeletal muscle cancer of immature muscle cells. |
| Ribbon synapse | synapse characterized by presence of a ribbon—presynaptic electron-dense body build up by several scaffolding proteins, tethering a halo of SVs and necessary to, e.g., support large readily releasable pool of SVs. |
| Sarcolemma | surface membrane limiting striated (skeletal and cardiac) muscle fibers. |
| T-tubules | system of transverse tubular extensions of sarcolemma penetrating the interior of striated myofibers and important for propagation of action potentials and synchronization of the fibers’ contraction. |
| Triad | constitutes the anatomical basis for excitation-contraction coupling in striated muscle fibers and is formed by one T-tubule in the middle and two terminal cisternae of sarcoplasmic reticulum contacting it from both sides. |
| A/J mice | mouse model of dysferlinopathy, carrying an ETn retrotransposon insertion in intron 4 of Dysf, which results in the complete lack of dysferlin expression. |
| SJL/J mice | mouse model of dysferlinopathy characterized by residual expression of dysferlin carrying a 57 amino acids deletion in the C2E domain due to a splice site mutation in exon 45. |
References
- Hofhuis, J.; Bersch, K.; Büssenschütt, R.; Drzymalski, M.; Liebetanz, D.; Nikolaev, V.O.; Wagner, S.; Maier, L.S.; Gärtner, J.; Klinge, L.; et al. Dysferlin mediates membrane tubulation and links T-tubule biogenesis to muscular dystrophy. J. Cell. Sci. 2017, 130, 841–852. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.P. Emerging Functional Differences between the Synaptotagmin and Ferlin Calcium Sensor Families. Biochemistry 2017, 56, 6413–6417. [Google Scholar] [CrossRef] [Green Version]
- Pangrsic, T.; Vogl, C. Balancing presynaptic release and endocytic membrane retrieval at hair cell ribbon synapses. FEBS Lett. 2018, 592, 3633–3650. [Google Scholar] [CrossRef]
- Bashir, R.; Britton, S.; Strachan, T.; Keers, S.; Vafiadaki, E.; Lako, M.; Richard, I.; Marchand, S.; Bourg, N.; Argov, Z.; et al. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 1998, 20, 37–42. [Google Scholar] [CrossRef]
- Liu, J.; Aoki, M.; Illa, I.; Wu, C.; Fardeau, M.; Angelini, C.; Serrano, C.; Urtizberea, J.A.; Hentati, F.; Hamida, M.B.; et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 1998, 20, 31–36. [Google Scholar] [CrossRef]
- Yasunaga, S.; Grati, M.; Cohen-Salmon, M.; El-Amraoui, A.; Mustapha, M.; Salem, N.; El-Zir, E.; Loiselet, J.; Petit, C. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat. Genet. 1999, 21, 363–369. [Google Scholar] [CrossRef]
- Kiselev, A.; Vaz, R.; Knyazeva, A.; Sergushichev, A.; Dmitrieva, R.; Khudiakov, A.; Jorholt, J.; Smolina, N.; Sukhareva, K.; Fomicheva, Y.; et al. Truncating Variant in Myof Gene Is Associated With Limb-Girdle Type Muscular Dystrophy and Cardiomyopathy. Front. Genet. 2019, 10, 608. [Google Scholar] [CrossRef]
- Peulen, O.; Rademaker, G.; Anania, S.; Turtoi, A.; Bellahcène, A.; Castronovo, V. Ferlin Overview: From Membrane to Cancer Biology. Cells 2019, 8, E954. [Google Scholar] [CrossRef] [Green Version]
- Lek, A.; Lek, M.; North, K.N.; Cooper, S.T. Phylogenetic analysis of ferlin genes reveals ancient eukaryotic origins. BMC Evol. Biol. 2010, 10, 231. [Google Scholar] [CrossRef] [Green Version]
- Pangršič, T.; Reisinger, E.; Moser, T. Otoferlin: A multi-C2 domain protein essential for hearing. Trends Neurosci. 2012, 35, 671–680. [Google Scholar] [CrossRef]
- Corbalan-Garcia, S.; Gómez-Fernández, J.C. Signaling through C2 domains: More than one lipid target. Biochim. Biophys. Acta 2014, 1838, 1536–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens, S.; McMahon, H.T. Mechanisms of membrane fusion: Disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008, 9, 543–556. [Google Scholar] [CrossRef]
- Butola, T.; Wichmann, C.; Moser, T. Piccolo Promotes Vesicle Replenishment at a Fast Central Auditory Synapse. Front. Synaptic Neurosci. 2017, 9, 14. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Babai, N.; Kaeser, P.; Südhof, T.C.; Schneggenburger, R. RIM1 and RIM2 redundantly determine Ca2+ channel density and readily releasable pool size at a large hindbrain synapse. J. Neurophysiol. 2015, 113, 255–263. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, K.; Yang, X.; Gerber, S.H.; Kwon, H.-B.; Ho, A.; Castillo, P.E.; Liu, X.; Südhof, T.C. Piccolo and bassoon maintain synaptic vesicle clustering without directly participating in vesicle exocytosis. Proc. Natl. Acad. Sci. USA 2010, 107, 6504–6509. [Google Scholar] [CrossRef] [Green Version]
- Camacho, M.; Basu, J.; Trimbuch, T.; Chang, S.; Pulido-Lozano, C.; Chang, S.-S.; Duluvova, I.; Abo-Rady, M.; Rizo, J.; Rosenmund, C. Heterodimerization of Munc13 C2A domain with RIM regulates synaptic vesicle docking and priming. Nat. Commun. 2017, 8, 15293. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.; Choi, U.B.; Leitz, J.; Rhee, H.J.; Lee, C.; Altas, B.; Zhao, M.; Pfuetzner, R.A.; Wang, A.L.; Brose, N.; et al. Molecular Mechanisms of Synaptic Vesicle Priming by Munc13 and Munc18. Neuron 2017, 95, 591–607.e10. [Google Scholar] [CrossRef]
- Luo, F.; Südhof, T.C. Synaptotagmin-7-Mediated Asynchronous Release Boosts High-Fidelity Synchronous Transmission at a Central Synapse. Neuron 2017, 94, 826–839.e3. [Google Scholar] [CrossRef]
- Osterberg, J.R.; Chon, N.L.; Boo, A.; Maynard, F.A.; Lin, H.; Knight, J.D. Membrane Docking of the Synaptotagmin 7 C2A Domain: Electron Paramagnetic Resonance Measurements Show Contributions from Two Membrane Binding Loops. Biochemistry 2015, 54, 5684–5695. [Google Scholar] [CrossRef] [Green Version]
- Parisotto, D.; Malsam, J.; Scheutzow, A.; Krause, J.M.; Söllner, T.H. SNAREpin assembly by Munc18-1 requires previous vesicle docking by synaptotagmin 1. J. Biol. Chem. 2012, 287, 31041–31049. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, P.S.; Houy, S.; Sørensen, J.B. C2-domain containing calcium sensors in neuroendocrine secretion. J. Neurochem. 2016, 139, 943–958. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Khvotchev, M.; Li, Y.C.; Chanaday, N.L.; Kavalali, E.T. Copine-6 Binds to SNAREs and Selectively Suppresses Spontaneous Neurotransmission. J. Neurosci. 2018, 38, 5888–5899. [Google Scholar] [CrossRef]
- Kikuma, K.; Li, X.; Kim, D.; Sutter, D.; Dickman, D.K. Extended Synaptotagmin Localizes to Presynaptic ER and Promotes Neurotransmission and Synaptic Growth in Drosophila. Genetics 2017, 207, 993–1006. [Google Scholar] [CrossRef] [Green Version]
- Genç, Ö.; Dickman, D.K.; Ma, W.; Tong, A.; Fetter, R.D.; Davis, G.W. MCTP is an ER-resident calcium sensor that stabilizes synaptic transmission and homeostatic plasticity. Elife 2017, 6, e22904. [Google Scholar] [CrossRef]
- Roux, I.; Safieddine, S.; Nouvian, R.; Grati, M.; Simmler, M.-C.; Bahloul, A.; Perfettini, I.; Le Gall, M.; Rostaing, P.; Hamard, G.; et al. Otoferlin, defective in a human deafness form, is essential for exocytosis at the auditory ribbon synapse. Cell 2006, 127, 277–289. [Google Scholar] [CrossRef]
- Chatterjee, P.; Padmanarayana, M.; Abdullah, N.; Holman, C.L.; LaDu, J.; Tanguay, R.L.; Johnson, C.P. Otoferlin deficiency in zebrafish results in defects in balance and hearing: Rescue of the balance and hearing phenotype with full-length and truncated forms of mouse otoferlin. Mol. Cell. Biol. 2015, 35, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Saheki, Y.; De Camilli, P. The Extended-Synaptotagmins. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1490–1493. [Google Scholar] [CrossRef]
