Analysis of Killer Immunoglobulin-Like Receptor Genes in Colorectal Cancer
Abstract
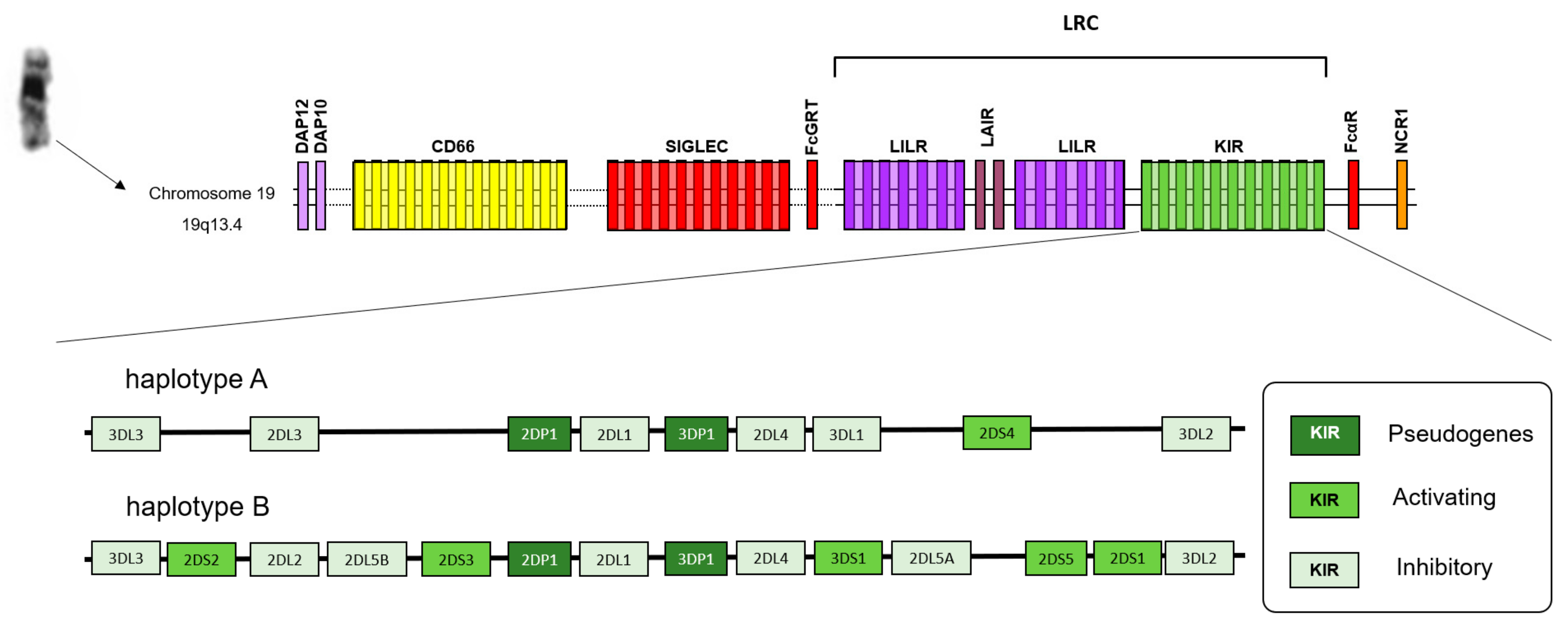
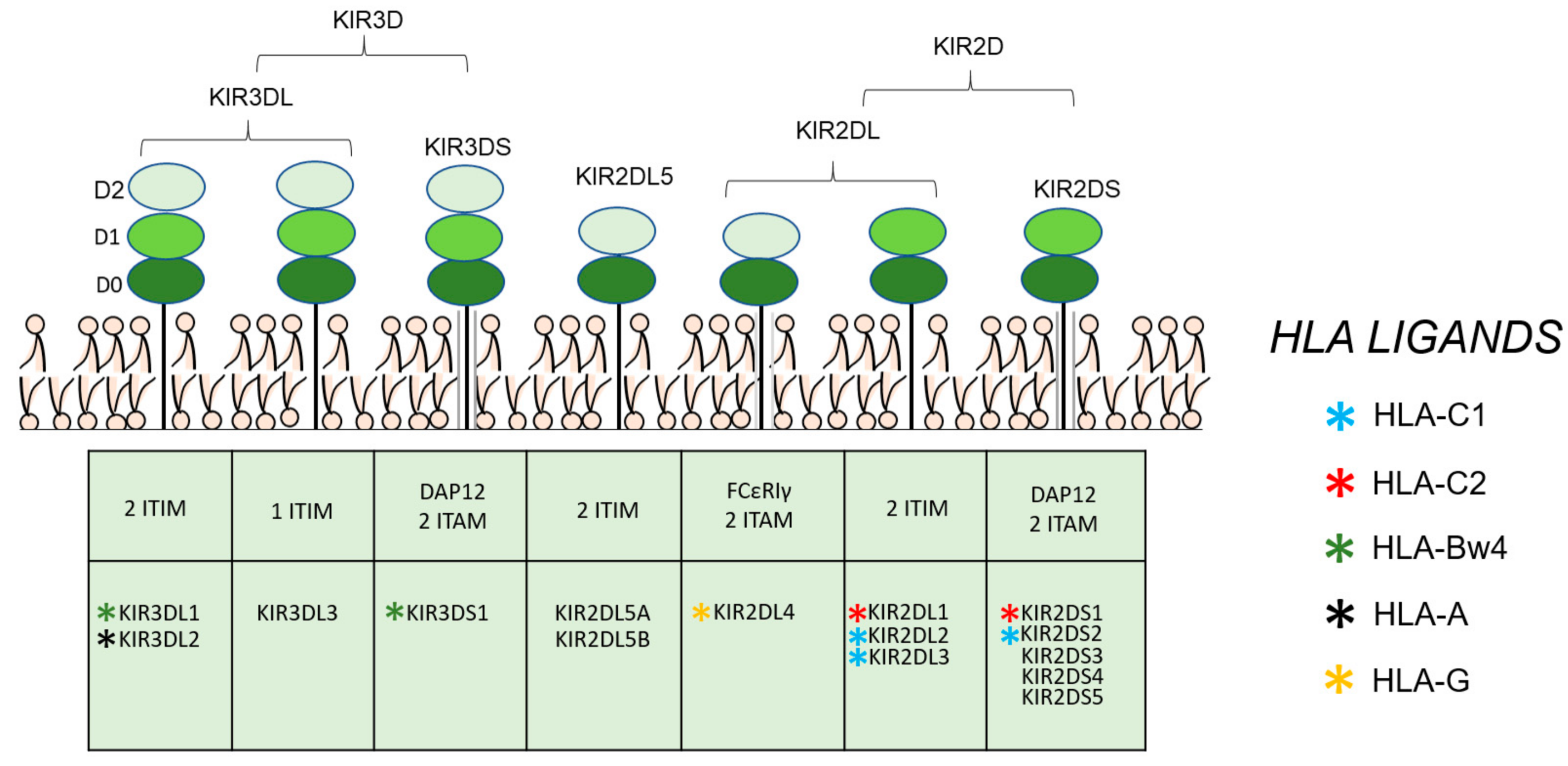
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Genotyping and Imputation
2.3. Statistical Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet (Lond. Engl.) 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Schmit, S.L.; Edlund, C.K.; Schumacher, F.R.; Gong, J.; Harrison, T.A.; Huyghe, J.R.; Qu, C.; Melas, M.; Van Den Berg, D.J.; Wang, H.; et al. Novel Common Genetic Susceptibility Loci for Colorectal Cancer. J. Natl. Cancer Inst. 2019, 111, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Law, P.J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; Farrington, S.; Svinti, V.; Palles, C.; Orlando, G.; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat. Commun. 2019, 10, 2154. [Google Scholar] [CrossRef] [PubMed]
- Guillerey, C.; Huntington, N.D.; Smyth, M.J. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 2016, 17, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Parham, P.; Guethlein, L.A. Genetics of Natural Killer Cells in Human Health, Disease, and Survival. Annu. Rev. Immunol. 2018, 36. [Google Scholar] [CrossRef]
- Lanier, L.L.; Cortiss, B.C.; Wu, J.; Leong, C.; Phillips, J.H. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 1998, 391, 703–707. [Google Scholar] [CrossRef]
- Olcese, L.; Cambiaggi, A.; Semenzato, G.; Bottino, C.; Moretta, A.; Vivier, E. Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by natural killer cells. J. Immunol. 1997, 158, 5083–5086. [Google Scholar]
- Shahjahan Miah, S.M.; Hughes, T.L.; Campbell, K.S. KIR2DL4 Differentially Signals Downstream Functions in Human NK Cells through Distinct Structural Modules. J. Immunol. 2008, 180, 2922–2932. [Google Scholar] [CrossRef]
- Winter, C.C.; Gumperz, J.E.; Parham, P.; Long, E.O.; Wagtmann, N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 1998, 161, 571–577. [Google Scholar]
- Cella, M.; Longo, A.; Ferrara, G.B.; Strominger, J.L.; Colonna, M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 1994, 180, 1235–1242. [Google Scholar] [CrossRef]
- Hansasuta, P.; Dong, T.; Thananchai, H.; Weekes, M.; Willberg, C.; Aldemir, H.; Rowland-Jones, S.; Braud, V.M. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur. J. Immunol. 2004. [Google Scholar] [CrossRef] [PubMed]
