An Alternative Splice Variant of HIPK2 with Intron Retention Contributes to Cytokinesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source and Bioinformatic Tools
2.2. Cells and Reagents
2.3. Patient Specimens
2.4. RNA Purification, Reverse Transcriptase RT-PCR, and Quantitative Real-Time qPCR Analyses
2.5. Engineering of Expression Vectors and Transfection
2.6. RNA Interference
2.7. Immunoprecipitation and In Vitro Kinase Assay
2.8. Mono-Dimensional and 2D Gel Electrophoresis and Western Blotting
2.9. Immunofluorescence Microscopy
2.10. Live-Cell Imaging
2.11. Anti-HIPK2-S Rabbit Polyclonal Ab Production and Purification
2.12. Statistics
3. Results
3.1. Detection in Human Cells of the Predicted HIPK2 Alternative Splice Variant with Intron 13 Retention
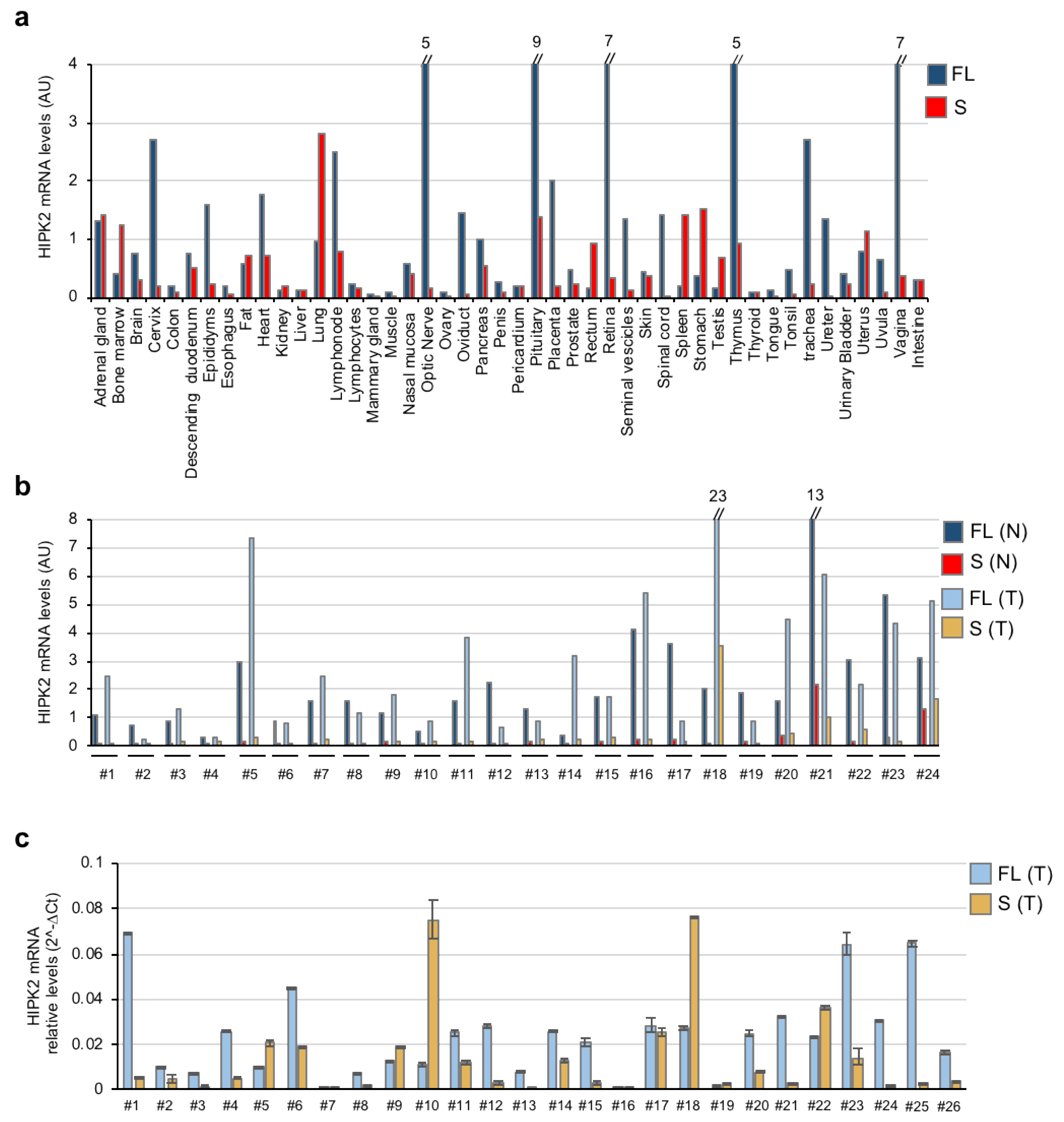
3.2. Tissue and Tumor Expression of HIPK2-FL and HIPK2-S Isoforms
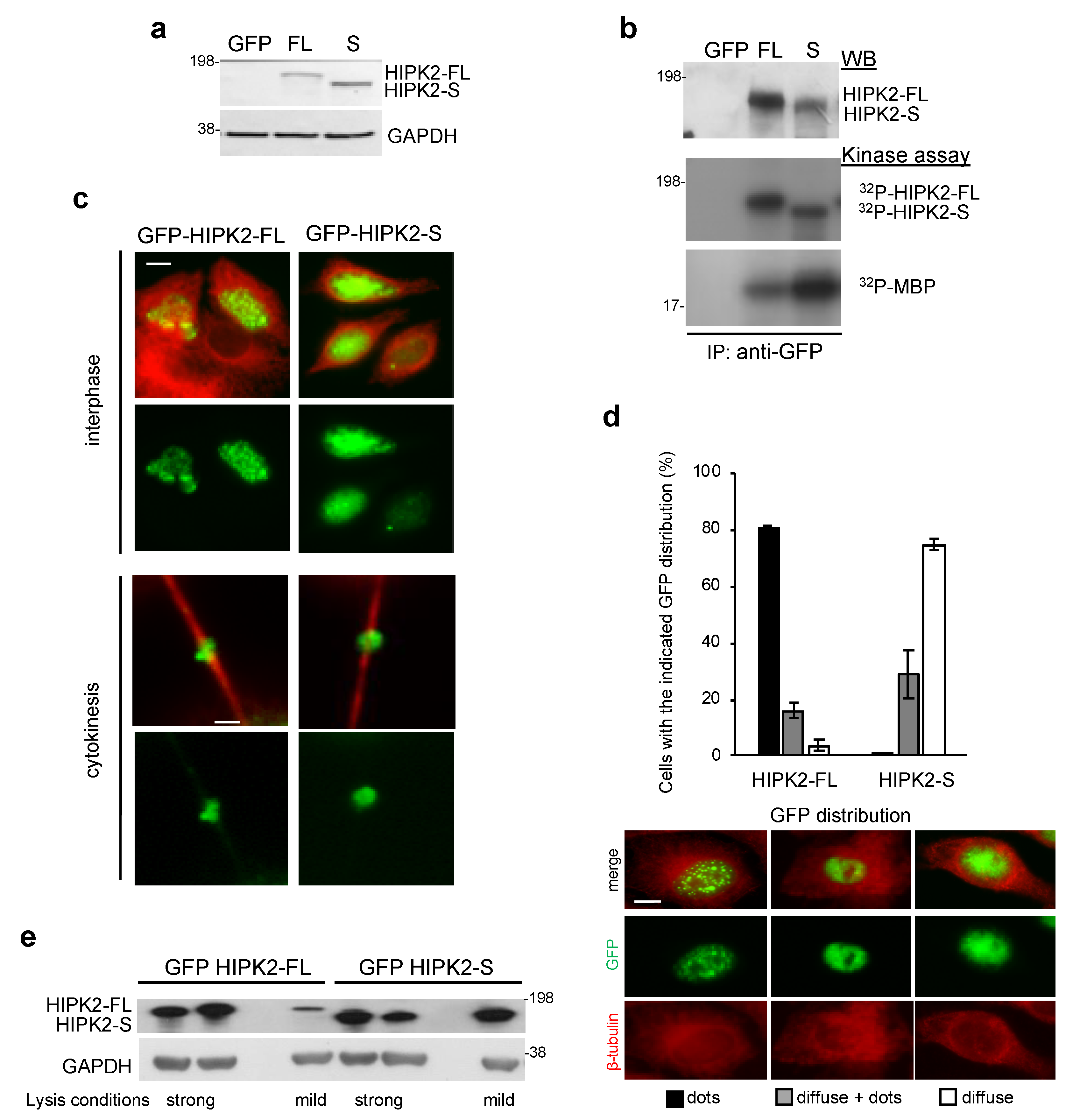
3.3. Cloning and Expression of the HIPK2-S Isoform
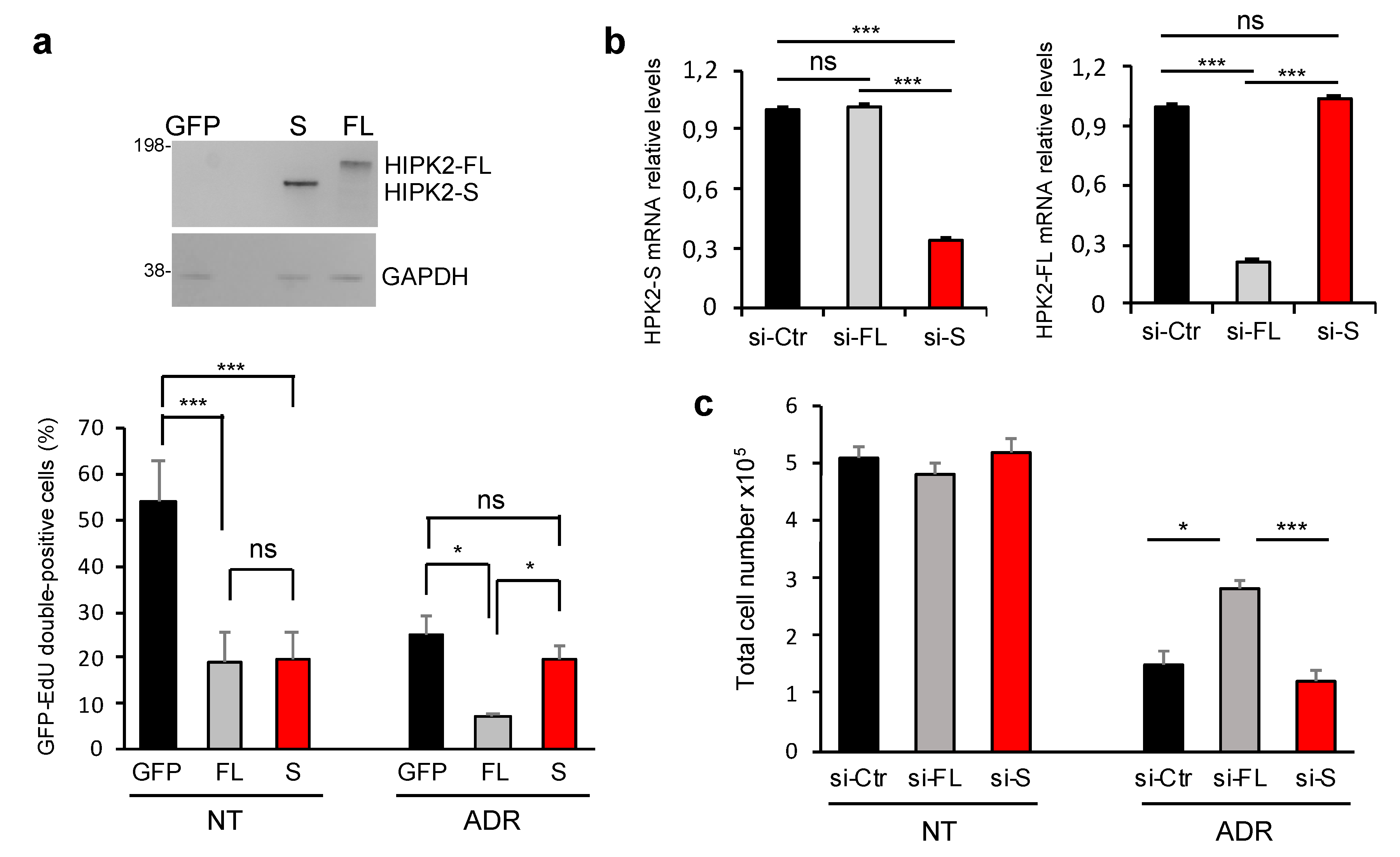
3.4. HIPK2-S Isoform Is Dispensable for DNA Damage Response
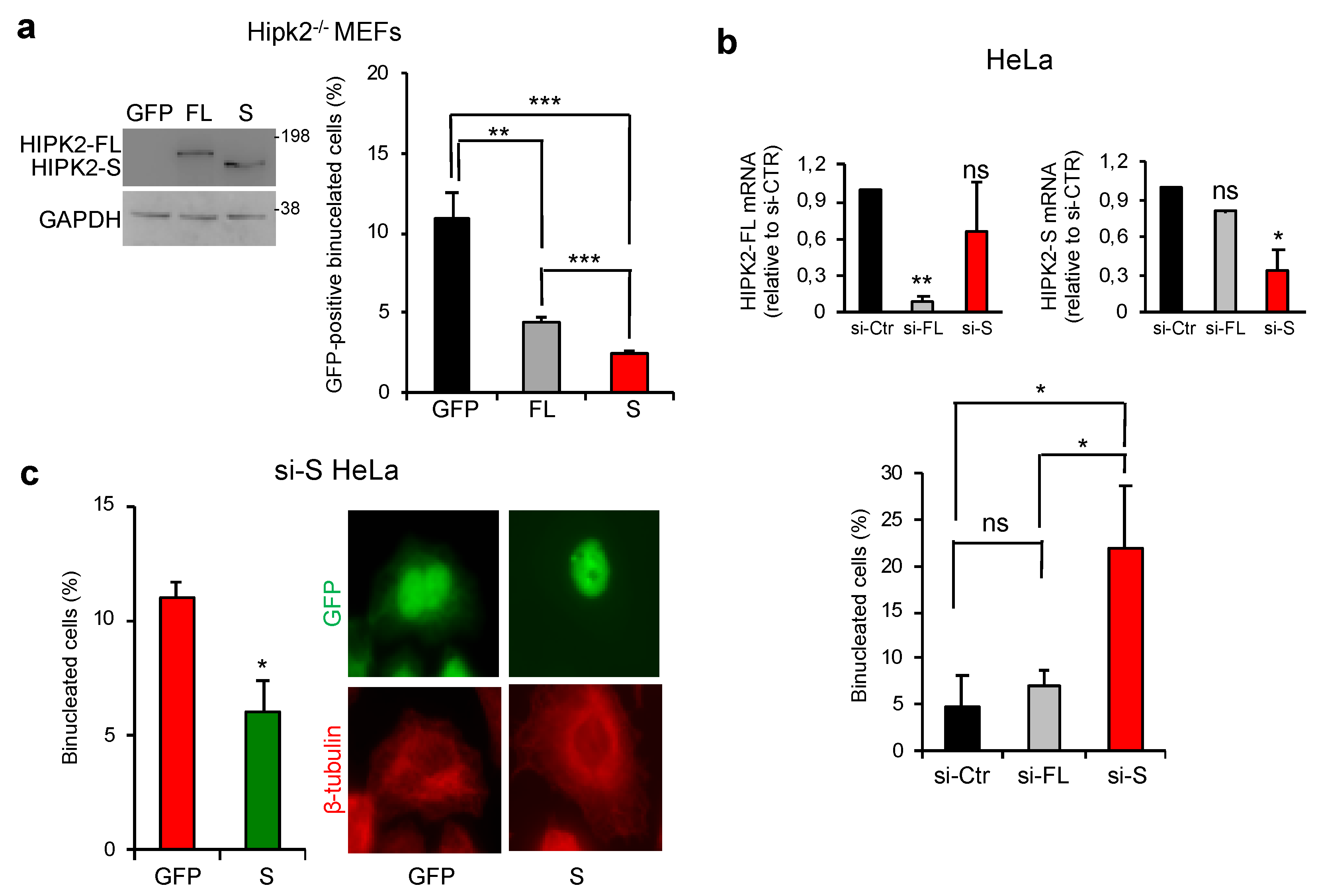
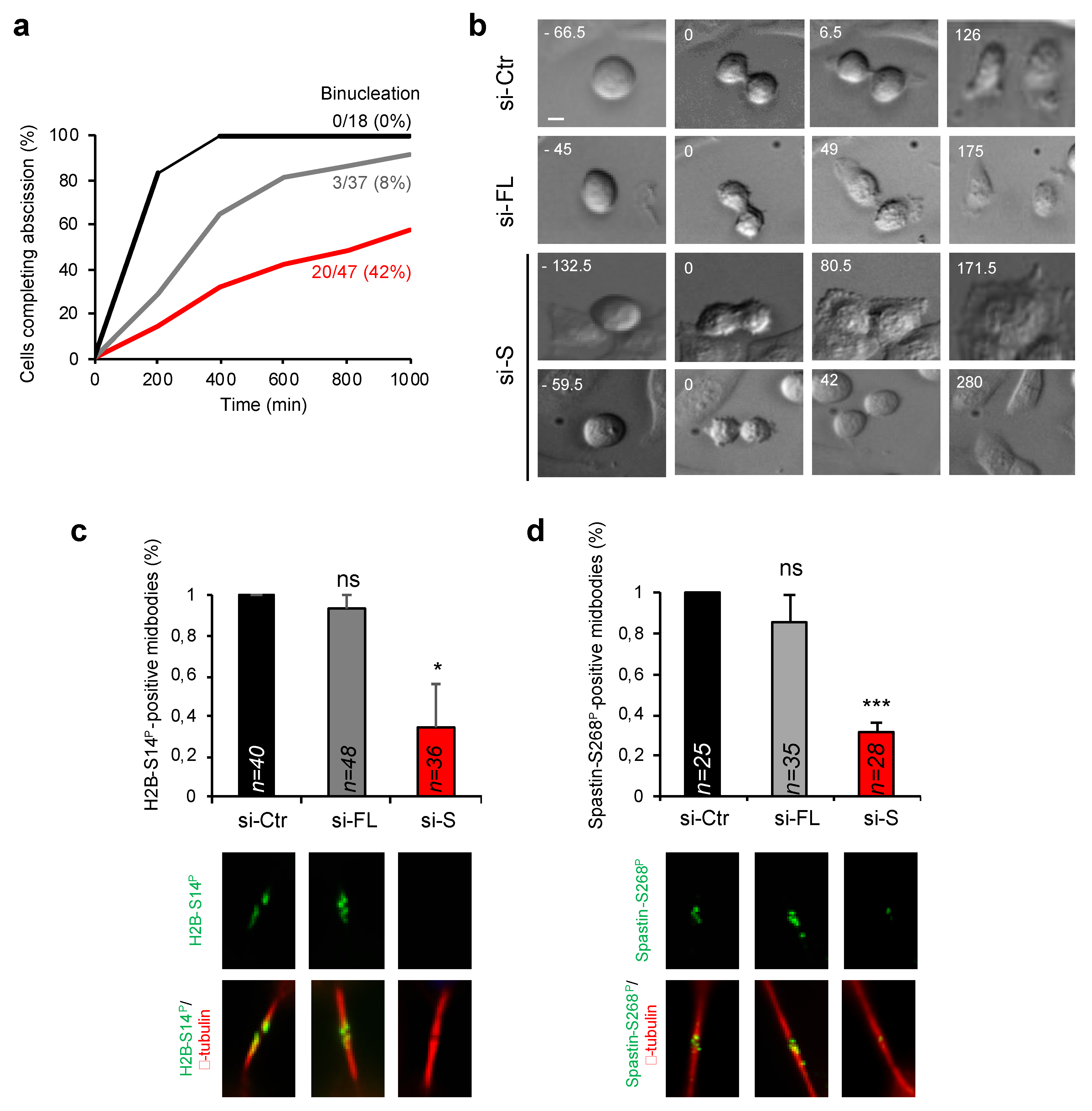
3.5. HIPK2-S Isoform Is Required for Faithful Cytokinesis
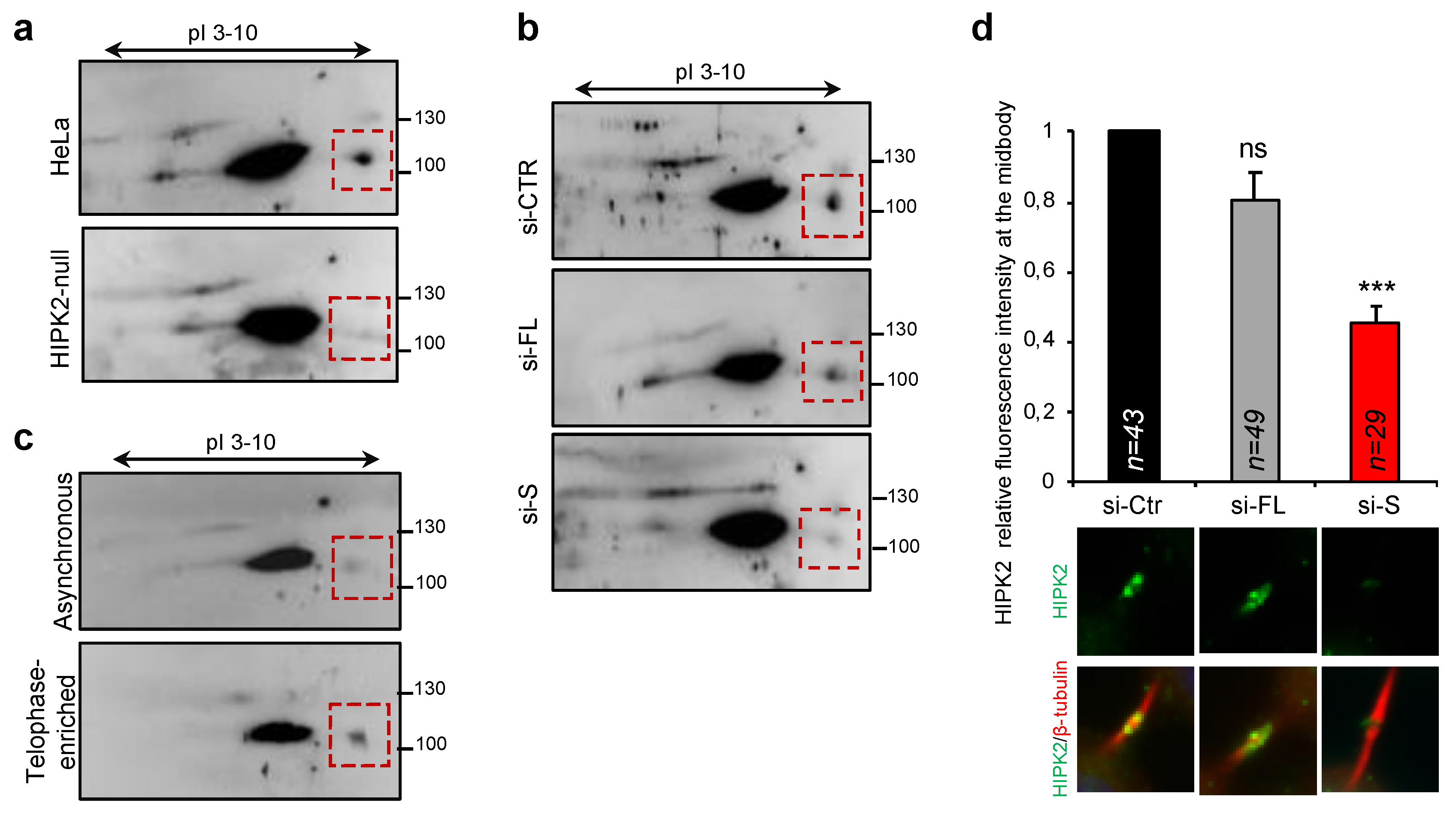
3.6. Endogenous HIPK2-S Is Enriched in Telophase and Localizes at the ICB of Dividing Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siepi, F.; Gatti, V.; Camerini, S.; Crescenzi, M.; Soddu, S. HIPK2 catalytic activity and subcellular localization are regulated by activation-loop Y354 autophosphorylation. Biochim. Biophys. Acta 2013, 1833, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Saul, V.V.; de la Vega, L.; Milanovic, M.; Kruger, M.; Braun, T.; Fritz-Wolf, K.; Becker, K.; Schmitz, M.L. HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J. Mol. Cell Biol. 2013, 5, 27–38. [Google Scholar] [CrossRef]