- Chen, H.; Cheng, Z.-Y.; Pan, Y.; Wang, Z.; Liu, Y.; Zhang, J.-Q. RASAL1 influences the proliferation and invasion of gastric cancer cells by regulating the RAS/ERK signaling pathway. Hum. Cell 2014, 27, 103–110. [Google Scholar] [CrossRef]
- Perestenko, P.; Watanabe, M.; Beusnard-Bee, T.; Guna, P.; McIlhinney, J. The second C2-domain of copine-2, copine-6 and copine-7 is responsible for their calcium-dependent membrane association. FEBS J. 2015, 282, 3722–3736. [Google Scholar] [CrossRef]
- Seo, Y.-M.; Park, S.-J.; Lee, H.-K.; Park, J.-C. Copine-7 binds to the cell surface receptor, nucleolin, and regulates ciliogenesis and Dspp expression during odontoblast differentiation. Sci. Rep. 2017, 7, 11283. [Google Scholar] [CrossRef] [Green Version]
- Margaria, J.P.; Ratto, E.; Gozzelino, L.; Li, H.; Hirsch, E. Class II PI3Ks at the Intersection between Signal Transduction and Membrane Trafficking. Biomolecules 2019, 9, E104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Sun, W.; Ye, G.; Ding, X.; Liu, Z.; Zhang, S.; Xia, T.; Xiao, B.; Xi, Y.; Guo, J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J. Transl. Med. 2013, 11, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redpath, G.M.I.; Sophocleous, R.A.; Turnbull, L.; Whitchurch, C.B.; Cooper, S.T. Ferlins Show Tissue-Specific Expression and Segregate as Plasma Membrane/Late Endosomal or Trans-Golgi/Recycling Ferlins. Traffic 2016, 17, 245–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonventre, J.A.; Holman, C.; Manchanda, A.; Codding, S.J.; Chau, T.; Huegel, J.; Barton, C.; Tanguay, R.; Johnson, C.P. Fer1l6 is essential for the development of vertebrate muscle tissue in zebrafish. Mol. Biol. Cell 2019, 30, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Harsini, F.M.; Chebrolu, S.; Fuson, K.L.; White, M.A.; Rice, A.M.; Sutton, R.B. FerA is a Membrane-Associating Four-Helix Bundle Domain in the Ferlin Family of Membrane-Fusion Proteins. Sci. Rep. 2018, 8, 10949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Padmanarayana, M.; Hams, N.; Speight, L.C.; Petersson, E.J.; Mehl, R.A.; Johnson, C.P. Characterization of the lipid binding properties of Otoferlin reveals specific interactions between PI(4,5)P2 and the C2C and C2F domains. Biochemistry 2014, 53, 5023–5033. [Google Scholar] [CrossRef]
- Johnson, C.P.; Chapman, E.R. Otoferlin is a calcium sensor that directly regulates SNARE-mediated membrane fusion. J. Cell Biol. 2010, 191, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Helfmann, S.; Neumann, P.; Tittmann, K.; Moser, T.; Ficner, R.; Reisinger, E. The crystal structure of the C₂A domain of otoferlin reveals an unconventional top loop region. J. Mol. Biol. 2011, 406, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, N.; Padmanarayana, M.; Marty, N.J.; Johnson, C.P. Quantitation of the calcium and membrane binding properties of the C2 domains of dysferlin. Biophys. J. 2014, 106, 382–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marty, N.J.; Holman, C.L.; Abdullah, N.; Johnson, C.P. The C2 domains of otoferlin, dysferlin, and myoferlin alter the packing of lipid bilayers. Biochemistry 2013, 52, 5585–5592. [Google Scholar] [CrossRef] [Green Version]
- Therrien, C.; Di Fulvio, S.; Pickles, S.; Sinnreich, M. Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry 2009, 48, 2377–2384. [Google Scholar] [CrossRef]
- Xu, L.; Pallikkuth, S.; Hou, Z.; Mignery, G.A.; Robia, S.L.; Han, R. Dysferlin forms a dimer mediated by the C2 domains and the transmembrane domain in vitro and in living cells. PLoS ONE 2011, 6, e27884. [Google Scholar] [CrossRef] [Green Version]
- Evesson, F.J.; Peat, R.A.; Lek, A.; Brilot, F.; Lo, H.P.; Dale, R.C.; Parton, R.G.; North, K.N.; Cooper, S.T. Reduced plasma membrane expression of dysferlin mutants is attributed to accelerated endocytosis via a syntaxin-4-associated pathway. J. Biol. Chem. 2010, 285, 28529–28539. [Google Scholar] [CrossRef] [Green Version]
- Espinoza-Fonseca, L.M. Pathogenic mutation R959W alters recognition dynamics of dysferlin inner DysF domain. Mol Biosyst 2016, 12, 973–981. [Google Scholar] [CrossRef]
- Middel, V.; Zhou, L.; Takamiya, M.; Beil, T.; Shahid, M.; Roostalu, U.; Grabher, C.; Rastegar, S.; Reischl, M.; Nienhaus, G.U.; et al. Dysferlin-mediated phosphatidylserine sorting engages macrophages in sarcolemma repair. Nat Commun 2016, 7, 12875. [Google Scholar] [CrossRef]
- Posey, A.D.; Demonbreun, A.; McNally, E.M. Ferlin proteins in myoblast fusion and muscle growth. Curr. Top. Dev. Biol. 2011, 96, 203–230. [Google Scholar]
- Sampath, S.C.; Sampath, S.C.; Millay, D.P. Myoblast fusion confusion: The resolution begins. Skelet Muscle 2018, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuson, K.; Rice, A.; Mahling, R.; Snow, A.; Nayak, K.; Shanbhogue, P.; Meyer, A.G.; Redpath, G.M.I.; Hinderliter, A.; Cooper, S.T.; et al. Alternate splicing of dysferlin C2A confers Ca2+-dependent and Ca2+-independent binding for membrane repair. Structure 2014, 22, 104–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harsini, F.M.; Bui, A.A.; Rice, A.M.; Chebrolu, S.; Fuson, K.L.; Turtoi, A.; Bradberry, M.; Chapman, E.R.; Sutton, R.B. Structural Basis for the Distinct Membrane Binding Activity of the Homologous C2A Domains of Myoferlin and Dysferlin. J. Mol. Biol. 2019, 431, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Harris, R.; Geddes, S.M.; Strehle, E.-M.; Watson, J.D.; Bashir, R.; Bushby, K.; Driscoll, P.C.; Keep, N.H. Solution structure of the inner DysF domain of myoferlin and implications for limb girdle muscular dystrophy type 2b. J. Mol. Biol. 2008, 379, 981–990. [Google Scholar] [CrossRef] [Green Version]
- Sula, A.; Cole, A.R.; Yeats, C.; Orengo, C.; Keep, N.H. Crystal structures of the human Dysferlin inner DysF domain. BMC Struct. Biol. 2014, 14, 3. [Google Scholar] [CrossRef] [Green Version]
- Anderson, L.V.; Davison, K.; Moss, J.A.; Young, C.; Cullen, M.J.; Walsh, J.; Johnson, M.A.; Bashir, R.; Britton, S.; Keers, S.; et al. Dysferlin is a plasma membrane protein and is expressed early in human development. Hum. Mol. Genet. 1999, 8, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Davis, D.B.; Delmonte, A.J.; Ly, C.T.; McNally, E.M. Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum. Mol. Genet. 2000, 9, 217–226. [Google Scholar] [CrossRef] [Green Version]
- Leung, C.; Shaheen, F.; Bernatchez, P.; Hackett, T.-L. Expression of myoferlin in human airway epithelium and its role in cell adhesion and zonula occludens-1 expression. PLoS ONE 2012, 7, e40478. [Google Scholar] [CrossRef] [Green Version]
- Bernatchez, P.N.; Acevedo, L.; Fernandez-Hernando, C.; Murata, T.; Chalouni, C.; Kim, J.; Erdjument-Bromage, H.; Shah, V.; Gratton, J.-P.; McNally, E.M.; et al. Myoferlin regulates vascular endothelial growth factor receptor-2 stability and function. J. Biol. Chem. 2007, 282, 30745–30753. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.M.; Ackerman, W.E.; Behrendt, N.J.; Vandre, D.D. While dysferlin and myoferlin are coexpressed in the human placenta, only dysferlin expression is responsive to trophoblast fusion in model systems. Biol. Reprod. 2009, 81, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Turtoi, A.; Blomme, A.; Bellahcène, A.; Gilles, C.; Hennequière, V.; Peixoto, P.; Bianchi, E.; Noel, A.; De Pauw, E.; Lifrange, E.; et al. Myoferlin is a key regulator of EGFR activity in breast cancer. Cancer Res. 2013, 73, 5438–5448. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-S.; Liu, X.-H.; Liu, L.-X.; Lou, W.-H.; Jin, D.-Y.; Yang, P.-Y.; Wang, X.-L. iTRAQ-based quantitative proteomics reveals myoferlin as a novel prognostic predictor in pancreatic adenocarcinoma. J Proteomics 2013, 91, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, C.; Hampl, V.; Holzer, K.; Aigner, A.; Penkava, J.; Frank, N.; Martin, D.E.; Maier, K.C.; Waldburger, N.; Roessler, S.; et al. The novel MKL target gene myoferlin modulates expansion and senescence of hepatocellular carcinoma. Oncogene 2017, 36, 3464–3476. [Google Scholar] [CrossRef] [PubMed]