- Augusto, D.G. The impact of KIR polymorphism on the risk of developing cancer: Not as strong as imagined? Front. Genet. 2016, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Menon, A.G.; Morreau, H.; Tollenaar, R.A.E.M.; Alphenaar, E.; Van Puijenbroek, M.; Putter, H.; Janssen-Van Rhijn, C.M.; Van De Velde, C.J.H.; Fleuren, G.J.; Kuppen, P.J.K. Down-regulation of HLA-A expression correlates with a better prognosis in colorectal cancer patients. Lab. Investig. A J. Tech. Methods Pathol. 2002, 82, 1725–1733. [Google Scholar] [CrossRef]
- Watson, N.F.S.; Ramage, J.M.; Madjd, Z.; Spendlove, I.; Ellis, I.O.; Scholefield, J.H.; Durrant, L.G. Immunosurveillance is active in colorectal cancer as downregulation but not complete loss of MHC class I expression correlates with a poor prognosis. Int. J. Cancer 2006, 118, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Shen, J.; Cox, C.; Wakefield, J.C.; Ehm, M.G.; Nelson, M.R.; Weir, B.S. HIBAG—HLA genotype imputation with attribute bagging. Pharm. J. 2014, 14, 192–200. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, J.; Gurdasani, D.; Delaneau, O.; Pirastu, N.; Ulivi, S.; Cocca, M.; Traglia, M.; Huang, J.; Huffman, J.E.; Rudan, I.; et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014, 10, e1004234. [Google Scholar] [CrossRef] [PubMed]
- Vukcevic, D.; Traherne, J.A.; Næss, S.; Ellinghaus, E.; Kamatani, Y.; Dilthey, A.; Lathrop, M.; Karlsen, T.H.; Franke, A.; Moffatt, M.; et al. Imputation of KIR Types from SNP Variation Data. Am. J. Hum. Genet. 2015, 97, 593–607. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Fan, Y.; Song, Y.-Q. PyHLA: Tests for the association between HLA alleles and diseases. BMC Bioinform. 2017, 18, 90. [Google Scholar] [CrossRef]
- Middleton, D.; Gonzelez, F. The extensive polymorphism of KIR genes. Immunology 2010, 129, 8–19. [Google Scholar] [CrossRef]
- Middleton, D.; Vilchez, J.R.; Cabrera, T.; Meenagh, A.; Williams, F.; Halfpenny, I.; Maleno, I.; Ruiz-Cabello, F.; Lopez-Nevot, M.A.; Garrido, F. Analysis of KIR gene frequencies in HLA class I characterised bladder, colorectal and laryngeal tumours. Tissue Antigens 2007, 69, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Al Omar, S.; Middleton, D.; Marshall, E.; Porter, D.; Xinarianos, G.; Raji, O.; Field, J.K.; Christmas, S.E. Associations between genes for killer immunoglobulin-like receptors and their ligands in patients with solid tumors. Hum. Immunol. 