- Kim, Y.H.; Choi, C.Y.; Lee, S.J.; Conti, M.A.; Kim, Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 1998, 273, 25875–25879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pho, V.; Bonasera, S.J.; Holtzman, J.; Tang, A.T.; Hellmuth, J.; Tang, S.; Janak, P.H.; Tecott, L.H.; Huang, E.J. Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat. Neurosci. 2007, 10, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Ku, S.; Ma, G.K.; Saito, S.; Tang, A.A.; Zhang, J.; Mao, J.H.; Appella, E.; Balmain, A.; Huang, E.J. HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 13040–13045. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, E.M.; Swarup, S.; Lee, W. Hipk proteins dually regulate Wnt/Wingless signal transduction. Fly (Austin) 2012, 6, 126–131. [Google Scholar] [CrossRef][Green Version]
- Rinaldo, C.; Prodosmo, A.; Siepi, F.; Soddu, S. HIPK2: A multitalented partner for transcription factors in DNA damage response and development. Biochem. Cell. Biol. 2007, 85, 411–418. [Google Scholar] [CrossRef]
- Poon, C.L.; Zhang, X.; Lin, J.I.; Manning, S.A.; Harvey, K.F. Homeodomain-interacting protein kinase regulates Hippo pathway-dependent tissue growth. Curr. Biol. 2012, 22, 1587–1594. [Google Scholar] [CrossRef]
- Lee, W.; Swarup, S.; Chen, J.; Ishitani, T.; Verheyen, E.M. Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of beta-catenin/Arm and stimulation of target gene expression. Development 2009, 136, 241–251. [Google Scholar] [CrossRef]
- Chalazonitis, A.; Tang, A.A.; Shang, Y.; Pham, T.D.; Hsieh, I.; Setlik, W.; Gershon, M.D.; Huang, E.J. Homeodomain interacting protein kinase 2 regulates postnatal development of enteric dopaminergic neurons and glia via BMP signaling. J. Neurosci. 2011, 31, 13746–13757. [Google Scholar] [CrossRef]