- Schug, N.; Braig, C.; Zimmermann, U.; Engel, J.; Winter, H.; Ruth, P.; Blin, N.; Pfister, M.; Kalbacher, H.; Knipper, M. Differential expression of otoferlin in brain, vestibular system, immature and mature cochlea of the rat. Eur. J. Neurosci. 2006, 24, 3372–3380. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Ren, Z. Overexpression of LncRNA FER1L4 in endometrial carcinoma is associated with favorable survival outcome. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8113–8118. [Google Scholar] [PubMed]
- Ma, W.; Zhang, C.-Q.; Li, H.-L.; Gu, J.; Miao, G.-Y.; Cai, H.-Y.; Wang, J.-K.; Zhang, L.-J.; Song, Y.-M.; Tian, Y.-H.; et al. LncRNA FER1L4 suppressed cancer cell growth and invasion in esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2638–2645. [Google Scholar]
- Gao, X.; Wang, N.; Wu, S.; Cui, H.; An, X.; Yang, Y. Long non-coding RNA FER1L4 inhibits cell proliferation and metastasis through regulation of the PI3K/AKT signaling pathway in lung cancer cells. Mol. Med. Rep. 2019, 20, 182–190. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Zheng, G.; Li, C.; Liu, C. Long non-coding RNA Fer-1-like family member 4 suppresses hepatocellular carcinoma cell proliferation by regulating PTEN in vitro and in vivo. Mol. Med. Rep. 2019, 19, 685–692. [Google Scholar] [CrossRef] [Green Version]
- Posey, A.D.; Pytel, P.; Gardikiotes, K.; Demonbreun, A.R.; Rainey, M.; George, M.; Band, H.; McNally, E.M. Endocytic recycling proteins EHD1 and EHD2 interact with fer-1-like-5 (Fer1L5) and mediate myoblast fusion. J. Biol. Chem. 2011, 286, 7379–7388. [Google Scholar] [CrossRef] [Green Version]
- Millay, D.P.; Maillet, M.; Roche, J.A.; Sargent, M.A.; McNally, E.M.; Bloch, R.J.; Molkentin, J.D. Genetic manipulation of dysferlin expression in skeletal muscle: Novel insights into muscular dystrophy. Am. J. Pathol. 2009, 175, 1817–1823. [Google Scholar] [CrossRef] [Green Version]
- Davis, D.B.; Doherty, K.R.; Delmonte, A.J.; McNally, E.M. Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J. Biol. Chem. 2002, 277, 22883–22888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haslett, J.N.; Sanoudou, D.; Kho, A.T.; Bennett, R.R.; Greenberg, S.A.; Kohane, I.S.; Beggs, A.H.; Kunkel, L.M. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc. Natl. Acad. Sci. USA 2002, 99, 15000–15005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demonbreun, A.R.; Lapidos, K.A.; Heretis, K.; Levin, S.; Dale, R.; Pytel, P.; Svensson, E.C.; McNally, E.M. Myoferlin regulation by NFAT in muscle injury, regeneration and repair. J. Cell. Sci. 2010, 123, 2413–2422. [Google Scholar] [CrossRef] [Green Version]
- Adams, G.R.; Haddad, F.; Bodell, P.W.; Tran, P.D.; Baldwin, K.M. Combined isometric, concentric, and eccentric resistance exercise prevents unloading-induced muscle atrophy in rats. J. Appl. Physiol. 2007, 103, 1644–1654. [Google Scholar] [CrossRef] [PubMed]
- De Luna, N.; Gallardo, E.; Illa, I. In vivo and in vitro dysferlin expression in human muscle satellite cells. J. Neuropathol. Exp. Neurol. 2004, 63, 1104–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belanto, J.J.; Diaz-Perez, S.V.; Magyar, C.E.; Maxwell, M.M.; Yilmaz, Y.; Topp, K.; Boso, G.; Jamieson, C.H.; Cacalano, N.A.; Jamieson, C.A.M. Dexamethasone induces dysferlin in myoblasts and enhances their myogenic differentiation. Neuromuscul. Disord. 2010, 20, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Waddell, L.B.; Lemckert, F.A.; Zheng, X.F.; Tran, J.; Evesson, F.J.; Hawkes, J.M.; Lek, A.; Street, N.E.; Lin, P.; Clarke, N.F.; et al. Dysferlin, annexin A1, and mitsugumin 53 are upregulated in muscular dystrophy and localize to longitudinal tubules of the T-system with stretch. J. Neuropathol. Exp. Neurol. 2011, 70, 302–313. [Google Scholar] [CrossRef] [Green Version]
- Klinge, L.; Harris, J.; Sewry, C.; Charlton, R.; Anderson, L.; Laval, S.; Chiu, Y.-H.; Hornsey, M.; Straub, V.; Barresi, R.; et al. Dysferlin associates with the developing T-tubule system in rodent and human skeletal muscle. Muscle Nerve 2010, 41, 166–173. [Google Scholar] [CrossRef]
- Roche, J.A.; Ru, L.W.; O’Neill, A.M.; Resneck, W.G.; Lovering, R.M.; Bloch, R.J. Unmasking potential intracellular roles for dysferlin through improved immunolabeling methods. J. Histochem. Cytochem. 2011, 59, 964–975. [Google Scholar] [CrossRef] [Green Version]
- Ampong, B.N.; Imamura, M.; Matsumiya, T.; Yoshida, M.; Takeda, S. Intracellular localization of dysferlin and its association with the dihydropyridine receptor. Acta Myol. 2005, 24, 134–144. [Google Scholar]
- Kerr, J.P.; Ziman, A.P.; Mueller, A.L.; Muriel, J.M.; Kleinhans-Welte, E.; Gumerson, J.D.; Vogel, S.S.; Ward, C.W.; Roche, J.A.; Bloch, R.J. Dysferlin stabilizes stress-induced Ca2+ signaling in the transverse tubule membrane. Proc. Natl. Acad. Sci. USA 2013, 110, 20831–20836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDade, J.R.; Archambeau, A.; Michele, D.E. Rapid actin-cytoskeleton-dependent recruitment of plasma membrane-derived dysferlin at wounds is critical for muscle membrane repair. FASEB J. 2014, 28, 3660–3670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, D.; Miyake, K.; Vogel, S.S.; Groh, S.; Chen, C.-C.; Williamson, R.; McNeil, P.L.; Campbell, K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 2003, 423, 168–172. [Google Scholar] [CrossRef]
- Barthélémy, F.; Defour, A.; Lévy, N.; Krahn, M.; Bartoli, M. Muscle Cells Fix Breaches by Orchestrating a Membrane Repair Ballet. J. Neuromuscul. Dis. 2018, 5, 21–28. [Google Scholar] [CrossRef] [Green Version]
- Lennon, N.J.; Kho, A.; Bacskai, B.J.; Perlmutter, S.L.; Hyman, B.T.; Brown, R.H. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 2003, 278, 50466–50473. [Google Scholar] [CrossRef] [Green Version]
- Demonbreun, A.R.; Quattrocelli, M.; Barefield, D.Y.; Allen, M.V.; Swanson, K.E.; McNally, E.M. An actin-dependent annexin complex mediates plasma membrane repair in muscle. J. Cell Biol. 2016, 213, 705–718. [Google Scholar] [CrossRef]
- Hernández-Deviez, D.J.; Howes, M.T.; Laval, S.H.; Bushby, K.; Hancock, J.F.; Parton, R.G. Caveolin regulates endocytosis of the muscle repair protein, dysferlin. J. Biol. Chem. 2008, 283, 6476–6488. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Weisleder, N.; Ko, J.-K.; Komazaki, S.; Sunada, Y.; Nishi, M.; Takeshima, H.; Ma, J. Membrane repair defects in muscular dystrophy are linked to altered interaction between MG53, caveolin-3, and dysferlin. J. Biol. Chem. 2009, 284, 15894–15902. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Laval, S.H.; van Remoortere, A.; Baudier, J.; Benaud, C.; Anderson, L.V.B.; Straub, V.; Deelder, A.; Frants, R.R.; den Dunnen, J.T.; et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 2007, 21, 732–742. [Google Scholar] [CrossRef] [Green Version]
- Park, J.W.; Kim, I.Y.; Choi, J.W.; Lim, H.J.; Shin, J.H.; Kim, Y.N.; Lee, S.H.; Son, Y.; Sohn, M.; Woo, J.K.; et al. AHNAK Loss in Mice Promotes Type II Pneumocyte Hyperplasia and Lung Tumor Development. Mol. Cancer Res. 2018, 16, 1287–1298. [Google Scholar] [CrossRef] [Green Version]