2010, 71, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Choi, H.B.; Jang, J.P.; Baek, I.C.; Choi, E.J.; Park, M.; Kim, T.G.; Oh, S.T. HLA-Cw polypmorphism and killer cell immunoglobulin-like receptor (KIR) gene analysis in Korean colorectal cancer patients. Int. J. Surg. 2014, 12, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Al Omar, S.Y.; Mansour, L.; Dar, J.A.; Alwasel, S.; Alkhuriji, A.; Arafah, M.; Al Obeed, O.; Christmas, S. The Relationship between Killer Cell Immunoglobulin-Like Receptors and HLA-C Polymorphisms in Colorectal Cancer in a Saudi Population. Genet. Test. Mol. Biomark. 2015, 19, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Ghanadi, K.; Shayanrad, B.; Ahmadi, S.A.Y.; Shahsavar, F.; Eliasy, H. Colorectal cancer and the KIR genes in the human genome: A meta-analysis. Genom. Data 2016, 10, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Portela, P.; Merzoni, J.; Lindenau, J.D.; Damin, D.C.; Wilson, T.J.; Roesler, R.; Schwartsmann, G.; Jobim, L.F.; Jobim, M. KIR genes and HLA class I ligands in a Caucasian Brazilian population with colorectal cancer. Hum. Immunol. 2017, 78, 263–268. [Google Scholar] [CrossRef]
- Barani, S.; Hosseini, S.V.; Ghaderi, A. Activating and inhibitory killer cell immunoglobulin like receptors (KIR) genes are involved in an increased susceptibility to colorectal adenocarcinoma and protection against invasion and metastasis. Immunobiology 2019, 224, 681–686. [Google Scholar] [CrossRef]
- Beksac, K.; Beksac, M.; Dalva, K.; Karaagaoglu, E.; Tirnaksiz, M.B. Impact of “Killer Immunoglobulin-Like Receptor /Ligand” Genotypes on Outcome following Surgery among Patients with Colorectal Cancer: Activating KIRs Are Associated with Long-Term Disease Free Survival. PLoS One 2015, 10, e0132526. [Google Scholar] [CrossRef]
- Dring, M.M.; Morrison, M.H.; McSharry, B.P.; Guinan, K.J.; Hagan, R.; O’Farrelly, C.; Gardiner, C.M. Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc. Natl. Acad. Sci. United States Am. 2011, 108, 5736–5741. [Google Scholar] [CrossRef]
- Wauquier, N.; Padilla, C.; Becquart, P.; Leroy, E.; Vieillard, V. Association of KIR2DS1 and KIR2DS3 with fatal outcome in Ebola virus infection. Immunogenetics 2010, 62, 767–771. [Google Scholar] [CrossRef]
- VandenBussche, C.J.; Mulrooney, T.J.; Frazier, W.R.; Dakshanamurthy, S.; Hurley, C.K. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 2009, 10, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Sconocchia, G.; Eppenberger-Castori, S.; Zlobec, I.; Karamitopoulou, E.; Arriga, R.; Coppola, A.; Caratelli, S.; Spagnoli, G.C.; Lauro, D.; Lugli, A.; et al. HLA class II antigen expression in colorectal carcinoma tumors as a favorable prognostic marker. Neoplasia (United States) 2014, 16, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Garrity-Park, M.M.; Loftus, E.V.; Sandborn, W.J.; Bryant, S.C.; Smyrk, T.C. MHC class II alleles in ulcerative colitis-associated colorectal cancer. Gut 2009, 58, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Aureli, A.; Canossi, A.; Del Beato, T.; Franceschilli, L.; Buonomo, O.; Papola, F.; De Sanctis, F.; Lanzilli, G.; Sileri, P.; Coppola, A.; et al. HLA-DRB1∗13:01 allele in the genetic susceptibility to colorectal carcinoma. Int. J. Cancer 2015, 136, 2464–2468. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, S.; Cariappa, A.; Tang, M.; Bell, D.; Haber, D.A.; Isselbacher, K.J.; Finkelstein, D.; Forcione, D.; Pillai, S. Genetic susceptibility to breast cancer: HLA DQB*03032 and HLA DRB1*11 may represent protective alleles. Proc. Natl. Acad. Sci. United States Am. 2000, 97, 11451–11454. [Google Scholar] [CrossRef]
- Arons, E.; Adams, S.; Venzon, D.J.; Pastan, I.; Kreitman, R.J. Class II human leucocyte antigen DRB1*11 in hairy cell leukaemia patients with and without haemolytic uraemic syndrome. Br. J. Haematol. 2014, 166, 729–738. [Google Scholar] [CrossRef]
- Meyer, D.; Vitor, V.R.; Bitarello, B.D.; Débora, D.Y.; Nunes, K. A genomic perspective on HLA evolution. Immunogenetics 2018, 70, 5–27. [Google Scholar] [CrossRef]
| CRC Patients (n = 943) | Healthy Controls (n = 1076) | |
|---|---|---|
| Age (median, range) | 69 (23–91) | 64.15 (24–92) |
| Gender Female Male | 338 (35.8%) 605 (64.2%) | 527 (48.98%) 549 (51.02%) |
| Smoke | ||
| Current smoker Former smoker Never smoker Unknown | 119 (12.6%) 386 (40.9%) 413 (43.8%) 26 (2.8%) | 180 (16.73%) 335 (31.13%) 553 (51.39%) 8 (0.74%) |
| TNM stage at diagnosis | ||
| I–II III IV Unknown | 418 (44.3%) 342 (36.3%) 131 (13.9%) 52 (5.5%) | |
| Tumor location | ||
| Rectum Sigma Cecum Ascending colon Sigmoid colon Descending colon Others* Unknown | 311 (33%) 168 (17.8%) 98 (10.4%) 81 (8.6%) 70 (7.4%) 52 (5.5%) 152 (16.1%) 11 (1.2%) | |
| Chr | SNP | Position | Associated Gene | A1 | MAF CRC Cases | MAF Healthy Controls | p Value | OR |
|---|---|---|---|---|---|---|---|---|
| 6 | rs16896742 | 29922740 | Intergenic | G | 0.31 | 0.3669 | 3.97 × 10−4 | 0.79 |
| 6 | rs28367832 | 31305731 | Intergenic | A | 0.42 | 0.4845 | 6.57 × 10−5 | 0.77 |
| 6 | rs9277952 | 33204274 | Intergenic | A | 0.15 | 0.1097 | 4.01 × 10−5 | 1.47 |
| HLA Allele | AF CRC Cases | AF Healthy Controls | P value | OR | 95% CI | Padj |