- Cao, L.; Yang, G.; Gao, S.; Jing, C.; Montgomery, R.R.; Yin, Y.; Wang, P.; Fikrig, E.; You, F. HIPK2 is necessary for type I interferon-mediated antiviral immunity. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Calzado, M.A.; Renner, F.; Roscic, A.; Schmitz, M.L. HIPK2: A versatile switchboard regulating the transcription machinery and cell death. Cell Cycle 2007, 6, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Ann, E.J.; Kim, M.Y.; Yoon, J.H.; Ahn, J.S.; Jo, E.H.; Lee, H.J.; Lee, H.W.; Kang, H.G.; Choi, D.W.; Chun, K.H.; et al. Tumor Suppressor HIPK2 Regulates Malignant Growth via Phosphorylation of Notch1. Cancer Res. 2016, 76, 4728–4740. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Doan, C.N.; Arnold, T.D.; Lee, S.; Tang, A.A.; Reichardt, L.F.; Huang, E.J. Transcriptional corepressors HIPK1 and HIPK2 control angiogenesis via TGF-beta-TAK1-dependent mechanism. PLoS Biol. 2013, 11, e1001527. [Google Scholar] [CrossRef]
- Blaquiere, J.A.; Verheyen, E.M. Homeodomain-Interacting Protein Kinases: Diverse and Complex Roles in Development and Disease. Curr. Top. Dev. Biol. 2017, 123, 73–103. [Google Scholar] [CrossRef]
- Sombroek, D.; Hofmann, T.G. How cells switch HIPK2 on and off. Cell Death Differ. 2009, 16, 187–194. [Google Scholar] [CrossRef]
- Saul, V.V.; Schmitz, M.L. Posttranslational modifications regulate HIPK2, a driver of proliferative diseases. J. Mol. Med. 2013, 91, 1051–1058. [Google Scholar] [CrossRef]
- Wook Choi, D.; Yong Choi, C. HIPK2 modification code for cell death and survival. Mol. Cell. Oncol. 2014, 1, e955999. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoshimatsu, Y.; Hildebrand, J.; Frisch, S.M.; Goodman, R.H. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 2003, 115, 177–186. [Google Scholar] [CrossRef]
- Qin, Y.; Hu, Q.; Ji, S.; Xu, J.; Dai, W.; Liu, W.; Xu, W.; Sun, Q.; Zhang, Z.; Ni, Q.; et al. Homeodomain-interacting protein kinase 2 suppresses proliferation and aerobic glycolysis via ERK/cMyc axis in pancreatic cancer. Cell Prolif. 2019, 52, e12603. [Google Scholar] [CrossRef]