- de Morrée, A.; Hensbergen, P.J.; van Haagen, H.H.H.B.M.; Dragan, I.; Deelder, A.M.; ’t Hoen, P.A.C.; Frants, R.R.; van der Maarel, S.M. Proteomic analysis of the dysferlin protein complex unveils its importance for sarcolemmal maintenance and integrity. PLoS ONE 2010, 5, e13854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuda, C.; Kameyama, K.; Tagawa, K.; Ogawa, M.; Suzuki, A.; Yamaji, S.; Okamoto, H.; Nishino, I.; Hayashi, Y.K. Dysferlin interacts with affixin (beta-parvin) at the sarcolemma. J. Neuropathol. Exp. Neurol. 2005, 64, 334–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flix, B.; de la Torre, C.; Castillo, J.; Casal, C.; Illa, I.; Gallardo, E. Dysferlin interacts with calsequestrin-1, myomesin-2 and dynein in human skeletal muscle. Int. J. Biochem. Cell Biol. 2013, 45, 1927–1938. [Google Scholar] [CrossRef] [PubMed]
- Defour, A.; Van der Meulen, J.H.; Bhat, R.; Bigot, A.; Bashir, R.; Nagaraju, K.; Jaiswal, J.K. Dysferlin regulates cell membrane repair by facilitating injury-triggered acid sphingomyelinase secretion. Cell Death Dis. 2014, 5, e1306. [Google Scholar] [CrossRef] [Green Version]
- Moe, A.M.; Golding, A.E.; Bement, W.M. Cell healing: Calcium, repair and regeneration. Semin. Cell Dev. Biol. 2015, 45, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Roostalu, U.; Strähle, U. In vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell 2012, 22, 515–529. [Google Scholar] [CrossRef] [Green Version]
- Piccolo, F.; Moore, S.A.; Ford, G.C.; Campbell, K.P. Intracellular accumulation and reduced sarcolemmal expression of dysferlin in limb--girdle muscular dystrophies. Ann. Neurol. 2000, 48, 902–912. [Google Scholar] [CrossRef]
- Selcen, D.; Stilling, G.; Engel, A.G. The earliest pathologic alterations in dysferlinopathy. Neurology 2001, 56, 1472–1481. [Google Scholar] [CrossRef]
- McDade, J.R.; Michele, D.E. Membrane damage-induced vesicle-vesicle fusion of dysferlin-containing vesicles in muscle cells requires microtubules and kinesin. Hum. Mol. Genet. 2014, 23, 1677–1686. [Google Scholar] [CrossRef] [Green Version]
- Tam, C.; Idone, V.; Devlin, C.; Fernandes, M.C.; Flannery, A.; He, X.; Schuchman, E.; Tabas, I.; Andrews, N.W. Exocytosis of acid sphingomyelinase by wounded cells promotes endocytosis and plasma membrane repair. J. Cell Biol. 2010, 189, 1027–1038. [Google Scholar] [CrossRef] [Green Version]
- Corrotte, M.; Almeida, P.E.; Tam, C.; Castro-Gomes, T.; Fernandes, M.C.; Millis, B.A.; Cortez, M.; Miller, H.; Song, W.; Maugel, T.K.; et al. Caveolae internalization repairs wounded cells and muscle fibers. Elife 2013, 2, e00926. [Google Scholar] [CrossRef]
- Draeger, A.; Babiychuk, E.B. Ceramide in plasma membrane repair. Handb. Exp. Pharmacol. 2013, 216, 341–353. [Google Scholar]
- Borgonovo, B.; Cocucci, E.; Racchetti, G.; Podini, P.; Bachi, A.; Meldolesi, J. Regulated exocytosis: A novel, widely expressed system. Nat. Cell Biol. 2002, 4, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Klinge, L.; Laval, S.; Keers, S.; Haldane, F.; Straub, V.; Barresi, R.; Bushby, K. From T-tubule to sarcolemma: Damage-induced dysferlin translocation in early myogenesis. FASEB J. 2007, 21, 1768–1776. [Google Scholar] [CrossRef] [PubMed]
- Lek, A.; Evesson, F.J.; Lemckert, F.A.; Redpath, G.M.I.; Lueders, A.-K.; Turnbull, L.; Whitchurch, C.B.; North, K.N.; Cooper, S.T. Calpains, cleaved mini-dysferlinC72, and L-type channels underpin calcium-dependent muscle membrane repair. J. Neurosci. 2013, 33, 5085–5094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, P.; Zhu, H.; Cai, C.; Wang, X.; Cao, C.; Xiao, R.; Pan, Z.; Weisleder, N.; Takeshima, H.; Ma, J. Nonmuscle myosin IIA facilitates vesicle trafficking for MG53-mediated cell membrane repair. FASEB J. 2012, 26, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Boye, T.L.; Maeda, K.; Pezeshkian, W.; Sønder, S.L.; Haeger, S.C.; Gerke, V.; Simonsen, A.C.; Nylandsted, J. Annexin A4 and A6 induce membrane curvature and constriction during cell membrane repair. Nat. Commun. 2017, 8, 1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demonbreun, A.R.; Rossi, A.E.; Alvarez, M.G.; Swanson, K.E.; Deveaux, H.K.; Earley, J.U.; Hadhazy, M.; Vohra, R.; Walter, G.A.; Pytel, P.; et al. Dysferlin and myoferlin regulate transverse tubule formation and glycerol sensitivity. Am. J. Pathol. 2014, 184, 248–259. [Google Scholar] [CrossRef] [Green Version]
- Lukyanenko, V.; Muriel, J.M.; Bloch, R.J. Coupling of excitation to Ca2+ release is modulated by dysferlin. J. Physiol. (Lond.) 2017, 595, 5191–5207. [Google Scholar] [CrossRef] [Green Version]
- Galbiati, F.; Engelman, J.A.; Volonte, D.; Zhang, X.L.; Minetti, C.; Li, M.; Hou, H.; Kneitz, B.; Edelmann, W.; Lisanti, M.P. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J. Biol. Chem. 2001, 276, 21425–21433. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Marcucci, M.; Daniell, L.; Pypaert, M.; Weisz, O.A.; Ochoa, G.-C.; Farsad, K.; Wenk, M.R.; De Camilli, P. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science 2002, 297, 1193–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, G.W.; Mekhedov, E.; Blank, P.S.; de Morree, A.; Pekkurnaz, G.; Nagaraju, K.; Zimmerberg, J. GREG cells, a dysferlin-deficient myogenic mouse cell line. Exp. Cell Res. 2012, 318, 127–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippi, S.; Bigot, A.; Marg, A.; Mouly, V.; Spuler, S.; Zacharias, U. Dysferlin-deficient immortalized human myoblasts and myotubes as a useful tool to study dysferlinopathy. PLoS Curr. 2012, 4, RRN1298. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.A.; McGeachie, J.K.; Grounds, M.D. Cellular differences in the regeneration of murine skeletal muscle: A quantitative histological study in SJL/J and BALB/c mice. Cell Tissue Res. 1992, 269, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Maley, M.A.; Fan, Y.; Beilharz, M.W.; Grounds, M.D. Intrinsic differences in MyoD and myogenin expression between primary cultures of SJL/J and BALB/C skeletal muscle. Exp. Cell Res. 1994, 211, 99–107. [Google Scholar] [CrossRef]
- Ishiba, R.; Santos, A.L.F.; Almeida, C.F.; Caires, L.C.; Ribeiro, A.F.; Ayub-Guerrieri, D.; Fernandes, S.A.; Souza, L.S.; Vainzof, M. Faster regeneration associated to high expression of Fam65b and Hdac6 in dysferlin-deficient mouse. J. Mol. Histol. 2019, 50, 375–387. [Google Scholar] [CrossRef]
- de Luna, N.; Gallardo, E.; Soriano, M.; Dominguez-Perles, R.; de la Torre, C.; Rojas-García, R.; García-Verdugo, J.M.; Illa, I. Absence of dysferlin alters myogenin expression and delays human muscle differentiation “in vitro”. J. Biol. Chem. 2006, 281, 17092–17098. [Google Scholar] [CrossRef] [Green Version]
- Cohen, T.V.; Cohen, J.E.; Partridge, T.A. Myogenesis in dysferlin-deficient myoblasts is inhibited by an intrinsic inflammatory response. Neuromuscul. Disord. 2012, 22, 648–658. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Vashisht, A.A.; O’Rourke, J.; Corbel, S.Y.; Moran, R.; Romero, A.; Miraglia, L.; Zhang, J.; Durrant, E.; Schmedt, C.; et al. The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 2017, 8, 15664. [Google Scholar] [CrossRef] [Green Version]
- Demonbreun, A.R.; Fahrenbach, J.P.; Deveaux, K.; Earley, J.U.; Pytel, P.; McNally, E.M. Impaired muscle growth and response to insulin-like growth factor 1 in dysferlin-mediated muscular dystrophy. Hum. Mol. Genet. 2011, 20, 779–789. [Google Scholar] [CrossRef]