|---|---|---|---|---|---|---|
| DRB1*11:01 | 0.1002 | 0.0785 | 0.0137 | 1.32 | 1.06–1.65 | 0.0961 |
| C*07:01 | 0.1304 | 0.1543 | 0.0321 | 0.82 | 0.69–0.98 | 0.1774 |
| C*12:03 | 0.0689 | 0.0534 | 0.0443 | 1.30 | 1.01–1.67 | 0.1774 |
| A*02:01 | 0.2264 | 0.2519 | 0.0576 | 0.87 | 0.75–1.00 | 0.3453 |
| A*29:02 | 0.0848 | 0.0730 | 0.1554 | 1.18 | 0.94–1.50 | 0.4662 |
| DRB1*04:01 | 0.0467 | 0.0562 | 0.1751 | 0.82 | 0.62–1.09 | 0.6128 |
| B*35:01 | 0.0583 | 0.0511 | 0.3096 | 1.15 | 0.88–1.52 | 0.8689 |
| C*16:01 | 0.0843 | 0.0757 | 0.3136 | 1.13 | 0.89–1.42 | 0.6060 |
| B*44:03 | 0.1071 | 0.0976 | 0.3240 | 1.11 | 0.90–1.35 | 0.8689 |
| C*07:02 | 0.0817 | 0.0897 | 0.3641 | 0.90 | 0.72–1.13 | 0.6060 |
| Healthy Controls | CRC Cases | ||||||
|---|---|---|---|---|---|---|---|
| KIR Gene | n | % | n | % | p-value * | Pbonf | OR (95% CI) |
| KIR2DL1 | 1075 | 99.91 | 942 | 99.89 | NS | NS | |
| KIR2DL2 | 490 | 45.54 | 418 | 44.33 | NS | NS | |
| KIR2DL3 | 925 | 85.97 | 818 | 86.74 | NS | NS | |
| KIR2DL4 | 1076 | 100 | 943 | 100 | NS | NS | |
| KIR2DL5 | 500 | 46.47 | 450 | 47.72 | NS | NS | |
| KIR3DL1ex4 | 1030 | 95.72 | 911 | 96.61 | NS | NS | |
| KIR3DL1ex9 | 1032 | 95.91 | 911 | 96.61 | NS | NS | |
| KIR3DL2 | 1076 | 100 | 943 | 100 | NS | NS | |
| KIR2DS1 | 386 | 35.87 | 346 | 36.69 | NS | NS | |
| KIR2DS2 | 530 | 49.26 | 480 | 50.9 | NS | NS | |
| KIR2DS3 | 350 | 32.53 | 370 | 39.24 | 0.002 | 0.036 | 1.34 (1.12–1.61) |
| KIR2DS4total | 1030 | 95.72 | 909 | 96.39 | NS | NS | |
| KIR2DS4wt | 366 | 34.01 | 306 | 32.45 | NS | NS | |
| KIR2DS4del | 905 | 84.11 | 801 | 84.94 | NS | NS | |
| KIR2DS5 | 286 | 26.58 | 257 | 27.25 | NS | NS | |
| KIR3DS1 | 337 | 31.32 | 319 | 33.83 | NS | NS | |
| KIR2DP1 | 1074 | 99.81 | 943 | 100 | NS | NS | |
| KIR3DP1 | 1076 | 100 | 943 | 100 | NS | NS | |
| KIR Genotype | n | % | n | % | p-value * | OR (95% CI) | |
| AA | 280 | 26.02 | 243 | 26.77 | NS | ||
| Bx | 796 | 73.98 | 700 | 74.23 | NS | ||
| Healthy Controls | CRC Cases | |||||
|---|---|---|---|---|---|---|
| HLA Ligands | N | % | n | % | p-Value | OR (95% CI) |
| Bw4 | 726 | 67.47 | 650 | 68.93 | NS | |
| Bw6 | 885 | 82.25 | 766 | 81.23 | NS | |
| Bw4/Bw4 | 191 | 17.75 | 177 | 18.77 | NS | |
| Bw4/Bw6 | 535 | 49.72 | 473 | 50.16 | NS | |
| Bw6/Bw6 | 350 | 32.53 | 293 | 31.07 | NS | |
| Bw4I80 | 416 | 38.66 | 361 | 38.28 | NS | |
| Bw4T80 | 402 | 37.36 | 373 | 39.55 | NS | |
| HLA-C1 | 890 | 82.71 | 752 | 79.75 | NS | |
| HLA-C2 | 725 | 67.38 | 640 | 67.87 | NS | |