- Puca, R.; Nardinocchi, L.; Gal, H.; Rechavi, G.; Amariglio, N.; Domany, E.; Notterman, D.A.; Scarsella, M.; Leonetti, C.; Sacchi, A.; et al. Reversible dysfunction of wild-type p53 following homeodomain-interacting protein kinase-2 knockdown. Cancer Res. 2008, 68, 3707–3714. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, C.; Prodosmo, A.; Siepi, F.; Rinaldo, C.; Galli, F.; Gentileschi, M.; Bartolazzi, A.; Costanzo, A.; Sacchi, A.; Guerrini, L.; et al. HIPK2 phosphorylates DeltaNp63alpha and promotes its degradation in response to DNA damage. Oncogene 2011, 30, 4802–4813. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, T.G.; Moller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Droge, W.; Will, H.; Schmitz, M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- D’Orazi, G.; Cecchinelli, B.; Bruno, T.; Manni, I.; Higashimoto, Y.; Saito, S.; Gostissa, M.; Coen, S.; Marchetti, A.; Del Sal, G.; et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002, 4, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R.; Lee, S.Y.; Shin, K.S.; Choi, C.Y.; Kang, S.J. p300-mediated acetylation increased the protein stability of HIPK2 and enhanced its tumor suppressor function. Sci. Rep. 2017, 7, 16136. [Google Scholar] [CrossRef]
- Donninger, H.; Calvisi, D.F.; Barnoud, T.; Clark, J.; Schmidt, M.L.; Vos, M.D.; Clark, G.J. NORE1A is a Ras senescence effector that controls the apoptotic/senescent balance of p53 via HIPK2. J. Cell Biol. 2015, 208, 777–789. [Google Scholar] [CrossRef]
- Mao, J.H.; Wu, D.; Kim, I.J.; Kang, H.C.; Wei, G.; Climent, J.; Kumar, A.; Pelorosso, F.G.; DelRosario, R.; Huang, E.J.; et al. Hipk2 cooperates with p53 to suppress gamma-ray radiation-induced mouse thymic lymphoma. Oncogene 2012, 31, 1176–1180. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Gong, H.; Zeng, Y.; Tao, L.; Wang, J.; Jiang, J.; Xu, D.; Bao, E.; Qiu, J.; Liu, Z. Downregulation of homeodomain-interacting protein kinase-2 contributes to bladder cancer metastasis by regulating Wnt signaling. J. Cell Biochem. 2014, 115, 1762–1767. [Google Scholar] [CrossRef]