- Zanou, N.; Gailly, P. Skeletal muscle hypertrophy and regeneration: Interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell. Mol. Life Sci. 2013, 70, 4117–4130. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-H.; Hornsey, M.A.; Klinge, L.; Jørgensen, L.H.; Laval, S.H.; Charlton, R.; Barresi, R.; Straub, V.; Lochmüller, H.; Bushby, K. Attenuated muscle regeneration is a key factor in dysferlin-deficient muscular dystrophy. Hum. Mol. Genet. 2009, 18, 1976–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blau, H.M.; Cosgrove, B.D.; Ho, A.T.V. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 2015, 21, 854–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doherty, K.R.; Cave, A.; Davis, D.B.; Delmonte, A.J.; Posey, A.; Earley, J.U.; Hadhazy, M.; McNally, E.M. Normal myoblast fusion requires myoferlin. Development 2005, 132, 5565–5575. [Google Scholar] [CrossRef] [Green Version]
- Demonbreun, A.R.; Posey, A.D.; Heretis, K.; Swaggart, K.A.; Earley, J.U.; Pytel, P.; McNally, E.M. Myoferlin is required for insulin-like growth factor response and muscle growth. FASEB J. 2010, 24, 1284–1295. [Google Scholar] [CrossRef] [Green Version]
- Posey, A.D.; Swanson, K.E.; Alvarez, M.G.; Krishnan, S.; Earley, J.U.; Band, H.; Pytel, P.; McNally, E.M.; Demonbreun, A.R. EHD1 mediates vesicle trafficking required for normal muscle growth and transverse tubule development. Dev. Biol. 2014, 387, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.; Kumar, B.; Lang, J.C.; Teknos, T.N.; Kumar, P. A muscle-specific protein “myoferlin” modulates IL-6/STAT3 signaling by chaperoning activated STAT3 to nucleus. Oncogene 2017, 36, 6374–6382. [Google Scholar] [CrossRef] [Green Version]
- Doherty, K.R.; Demonbreun, A.R.; Wallace, G.Q.; Cave, A.; Posey, A.D.; Heretis, K.; Pytel, P.; McNally, E.M. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J. Biol. Chem. 2008, 283, 20252–20260. [Google Scholar] [CrossRef] [Green Version]
- Lenhart, K.C.; O’Neill, T.J.; Cheng, Z.; Dee, R.; Demonbreun, A.R.; Li, J.; Xiao, X.; McNally, E.M.; Mack, C.P.; Taylor, J.M. GRAF1 deficiency blunts sarcolemmal injury repair and exacerbates cardiac and skeletal muscle pathology in dystrophin-deficient mice. Skelet Muscle 2015, 5, 27. [Google Scholar] [CrossRef] [Green Version]
- Lenhart, K.C.; Becherer, A.L.; Li, J.; Xiao, X.; McNally, E.M.; Mack, C.P.; Taylor, J.M. GRAF1 promotes ferlin-dependent myoblast fusion. Dev. Biol. 2014, 393, 298–311. [Google Scholar] [CrossRef] [Green Version]
- Melo, A.A.; Hegde, B.G.; Shah, C.; Larsson, E.; Isas, J.M.; Kunz, S.; Lundmark, R.; Langen, R.; Daumke, O. Structural insights into the activation mechanism of dynamin-like EHD ATPases. Proc. Natl. Acad. Sci. USA 2017, 114, 5629–5634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, C.; Yu, C.; Lin, M.I.; Tognon, C.; Bernatchez, P. Expression of myoferlin in human and murine carcinoma tumors: Role in membrane repair, cell proliferation, and tumorigenesis. Am. J. Pathol. 2013, 182, 1900–1909. [Google Scholar] [CrossRef] [PubMed]
- Bii, V.M.; Collins, C.P.; Hocum, J.D.; Trobridge, G.D. Replication-incompetent gammaretroviral and lentiviral vector-based insertional mutagenesis screens identify prostate cancer progression genes. Oncotarget 2018, 9, 15451–15463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, W.-Q.; Xia, M.; Xu, M.; Boini, K.M.; Ritter, J.K.; Li, N.-J.; Li, P.-L. Lysosome fusion to the cell membrane is mediated by the dysferlin C2A domain in coronary arterial endothelial cells. J. Cell. Sci. 2012, 125, 1225–1234. [Google Scholar] [CrossRef] [Green Version]
- Fahmy, K.; Gonzalez, A.; Arafa, M.; Peixoto, P.; Bellahcène, A.; Turtoi, A.; Delvenne, P.; Thiry, M.; Castronovo, V.; Peulen, O. Myoferlin plays a key role in VEGFA secretion and impacts tumor-associated angiogenesis in human pancreas cancer. Int. J. Cancer 2016, 138, 652–663. [Google Scholar] [CrossRef]
- Miyatake, Y.; Yamano, T.; Hanayama, R. Myoferlin-Mediated Lysosomal Exocytosis Regulates Cytotoxicity by Phagocytes. J. Immunol. 2018, 201, 3051–3057. [Google Scholar] [CrossRef]
- Bernatchez, P.N.; Sharma, A.; Kodaman, P.; Sessa, W.C. Myoferlin is critical for endocytosis in endothelial cells. Am. J. Physiol. Cell Physiol. 2009, 297, C484–C492. [Google Scholar] [CrossRef] [Green Version]
- Cárdenas, A.M.; González-Jamett, A.M.; Cea, L.A.; Bevilacqua, J.A.; Caviedes, P. Dysferlin function in skeletal muscle: Possible pathological mechanisms and therapeutical targets in dysferlinopathies. Exp. Neurol. 2016, 283, 246–254. [Google Scholar] [CrossRef]
- Nagaraju, K.; Rawat, R.; Veszelovszky, E.; Thapliyal, R.; Kesari, A.; Sparks, S.; Raben, N.; Plotz, P.; Hoffman, E.P. Dysferlin deficiency enhances monocyte phagocytosis: A model for the inflammatory onset of limb-girdle muscular dystrophy 2B. Am. J. Pathol. 2008, 172, 774–785. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Yu, C.; Leung, C.; Trane, A.; Lau, M.; Utokaparch, S.; Shaheen, F.; Sheibani, N.; Bernatchez, P. A new role for the muscle repair protein dysferlin in endothelial cell adhesion and angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 2196–2204. [Google Scholar] [CrossRef] [Green Version]
- de Morrée, A.; Flix, B.; Bagaric, I.; Wang, J.; van den Boogaard, M.; Grand Moursel, L.; Frants, R.R.; Illa, I.; Gallardo, E.; Toes, R.; et al. Dysferlin regulates cell adhesion in human monocytes. J. Biol. Chem. 2013, 288, 14147–14157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dulon, D.; Safieddine, S.; Jones, S.M.; Petit, C. Otoferlin is critical for a highly sensitive and linear calcium-dependent exocytosis at vestibular hair cell ribbon synapses. J. Neurosci. Off. J. Soc. Neurosci. 2009, 29, 10474–10487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safieddine, S.; Wenthold, R.J. SNARE complex at the ribbon synapses of cochlear hair cells: Analysis of synaptic vesicle- and synaptic membrane-associated proteins. Eur. J. Neurosci. 1999, 11, 803–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khimich, D.; Nouvian, R.; Pujol, R.; Tom Dieck, S.; Egner, A.; Gundelfinger, E.D.; Moser, T. Hair cell synaptic ribbons are essential for synchronous auditory signalling. Nature 2005, 434, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Beurg, M.; Michalski, N.; Safieddine, S.; Bouleau, Y.; Schneggenburger, R.; Chapman, E.R.; Petit, C.; Dulon, D. Control of exocytosis by synaptotagmins and otoferlin in auditory hair cells. J. Neurosci. 2010, 30, 13281–13290. [Google Scholar] [CrossRef] [Green Version]
- Gebhart, M.; Juhasz-Vedres, G.; Zuccotti, A.; Brandt, N.; Engel, J.; Trockenbacher, A.; Kaur, G.; Obermair, G.J.; Knipper, M.; Koschak, A.; et al. Modulation of Cav1.3 Ca2+ channel gating by Rab3 interacting molecule. Mol. Cell. Neurosci. 2010, 44, 246–259. [Google Scholar] [CrossRef]
- Johnson, S.L.; Franz, C.; Kuhn, S.; Furness, D.N.; Rüttiger, L.; Münkner, S.; Rivolta, M.N.; Seward, E.P.; Herschman, H.R.; Engel, J.; et al. Synaptotagmin IV determines the linear Ca2+ dependence of vesicle fusion at auditory ribbon synapses. Nat. Neurosci. 2010, 13, 45–52. [Google Scholar] [CrossRef] [Green Version]
- Vogl, C.; Cooper, B.H.; Neef, J.; Wojcik, S.M.; Reim, K.; Reisinger, E.; Brose, N.; Rhee, J.-S.; Moser, T.; Wichmann, C. Unconventional molecular regulation of synaptic vesicle replenishment in cochlear inner hair cells. J. Cell. Sci. 2015, 128, 638–644. [Google Scholar] [CrossRef] [Green Version]
- Pangrsic, T.; Lasarow, L.; Reuter, K.; Takago, H.; Schwander, M.; Riedel, D.; Frank, T.; Tarantino, L.M.; Bailey, J.S.; Strenzke, N.; et al. Hearing requires otoferlin-dependent efficient replenishment of synaptic vesicles in hair cells. Nat. Neurosci. 2010, 13, 869–876. [Google Scholar] [CrossRef]
- Buss, F.; Luzio, J.P.; Kendrick-Jones, J. Myosin VI, an actin motor for membrane traffic and cell migration. Traffic 2002, 3, 851–858. [Google Scholar] [CrossRef]