| HLA-C1C1 | 351 | 32.62 | 303 | 32.13 | NS | |
| HLA-C1C2 | 539 | 50.09 | 449 | 47.61 | NS | |
| HLA-C2C2 | 186 | 17.29 | 191 | 20.25 | NS | |
| KIR genotypes | N | % | n | % | p-value | OR (95% CI) |
| KIR3DL1/KIR3DL1 | 738 | 68.59 | 624 | 66.17 | NS | |
| KIR3DL1/KIR3DS1 | 294 | 27.32 | 287 | 30.43 | NS | |
| KIR3DS1/KIR3DS1 | 43 | 4.00 | 32 | 3.39 | NS | |
| KIR2DL2/KIR2DL2 | 149 | 13.85 | 125 | 13.26 | NS | |
| KIR2DL2/KIR2DL3 | 341 | 31.69 | 293 | 31.07 | NS | |
| KIR2DL3/KIR2DL3 | 584 | 54.28 | 525 | 55.67 | NS | |
| NS | ||||||
| KIR ligand associations | N | % | n | % | p-value | OR (95% CI) |
| KIR3DS1-Bw4I80 | 131 | 12.17 | 114 | 12.09 | NS | |
| KIR3DS1-Bw4T80 | 125 | 11.62 | 123 | 13.04 | NS | |
| KIR3DS1-Bw4 | 228 | 21.19 | 206 | 21.85 | NS | |
| KIR3DS1-Bw6 | 285 | 26.49 | 264 | 27.99 | NS | |
| KIR3DL1-Bw4I80 | 402 | 37.36 | 349 | 37.01 | NS | |
| KIR3DL1-Bw4T80 | 386 | 35.87 | 356 | 37.75 | NS | |
| KIR3DL1-Bw4 | 697 | 64.78 | 626 | 66.38 | NS | |
| KIR3DL1-Bw6 | 847 | 78.72 | 742 | 78.69 | NS | |
| KIR2DL1/HLA-C1C1 | 351 | 32.62 | 302 | 32.03 | NS | |
| KIR2DL1/HLA-C1C2 | 538 | 50.00 | 449 | 47.61 | NS | |
| KIR2DL1/HLA-C2C2 | 186 | 17.29 | 191 | 20.25 | NS | |
| KIR2DL2/HLA-C1C1 | 149 | 13.85 | 125 | 13.26 | NS | |
| KIR2DL2/HLA-C1C2 | 189 | 17.57 | 153 | 16.22 | NS | |
| KIR2DL2/HLA-C2C2 | 152 | 14.13 | 140 | 14.85 | NS | |
| KIR2DL3/HLA-C1C1 | 339 | 31.51 | 297 | 31.50 | NS | |
| KIR2DL3/HLA-C1C2 | 403 | 37.45 | 337 | 35.74 | NS | |
| KIR2DL3/HLA-C2C2 | 183 | 17.01 | 184 | 19.51 | NS | |
| KIR2DS1/HLA-C1C1 | 115 | 10.69 | 102 | 10.82 | NS | |
| KIR2DS1/HLA-C1C2 | 194 | 18.03 | 175 | 18.56 | NS | |
| KIR2DS1/HLA-C2C2 | 77 | 7.16 | 69 | 7.32 | NS | |
| Reference | Type of Experiment/Objective | Conclusions |
|---|---|---|
| [19] | 109 CRC patients (70 bladder and 34 laryngeal) and 100 controls. HLA and KIR genotyping. | No differences in KIR/HLA frequencies was observed between patients and controls. |
| [20] | 128 CRC patients and 255 controls. KIR and HLA genotyping. | The data showed no significant differences between KIR gene frequencies in CRC patients versus controls. |
| [21] | 241 CRC patients and 159 controls from Korean populations. KIR and HLA-C genotyping | The activating KIR2DS5 was more frequent in Korean CRC patients, showing a risk for the disease. The frequencies of KIR3DL1, KIR2DS2 and KIR2DS4 were lower in the rectal cancer subgroup, and they could have a protective effect against CRC. Also, the lower frequency of KIR2DS2 in patients with HLA-C1 homozygote, may be a protective effect too. |