- Li, X.L.; Arai, Y.; Harada, H.; Shima, Y.; Yoshida, H.; Rokudai, S.; Aikawa, Y.; Kimura, A.; Kitabayashi, I. Mutations of the HIPK2 gene in acute myeloid leukemia and myelodysplastic syndrome impair AML1- and p53-mediated transcription. Oncogene 2007, 26, 7231–7239. [Google Scholar] [CrossRef]
- He, Y.; Roos, W.P.; Wu, Q.; Hofmann, T.G.; Kaina, B. The SIAH1-HIPK2-p53ser46 Damage Response Pathway is Involved in Temozolomide-Induced Glioblastoma Cell Death. Mol. Cancer Res. 2019, 17, 1129–1141. [Google Scholar] [CrossRef]
- D’Orazi, G.; Rinaldo, C.; Soddu, S. Updates on HIPK2: A resourceful oncosuppressor for clearing cancer. J. Exp. Clin. Cancer Res. 2012, 31, 63. [Google Scholar] [CrossRef] [PubMed]
- Mancini, F.; Pieroni, L.; Monteleone, V.; Luca, R.; Fici, L.; Luca, E.; Urbani, A.; Xiong, S.; Soddu, S.; Masetti, R.; et al. MDM4/HIPK2/p53 cytoplasmic assembly uncovers coordinated repression of molecules with anti-apoptotic activity during early DNA damage response. Oncogene 2016, 35, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Blaquiere, J.A.; Wong, K.K.L.; Kinsey, S.D.; Wu, J.; Verheyen, E.M. Homeodomain-interacting protein kinase promotes tumorigenesis and metastatic cell behavior. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, H.; Yeh, T.H.; Yu, J.; Sharma, M.K.; Perry, A.; Leonard, J.R.; Watson, M.A.; Gutmann, D.H.; Nagarajan, R. High-resolution, dual-platform aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene 2008, 27, 4745–4751. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Deshmukh, H.; Gutmann, R.J.; Emnett, R.J.; Rodriguez, F.J.; Watson, M.A.; Nagarajan, R.; Gutmann, D.H. Alterations of BRAF and HIPK2 loci predominate in sporadic pilocytic astrocytoma. Neurology 2009, 73, 1526–1531. [Google Scholar] [CrossRef]
- Kwon, M.J.; Kang, S.Y.; Nam, E.S.; Cho, S.J.; Rho, Y.S. HIPK2 Overexpression and Its Prognostic Role in Human Papillomavirus-Positive Tonsillar Squamous Cell Carcinoma. Biomed. Res. Int. 2017, 2017, 1056427. [Google Scholar] [CrossRef]