- Heidrych, P.; Zimmermann, U.; Bress, A.; Pusch, C.M.; Ruth, P.; Pfister, M.; Knipper, M.; Blin, N. Rab8b GTPase, a protein transport regulator, is an interacting partner of otoferlin, defective in a human autosomal recessive deafness form. Hum. Mol. Genet. 2008, 17, 3814–3821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidrych, P.; Zimmermann, U.; Kuhn, S.; Franz, C.; Engel, J.; Duncker, S.V.; Hirt, B.; Pusch, C.M.; Ruth, P.; Pfister, M.; et al. Otoferlin interacts with myosin VI: Implications for maintenance of the basolateral synaptic structure of the inner hair cell. Hum. Mol. Genet. 2009, 18, 2779–2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncker, S.V.; Franz, C.; Kuhn, S.; Schulte, U.; Campanelli, D.; Brandt, N.; Hirt, B.; Fakler, B.; Blin, N.; Ruth, P.; et al. Otoferlin couples to clathrin-mediated endocytosis in mature cochlear inner hair cells. J. Neurosci. 2013, 33, 9508–9519. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Maritzen, T.; Wichmann, C.; Jing, Z.; Neef, A.; Revelo, N.H.; Al-Moyed, H.; Meese, S.; Wojcik, S.M.; Panou, I.; et al. Disruption of adaptor protein 2μ (AP-2μ) in cochlear hair cells impairs vesicle reloading of synaptic release sites and hearing. EMBO J. 2015, 34, 2686–2702. [Google Scholar] [CrossRef]
- Kroll, J.; Jaime Tobón, L.M.; Vogl, C.; Neef, J.; Kondratiuk, I.; König, M.; Strenzke, N.; Wichmann, C.; Milosevic, I.; Moser, T. Endophilin-A regulates presynaptic Ca2+ influx and synaptic vesicle recycling in auditory hair cells. EMBO J. 2019, 38, e100116. [Google Scholar] [CrossRef]
- Strenzke, N.; Chakrabarti, R.; Al-Moyed, H.; Müller, A.; Hoch, G.; Pangrsic, T.; Yamanbaeva, G.; Lenz, C.; Pan, K.-T.; Auge, E.; et al. Hair cell synaptic dysfunction, auditory fatigue and thermal sensitivity in otoferlin Ile515Thr mutants. EMBO J. 2016, 35, 2519–2535. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, R.; Michanski, S.; Wichmann, C. Vesicle sub-pool organization at inner hair cell ribbon synapses. EMBO Rep. 2018, 19, e44937. [Google Scholar] [CrossRef]
- Meese, S.; Cepeda, A.P.; Gahlen, F.; Adams, C.M.; Ficner, R.; Ricci, A.J.; Heller, S.; Reisinger, E.; Herget, M. Activity-Dependent Phosphorylation by CaMKIIδ Alters the Ca2+ Affinity of the Multi-C2-Domain Protein Otoferlin. Front. Synaptic. Neurosci. 2017, 9, 13. [Google Scholar] [CrossRef] [Green Version]
- Takago, H.; Oshima-Takago, T.; Moser, T. Disruption of Otoferlin Alters the Mode of Exocytosis at the Mouse Inner Hair Cell Ribbon Synapse. Front. Mol. Neurosci. 2018, 11, 492. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, L.; Guo, A.; Liu, L.C.; Yu, F.; Diao, N.; Xu, C.; Wang, D. Overexpression of FER1L4 promotes the apoptosis and suppresses epithelial-mesenchymal transition and stemness markers via activating PI3K/AKT signaling pathway in osteosarcoma cells. Pathol. Res. Pract. 2019, 215, 152412. [Google Scholar] [CrossRef]
- Wang, X.; Dong, K.; Jin, Q.; Ma, Y.; Yin, S.; Wang, S. Upregulation of lncRNA FER1L4 suppresses the proliferation and migration of the hepatocellular carcinoma via regulating PI3K/AKT signal pathway. J. Cell. Biochem. 2019, 120, 6781–6788. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Nie, D.; Wang, G.; Sun, C.; Chen, G. FER1L4/miR-372/E2F1 works as a ceRNA system to regulate the proliferation and cell cycle of glioma cells. J. Cell. Mol. Med. 2019, 23, 3224–3233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, Q.; Li, H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem. Biophys. Res. Commun. 2016, 478, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Fei, D.; Zhang, X.; Liu, J.; Tan, L.; Xing, J.; Zhao, D.; Zhang, Y. Long Noncoding RNA FER1L4 Suppresses Tumorigenesis by Regulating the Expression of PTEN Targeting miR-18a-5p in Osteosarcoma. Cell. Physiol. Biochem. 2018, 51, 1364–1375. [Google Scholar] [CrossRef]
- Papa, A.; Pandolfi, P.P. The PTEN−PI3K Axis in Cancer. Biomolecules 2019, 9, 153. [Google Scholar] [CrossRef] [Green Version]
- Xia, T.; Chen, S.; Jiang, Z.; Shao, Y.; Jiang, X.; Li, P.; Xiao, B.; Guo, J. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci. Rep. 2015, 5, 13445. [Google Scholar] [CrossRef]
- Piper, A.-K.; Ross, S.E.; Redpath, G.M.; Lemckert, F.A.; Woolger, N.; Bournazos, A.; Greer, P.A.; Sutton, R.B.; Cooper, S.T. Enzymatic cleavage of myoferlin releases a dual C2-domain module linked to ERK signalling. Cell. Signal. 2017, 33, 30–40. [Google Scholar] [CrossRef]
- Redpath, G.M.I.; Woolger, N.; Piper, A.K.; Lemckert, F.A.; Lek, A.; Greer, P.A.; North, K.N.; Cooper, S.T. Calpain cleavage within dysferlin exon 40a releases a synaptotagmin-like module for membrane repair. Mol. Biol. Cell 2014, 25, 3037–3048. [Google Scholar] [CrossRef]
- Söllner, T.; Whiteheart, S.W.; Brunner, M.; Erdjument-Bromage, H.; Geromanos, S.; Tempst, P.; Rothman, J.E. SNAP receptors implicated in vesicle targeting and fusion. Nature 1993, 362, 318–324. [Google Scholar] [CrossRef]
- Foster, L.J.; Yeung, B.; Mohtashami, M.; Ross, K.; Trimble, W.S.; Klip, A. Binary interactions of the SNARE proteins syntaxin-4, SNAP23, and VAMP-2 and their regulation by phosphorylation. Biochemistry 1998, 37, 11089–11096. [Google Scholar] [CrossRef]
- Codding, S.J.; Marty, N.; Abdullah, N.; Johnson, C.P. Dysferlin Binds SNAREs (Soluble N-Ethylmaleimide-sensitive Factor (NSF) Attachment Protein Receptors) and Stimulates Membrane Fusion in a Calcium-sensitive Manner. J. Biol. Chem. 2016, 291, 14575–14584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, N.A.; Drescher, M.J.; Drescher, D.G. Direct interaction of otoferlin with syntaxin 1A, SNAP-25, and the L-type voltage-gated calcium channel Cav1.3. J. Biol. Chem. 2009, 284, 1364–1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouvian, R.; Neef, J.; Bulankina, A.V.; Reisinger, E.; Pangršič, T.; Frank, T.; Sikorra, S.; Brose, N.; Binz, T.; Moser, T. Exocytosis at the hair cell ribbon synapse apparently operates without neuronal SNARE proteins. Nat. Neurosci. 2011, 14, 411–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reisinger, E.; Bresee, C.; Neef, J.; Nair, R.; Reuter, K.; Bulankina, A.; Nouvian, R.; Koch, M.; Bückers, J.; Kastrup, L.; et al. Probing the functional equivalence of otoferlin and synaptotagmin 1 in exocytosis. J. Neurosci. 2011, 31, 4886–4895. [Google Scholar] [CrossRef] [PubMed]
- Washington, N.L.; Ward, S. FER-1 regulates Ca2+ -mediated membrane fusion during C. elegans spermatogenesis. J. Cell. Sci. 2006, 119, 2552–2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hams, N.; Padmanarayana, M.; Qiu, W.; Johnson, C.P. Otoferlin is a multivalent calcium-sensitive scaffold linking SNAREs and calcium channels. Proc. Natl. Acad. Sci. USA 2017, 114, 8023–8028. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.Y.; Jang, H.; Han, J.H.; Park, H.J.; Lee, J.H.; Kim, S.W.; Kim, S.M.; Park, Y.-E.; Kim, D.-S.; Bang, D.; et al. Targeted next-generation sequencing for the genetic diagnosis of dysferlinopathy. Neuromuscul. Disord. 2015, 25, 502–510. [Google Scholar] [CrossRef]
- Michalski, N.; Goutman, J.D.; Auclair, S.M.; Boutet de Monvel, J.; Tertrais, M.; Emptoz, A.; Parrin, A.; Nouaille, S.; Guillon, M.; Sachse, M.; et al. Otoferlin acts as a Ca2+ sensor for vesicle fusion and vesicle pool replenishment at auditory hair cell ribbon synapses. Elife 2017, 6, e31013. [Google Scholar] [CrossRef] [Green Version]
- Blandin, G.; Beroud, C.; Labelle, V.; Nguyen, K.; Wein, N.; Hamroun, D.; Williams, B.; Monnier, N.; Rufibach, L.E.; Urtizberea, J.A.; et al. UMD-DYSF, a novel locus specific database for the compilation and interactive analysis of mutations in the dysferlin gene. Hum. Mutat. 2012, 33, E2317–E2331. [Google Scholar] [CrossRef]
- Aoki, M. Dysferlinopathy. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Stephens, K., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 2015. [Google Scholar]