| [22] | 52 CRC patients and 70 controls from Saudi population. KIR and HLA-C genotyping. | Activating KIRs (2DS1, 2DS2, 2DS3, 2DS5 and 3DS1) was more frequent in CRC patients, suggesting their presence a risk for disease. |
| [23] | 470 CRC patients and 483 controls. KIR genotyping. | The presence of KIR2DS5 was associated with CRC like as a non-protective gene. This result explains the inflammatory basis of this cancer. |
| [24] | 154 CRC patients and 216 controls from Caucasian Brazilian population. KIR and HLA genotyping. | No associations between KIRs and HLA in CRC was observed. However, the Bx haplotype was more frequent in controls than in patients, being a possible mechanism of protection to CRC. |
| [25] | 165 colorectal adenocarcinoma patients and 165 controls. KIR genotyping. | The presence of activating KIRs (≥ 4) and KIR3DL1, 3DS1, 2DS1 and 2DS4, were associated with protection against metastasis in CRC patients. |
| [26] | 29 CRC recurrent patients (in 5 years) vs. 58 CRC non-recurrent patients (in 5 years) after surgery and 154 controls. KIR and HLA-class I genotyping. | The increment of activating KIRs (in particularly 2DS2 and 2DS3) and the lack of inhibitory KIRs (in particularly 2DL1) was associated with long term disease-free survival and this was independent of tumor localization or stage. Also, HLA-A-Bw4 was associated with recurrent disease. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diaz-Peña, R.; Mondelo-Macía, P.; Molina de la Torre, A.J.; Sanz-Pamplona, R.; Moreno, V.; Martín, V. Analysis of Killer Immunoglobulin-Like Receptor Genes in Colorectal Cancer. Cells 2020, 9, 514. https://doi.org/10.3390/cells9020514
Diaz-Peña R, Mondelo-Macía P, Molina de la Torre AJ, Sanz-Pamplona R, Moreno V, Martín V. Analysis of Killer Immunoglobulin-Like Receptor Genes in Colorectal Cancer. Cells. 2020; 9(2):514. https://doi.org/10.3390/cells9020514
Chicago/Turabian StyleDiaz-Peña, Roberto, Patricia Mondelo-Macía, Antonio José Molina de la Torre, Rebeca Sanz-Pamplona, Víctor Moreno, and Vicente Martín. 2020. "Analysis of Killer Immunoglobulin-Like Receptor Genes in Colorectal Cancer" Cells 9, no. 2: 514. https://doi.org/10.3390/cells9020514
APA StyleDiaz-Peña, R., Mondelo-Macía, P., Molina de la Torre, A. J., Sanz-Pamplona, R., Moreno, V., & Martín, V. (2020). Analysis of Killer Immunoglobulin-Like Receptor Genes in Colorectal Cancer. Cells, 9(2), 514. https://doi.org/10.3390/cells9020514