- Liu, F.; Li, N.; Liu, Y.; Zhang, J.; Zhang, J.; Wang, Z. Homeodomain interacting protein kinase-2 phosphorylates FOXM1 and promotes FOXM1-mediated tumor growth in renal cell carcinoma. J. Cell Biochem. 2019, 120, 10391–10401. [Google Scholar] [CrossRef]
- Torrente, L.; Sanchez, C.; Moreno, R.; Chowdhry, S.; Cabello, P.; Isono, K.; Koseki, H.; Honda, T.; Hayes, J.D.; Dinkova-Kostova, A.T.; et al. Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene 2017, 36, 6204–6212. [Google Scholar] [CrossRef]
- Jin, Y.; Ratnam, K.; Chuang, P.Y.; Fan, Y.; Zhong, Y.; Dai, Y.; Mazloom, A.R.; Chen, E.Y.; D’Agati, V.; Xiong, H.; et al. A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat. Med. 2012, 18, 580–588. [Google Scholar] [CrossRef]
- Garufi, A.; Traversi, G.; Cirone, M.; D’Orazi, G. HIPK2 role in the tumor-host interaction: Impact on fibroblasts transdifferentiation CAF-like. IUBMB Life 2019, 71, 2055–2061. [Google Scholar] [CrossRef]
- Hofmann, T.G.; Glas, C.; Bitomsky, N. HIPK2: A tumour suppressor that controls DNA damage-induced cell fate and cytokinesis. Bioessays 2013, 35, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, C.; Moncada, A.; Gradi, A.; Ciuffini, L.; D’Eliseo, D.; Siepi, F.; Prodosmo, A.; Giorgi, A.; Pierantoni, G.M.; Trapasso, F.; et al. HIPK2 controls cytokinesis and prevents tetraploidization by phosphorylating histone H2B at the midbody. Mol. Cell 2012, 47, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Barbiero, I.; Valente, D.; Chandola, C.; Magi, F.; Bergo, A.; Monteonofrio, L.; Tramarin, M.; Fazzari, M.; Soddu, S.; Landsberger, N.; et al. CDKL5 localizes at the centrosome and midbody and is required for faithful cell division. Sci. Rep. 2017, 7, 6228. [Google Scholar] [CrossRef]