- Weiler, T.; Bashir, R.; Anderson, L.V.; Davison, K.; Moss, J.A.; Britton, S.; Nylen, E.; Keers, S.; Vafiadaki, E.; Greenberg, C.R.; et al. Identical mutation in patients with limb girdle muscular dystrophy type 2B or Miyoshi myopathy suggests a role for modifier gene(s). Hum. Mol. Genet. 1999, 8, 871–877. [Google Scholar] [CrossRef] [Green Version]
- Illarioshkin, S.N.; Ivanova-Smolenskaya, I.A.; Greenberg, C.R.; Nylen, E.; Sukhorukov, V.S.; Poleshchuk, V.V.; Markova, E.D.; Wrogemann, K. Identical dysferlin mutation in limb-girdle muscular dystrophy type 2B and distal myopathy. Neurology 2000, 55, 1931–1933. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Matsuzaki, T.; Suehara, M.; Kanzato, N.; Takashima, H.; Higuchi, I.; Matsumura, T.; Goto, K.; Arahata, K.; Osame, M. Phenotypic variation in a large Japanese family with Miyoshi myopathy with nonsense mutation in exon 19 of dysferlin gene. J. Neurol. Sci. 2001, 184, 15–19. [Google Scholar] [CrossRef]
- Urtizberea, J.A.; Bassez, G.; Leturcq, F.; Nguyen, K.; Krahn, M.; Levy, N. Dysferlinopathies. Neurol. India 2008, 56, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Magri, F.; Nigro, V.; Angelini, C.; Mongini, T.; Mora, M.; Moroni, I.; Toscano, A.; D’angelo, M.G.; Tomelleri, G.; Siciliano, G.; et al. The italian limb girdle muscular dystrophy registry: Relative frequency, clinical features, and differential diagnosis. Muscle Nerve 2017, 55, 55–68. [Google Scholar] [CrossRef]
- Fanin, M.; Angelini, C. Progress and challenges in diagnosis of dysferlinopathy. Muscle Nerve 2016, 54, 821–835. [Google Scholar] [CrossRef]
- Hornsey, M.A.; Laval, S.H.; Barresi, R.; Lochmüller, H.; Bushby, K. Muscular dystrophy in dysferlin-deficient mouse models. Neuromuscul. Disord. 2013, 23, 377–387. [Google Scholar] [CrossRef]
- Sellers, S.L.; Milad, N.; White, Z.; Pascoe, C.; Chan, R.; Payne, G.W.; Seow, C.; Rossi, F.; Seidman, M.A.; Bernatchez, P. Increased nonHDL cholesterol levels cause muscle wasting and ambulatory dysfunction in the mouse model of LGMD2B. J. Lipid Res. 2018, 59, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Ho, M.; Post, C.M.; Donahue, L.R.; Lidov, H.G.W.; Bronson, R.T.; Goolsby, H.; Watkins, S.C.; Cox, G.A.; Brown, R.H. Disruption of muscle membrane and phenotype divergence in two novel mouse models of dysferlin deficiency. Hum. Mol. Genet. 2004, 13, 1999–2010. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, K.; Geier, C.; Qadri, F.; Hubner, N.; Schulz, H.; Erdmann, B.; Gross, V.; Bauer, D.; Dechend, R.; Dietz, R.; et al. Dysfunction of dysferlin-deficient hearts. J. Mol. Med. 2007, 85, 1203–1214. [Google Scholar] [CrossRef]
- Nishikawa, A.; Mori-Yoshimura, M.; Segawa, K.; Hayashi, Y.K.; Takahashi, T.; Saito, Y.; Nonaka, I.; Krahn, M.; Levy, N.; Shimizu, J.; et al. Respiratory and cardiac function in japanese patients with dysferlinopathy. Muscle Nerve 2016, 53, 394–401. [Google Scholar] [CrossRef]
- Rosales, X.Q.; Moser, S.J.; Tran, T.; McCarthy, B.; Dunn, N.; Habib, P.; Simonetti, O.P.; Mendell, J.R.; Raman, S.V. Cardiovascular magnetic resonance of cardiomyopathy in limb girdle muscular dystrophy 2B and 2I. J. Cardiovasc. Magn. Reson. Off. J. Soc. Cardiovasc. Magn. Reson. 2011, 13, 39. [Google Scholar] [CrossRef] [Green Version]
- Rubi, L.; Gawali, V.S.; Kubista, H.; Todt, H.; Hilber, K.; Koenig, X. Proper Voltage-Dependent Ion Channel Function in Dysferlin-Deficient Cardiomyocytes. Cell Physiol. Biochem. 2015, 36, 1049–1058. [Google Scholar] [CrossRef]
- Chase, T.H.; Cox, G.A.; Burzenski, L.; Foreman, O.; Shultz, L.D. Dysferlin deficiency and the development of cardiomyopathy in a mouse model of limb-girdle muscular dystrophy 2B. Am. J. Pathol. 2009, 175, 2299–2308. [Google Scholar] [CrossRef] [Green Version]
- Kitmitto, A.; Baudoin, F.; Cartwright, E.J. Cardiomyocyte damage control in heart failure and the role of the sarcolemma. J. Muscle Res. Cell. Motil. 2019, 40, 319–333. [Google Scholar] [CrossRef] [Green Version]
- Han, R.; Bansal, D.; Miyake, K.; Muniz, V.P.; Weiss, R.M.; McNeil, P.L.; Campbell, K.P. Dysferlin-mediated membrane repair protects the heart from stress-induced left ventricular injury. J. Clin. Invest. 2007, 117, 1805–1813. [Google Scholar] [CrossRef]
- Tzeng, H.-P.; Evans, S.; Gao, F.; Chambers, K.; Topkara, V.K.; Sivasubramanian, N.; Barger, P.M.; Mann, D.L. Dysferlin mediates the cytoprotective effects of TRAF2 following myocardial ischemia reperfusion injury. J. Am. Heart Assoc. 2014, 3, e000662. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Wei, H.; Jin, J.-P. Dysferlin deficiency blunts β-adrenergic-dependent lusitropic function of mouse heart. J. Physiol. 2015, 593, 5127–5144. [Google Scholar] [CrossRef] [Green Version]
- Lemckert, F.A.; Bournazos, A.; Eckert, D.M.; Kenzler, M.; Hawkes, J.M.; Butler, T.L.; Ceely, B.; North, K.N.; Winlaw, D.S.; Egan, J.R.; et al. Lack of MG53 in human heart precludes utility as a biomarker of myocardial injury or endogenous cardioprotective factor. Cardiovasc. Res. 2016, 110, 178–187. [Google Scholar] [CrossRef]
- Wang, C.; Wong, J.; Fung, G.; Shi, J.; Deng, H.; Zhang, J.; Bernatchez, P.; Luo, H. Dysferlin deficiency confers increased susceptibility to coxsackievirus-induced cardiomyopathy. Cell. Microbiol. 2015, 17, 1423–1430. [Google Scholar] [CrossRef]
- Fujita, E.; Kouroku, Y.; Isoai, A.; Kumagai, H.; Misutani, A.; Matsuda, C.; Hayashi, Y.K.; Momoi, T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: Ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Hum. Mol. Genet. 2007, 16, 618–629. [Google Scholar] [CrossRef]
- Voigt, T.; Sebald, H.-J.; Schoenauer, R.; Levano, S.; Girard, T.; Hoppeler, H.H.; Babiychuk, E.B.; Draeger, A. Annexin A1 is a biomarker of T-tubular repair in skeletal muscle of nonmyopathic patients undergoing statin therapy. FASEB J. 2013, 27, 2156–2164. [Google Scholar] [CrossRef]
- Hogarth, M.W.; Defour, A.; Lazarski, C.; Gallardo, E.; Diaz Manera, J.; Partridge, T.A.; Nagaraju, K.; Jaiswal, J.K. Fibroadipogenic progenitors are responsible for muscle loss in limb girdle muscular dystrophy 2B. Nat. Commun. 2019, 10, 2430. [Google Scholar] [CrossRef] [Green Version]
- Zhivotovsky, B.; Orrenius, S. Calcium and cell death mechanisms: A perspective from the cell death community. Cell Calcium. 2011, 50, 211–221. [Google Scholar] [CrossRef]
- Prosser, B.L.; Khairallah, R.J.; Ziman, A.P.; Ward, C.W.; Lederer, W.J. X-ROS signaling in the heart and skeletal muscle: Stretch-dependent local ROS regulates [Ca2+]i. J. Mol. Cell. Cardiol. 2013, 58, 172–181. [Google Scholar] [CrossRef] [Green Version]
- Kombairaju, P.; Kerr, J.P.; Roche, J.A.; Pratt, S.J.P.; Lovering, R.M.; Sussan, T.E.; Kim, J.-H.; Shi, G.; Biswal, S.; Ward, C.W. Genetic silencing of Nrf2 enhances X-ROS in dysferlin-deficient muscle. Front. Physiol. 2014, 5, 57. [Google Scholar] [CrossRef] [Green Version]
- Beringer, A.; Gouriou, Y.; Lavocat, F.; Ovize, M.; Miossec, P. Blockade of Store-Operated Calcium Entry Reduces IL-17/TNF Cytokine-Induced Inflammatory Response in Human Myoblasts. Front. Immunol. 2018, 9, 3170. [Google Scholar] [CrossRef] [Green Version]
- McNally, E.M.; Ly, C.T.; Rosenmann, H.; Mitrani Rosenbaum, S.; Jiang, W.; Anderson, L.V.; Soffer, D.; Argov, Z. Splicing mutation in dysferlin produces limb-girdle muscular dystrophy with inflammation. Am. J. Med. Genet. 2000, 91, 305–312. [Google Scholar] [CrossRef]
- Yin, X.; Wang, Q.; Chen, T.; Niu, J.; Ban, R.; Liu, J.; Mao, Y.; Pu, C. CD4+ cells, macrophages, MHC-I and C5b-9 involve the pathogenesis of dysferlinopathy. Int. J. Clin. Exp. Pathol. 2015, 8, 3069–3075. [Google Scholar]