- Monteonofrio, L.; Valente, D.; Ferrara, M.; Camerini, S.; Miscione, R.; Crescenzi, M.; Rinaldo, C.; Soddu, S. HIPK2 and extrachromosomal histone H2B are separately recruited by Aurora-B for cytokinesis. Oncogene 2018, 37, 3562–3574. [Google Scholar] [CrossRef] [PubMed]
- Monteonofrio, L.; Valente, D.; Rinaldo, C.; Soddu, S. Extrachromosomal Histone H2B Contributes to the Formation of the Abscission Site for Cell Division. Cells 2019, 8, 1391. [Google Scholar] [CrossRef] [PubMed]
- Pisciottani, A.; Biancolillo, L.; Ferrara, M.; Valente, D.; Sardina, F.; Monteonofrio, L.; Camerini, S.; Crescenzi, M.; Soddu, S.; Rinaldo, C. HIPK2 Phosphorylates the Microtubule-Severing Enzyme Spastin at S268 for Abscission. Cells 2019, 8, 684. [Google Scholar] [CrossRef]
- Valente, D.; Bossi, G.; Moncada, A.; Tornincasa, M.; Indelicato, S.; Piscuoglio, S.; Karamitopoulou, E.D.; Bartolazzi, A.; Pierantoni, G.M.; Fusco, A.; et al. HIPK2 deficiency causes chromosomal instability by cytokinesis failure and increases tumorigenicity. Oncotarget 2015, 6, 10320–10334. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef]
- Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26. [Google Scholar] [CrossRef]
- David, C.J.; Manley, J.L. Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 2010, 24, 2343–2364. [Google Scholar] [CrossRef]
- Murray-Zmijewski, F.; Lane, D.P.; Bourdon, J.C. p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006, 13, 962–972. [Google Scholar] [CrossRef]
- Kurokawa, K.; Akaike, Y.; Masuda, K.; Kuwano, Y.; Nishida, K.; Yamagishi, N.; Kajita, K.; Tanahashi, T.; Rokutan, K. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene 2014, 33, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Di Rocco, G.; Verdina, A.; Gatti, V.; Virdia, I.; Toietta, G.; Todaro, M.; Stassi, G.; Soddu, S. Apoptosis induced by a HIPK2 full-length-specific siRNA is due to off-target effects rather than prevalence of HIPK2-Deltae8 isoform. Oncotarget 2016, 7, 1675–1686. [Google Scholar] [CrossRef] [PubMed]