- Mariano, A.; Henning, A.; Han, R. Dysferlin-deficient muscular dystrophy and innate immune activation. FEBS J. 2013, 280, 4165–4176. [Google Scholar] [CrossRef]
- Defour, A.; Medikayala, S.; Van der Meulen, J.H.; Hogarth, M.W.; Holdreith, N.; Malatras, A.; Duddy, W.; Boehler, J.; Nagaraju, K.; Jaiswal, J.K. Annexin A2 links poor myofiber repair with inflammation and adipogenic replacement of the injured muscle. Hum. Mol. Genet. 2017, 26, 1979–1991. [Google Scholar] [CrossRef] [Green Version]
- Wenzel, K.; Zabojszcza, J.; Carl, M.; Taubert, S.; Lass, A.; Harris, C.L.; Ho, M.; Schulz, H.; Hummel, O.; Hubner, N.; et al. Increased susceptibility to complement attack due to down-regulation of decay-accelerating factor/CD55 in dysferlin-deficient muscular dystrophy. J. Immunol. (Baltim. Md. 1950) 2005, 175, 6219–6225. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, C.A.; Grounds, M.D.; Papadimitriou, J.M. The genotype of bone marrow-derived inflammatory cells does not account for differences in skeletal muscle regeneration between SJL/J and BALB/c mice. Cell Tissue Res. 1995, 280, 407–413. [Google Scholar] [CrossRef]
- Roche, J.A.; Tulapurkar, M.E.; Mueller, A.L.; van Rooijen, N.; Hasday, J.D.; Lovering, R.M.; Bloch, R.J. Myofiber damage precedes macrophage infiltration after in vivo injury in dysferlin-deficient A/J mouse skeletal muscle. Am. J. Pathol. 2015, 185, 1686–1698. [Google Scholar] [CrossRef]
- McElhanon, K.E.; Bhattacharya, S. Altered membrane integrity in the progression of muscle diseases. Life Sci. 2018, 192, 166–172. [Google Scholar] [CrossRef]
- Lostal, W.; Bartoli, M.; Roudaut, C.; Bourg, N.; Krahn, M.; Pryadkina, M.; Borel, P.; Suel, L.; Roche, J.A.; Stockholm, D.; et al. Lack of correlation between outcomes of membrane repair assay and correction of dystrophic changes in experimental therapeutic strategy in dysferlinopathy. PLoS ONE 2012, 7, e38036. [Google Scholar] [CrossRef]
- Santarelli, R.; Del Castillo, I.; Rodríguez-Ballesteros, M.; Scimemi, P.; Cama, E.; Arslan, E.; Starr, A. Abnormal cochlear potentials from deaf patients with mutations in the otoferlin gene. J. Assoc. Res. Otolaryngol. 2009, 10, 545–556. [Google Scholar] [CrossRef] [Green Version]
- Shearer, A.E.; Smith, R.J. OTOF-Related Deafness. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Stephens, K., Amemiya, A., Eds.; University of Washington, Seattle: Seattle, WA, USA, 1993. [Google Scholar]
- Varga, R.; Kelley, P.M.; Keats, B.J.; Starr, A.; Leal, S.M.; Cohn, E.; Kimberling, W.J. Non-syndromic recessive auditory neuropathy is the result of mutations in the otoferlin (OTOF) gene. J. Med. Genet. 2003, 40, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Varga, R.; Avenarius, M.R.; Kelley, P.M.; Keats, B.J.; Berlin, C.I.; Hood, L.J.; Morlet, T.G.; Brashears, S.M.; Starr, A.; Cohn, E.S.; et al. OTOF mutations revealed by genetic analysis of hearing loss families including a potential temperature sensitive auditory neuropathy allele. J. Med. Genet. 2006, 43, 576–581. [Google Scholar] [CrossRef] [Green Version]
- Marlin, S.; Feldmann, D.; Nguyen, Y.; Rouillon, I.; Loundon, N.; Jonard, L.; Bonnet, C.; Couderc, R.; Garabedian, E.N.; Petit, C.; et al. Temperature-sensitive auditory neuropathy associated with an otoferlin mutation: Deafening fever! Biochem. Biophys. Res. Commun. 2010, 394, 737–742. [Google Scholar] [CrossRef]
- Matsunaga, T.; Mutai, H.; Kunishima, S.; Namba, K.; Morimoto, N.; Shinjo, Y.; Arimoto, Y.; Kataoka, Y.; Shintani, T.; Morita, N.; et al. A prevalent founder mutation and genotype-phenotype correlations of OTOF in Japanese patients with auditory neuropathy. Clin. Genet. 2012, 82, 425–432. [Google Scholar] [CrossRef]
- Zhang, Q.; Lan, L.; Shi, W.; Yu, L.; Xie, L.-Y.; Xiong, F.; Zhao, C.; Li, N.; Yin, Z.; Zong, L.; et al. Temperature sensitive auditory neuropathy. Hear. Res. 2016, 335, 53–63. [Google Scholar] [CrossRef]
- Iwasa, Y.-I.; Nishio, S.-Y.; Sugaya, A.; Kataoka, Y.; Kanda, Y.; Taniguchi, M.; Nagai, K.; Naito, Y.; Ikezono, T.; Horie, R.; et al. OTOF mutation analysis with massively parallel DNA sequencing in 2,265 Japanese sensorineural hearing loss patients. PLoS ONE 2019, 14, e0215932. [Google Scholar] [CrossRef]
- Rodríguez-Ballesteros, M.; del Castillo, F.J.; Martín, Y.; Moreno-Pelayo, M.A.; Morera, C.; Prieto, F.; Marco, J.; Morant, A.; Gallo-Terán, J.; Morales-Angulo, C.; et al. Auditory neuropathy in patients carrying mutations in the otoferlin gene (OTOF). Hum. Mutat. 2003, 22, 451–456. [Google Scholar] [CrossRef]
- Gordon, C.J. The mouse thermoregulatory system: Its impact on translating biomedical data to humans. Physiol. Behav. 2017, 179, 55–66. [Google Scholar] [CrossRef]
- Eisenberg, M.C.; Kim, Y.; Li, R.; Ackerman, W.E.; Kniss, D.A.; Friedman, A. Mechanistic modeling of the effects of myoferlin on tumor cell invasion. Proc. Natl. Acad. Sci. USA 2011, 108, 20078–20083. [Google Scholar] [CrossRef] [Green Version]
- Sussman, J.; Stokoe, D.; Ossina, N.; Shtivelman, E. Protein kinase B phosphorylates AHNAK and regulates its subcellular localization. J. Cell Biol. 2001, 154, 1019–1030. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.H.; Sohn, M.; Lim, H.J.; Yoon, S.; Oh, H.; Shin, S.; Shin, J.H.; Oh, S.-H.; Kim, J.; Lee, D.K.; et al. Ahnak functions as a tumor suppressor via modulation of TGFβ/Smad signaling pathway. Oncogene 2014, 33, 4675–4684. [Google Scholar] [CrossRef]
- Yagami-Hiromasa, T.; Sato, T.; Kurisaki, T.; Kamijo, K.; Nabeshima, Y.; Fujisawa-Sehara, A. A metalloprotease-disintegrin participating in myoblast fusion. Nature 1995, 377, 652–656. [Google Scholar] [CrossRef]
- Mochizuki, S.; Okada, Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007, 98, 621–628. [Google Scholar] [CrossRef]
- Zhou, Y.; Xiong, L.; Zhang, Y.; Yu, R.; Jiang, X.; Xu, G. Quantitative proteomics identifies myoferlin as a novel regulator of A Disintegrin and Metalloproteinase 12 in HeLa cells. J. Proteom. 2016, 148, 94–104. [Google Scholar] [CrossRef]
- Gillies, A.R.; Lieber, R.L. Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve 2011, 44, 318–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Ackerman, W.E.; Mihai, C.; Volakis, L.I.; Ghadiali, S.; Kniss, D.A. Myoferlin depletion in breast cancer cells promotes mesenchymal to epithelial shape change and stalls invasion. PLoS ONE 2012, 7, e39766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sher, R.B.; Cox, G.A.; Mills, K.D.; Sundberg, J.P. Rhabdomyosarcomas in aging A/J mice. PLoS ONE 2011, 6, e23498. [Google Scholar] [CrossRef]

| Domain | Amino acids 1 | Mutations 2 | % Mutations 3 |
|---|---|---|---|
| C2A | 101 | 3 | 3.0 |
| C2A-C2B ICR | 121 | 6 | 5.0 |
| C2B | 96 | 12 | 12.5 |
| FerI | 71 | 4 | 5.6 |
| C2C | 114 | 9 | 7.9 |
| C2C-FerA ICR | 200 | 7 | 3.5 |
| FerA | 65 | 1 | 1.5 |
| FerB | 74 | 4 | 5.4 |
| DysF | 227 | 21 | 9.3 |
| C2D | 108 | 2 | 1.9 |
| C2D-C2E ICR | 77 | 2 | 2.6 |
| C2E | 99 | 7 | 7.1 |
| C2E-C2F ICR | 142 | 7 | 4.9 |
| C2F | 99 | 6 | 6.1 |
| C2F-C2G ICR | 134 | 12 | 9.0 |
| C2G | 129 | 12 | 9.3 |
| C2G-TM ICR | 104 | 6 | 5.8 |
| TM | 23 | 1 | 4.4 |
| Extracellular domain | 15 | 2 | 13.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulankina, A.V.; Thoms, S. Functions of Vertebrate Ferlins. Cells 2020, 9, 534. https://doi.org/10.3390/cells9030534
Bulankina AV, Thoms S. Functions of Vertebrate Ferlins. Cells. 2020; 9(3):534. https://doi.org/10.3390/cells9030534
Chicago/Turabian StyleBulankina, Anna V., and Sven Thoms. 2020. "Functions of Vertebrate Ferlins" Cells 9, no. 3: 534. https://doi.org/10.3390/cells9030534