- Derti, A.; Garrett-Engele, P.; Macisaac, K.D.; Stevens, R.C.; Sriram, S.; Chen, R.; Rohl, C.A.; Johnson, J.M.; Babak, T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012, 22, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Prodosmo, A.; De Amicis, A.; Nistico, C.; Gabriele, M.; Di Rocco, G.; Monteonofrio, L.; Piane, M.; Cundari, E.; Chessa, L.; Soddu, S. p53 centrosomal localization diagnoses ataxia-telangiectasia homozygotes and heterozygotes. J. Clin. Investig. 2013, 123, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Verdina, A.; Di Rocco, G.; Virdia, I.; Monteonofrio, L.; Gatti, V.; Policicchio, E.; Bruselles, A.; Tartaglia, M.; Soddu, S. HIPK2-T566 autophosphorylation diversely contributes to UV- and doxorubicin-induced HIPK2 activation. Oncotarget 2017, 8, 16744–16754. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Menon, R.P.; Gibson, T.J.; Pastore, A. The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. J. Mol. Biol. 2004, 343, 43–53. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Rodriguez-Gil, A.; Ritter, O.; Hornung, J.; Stekman, H.; Kruger, M.; Braun, T.; Kremmer, E.; Kracht, M.; Schmitz, M.L. HIPK family kinases bind and regulate the function of the CCR4-NOT complex. Mol. Biol. Cell 2016, 27, 1969–1980. [Google Scholar] [CrossRef]
- Gresko, E.; Roscic, A.; Ritterhoff, S.; Vichalkovski, A.; del Sal, G.; Schmitz, M.L. Autoregulatory control of the p53 response by caspase-mediated processing of HIPK2. EMBO J. 2006, 25, 1883–1894. [Google Scholar] [CrossRef]
- de la Vega, L.; Hornung, J.; Kremmer, E.; Milanovic, M.; Schmitz, M.L. Homeodomain-interacting protein kinase 2-dependent repression of myogenic differentiation is relieved by its caspase-mediated cleavage. Nucleic Acids Res. 2013, 41, 5731–5745. [Google Scholar] [CrossRef] [PubMed]
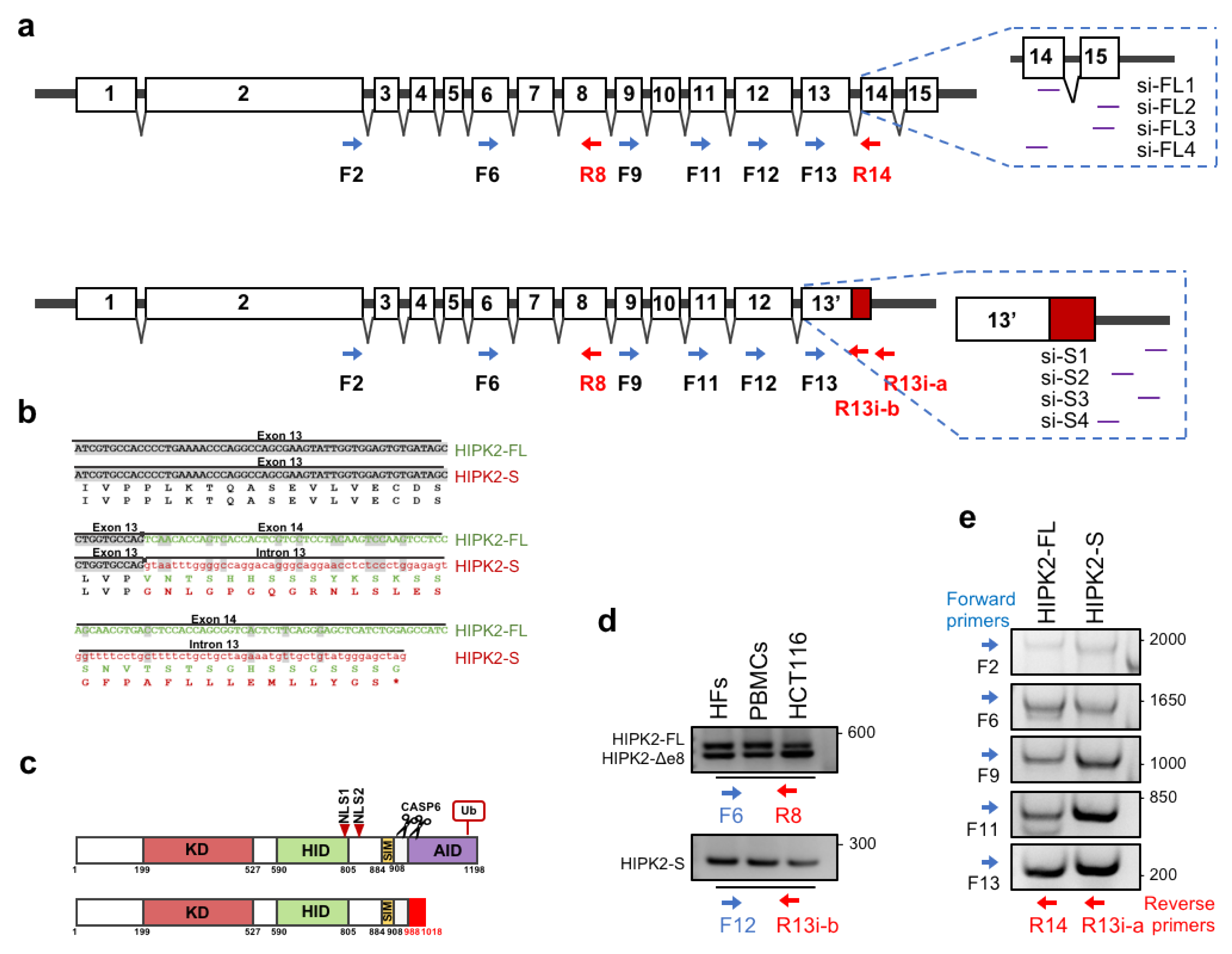
| RT-PCR primers | ||
| F2 | Forward | 5′-CAGCCAGCCACGTCTCCAAGG-3′ |
| F6 | Forward | 5′-GGCTGACCGGCGGGAGTT-3′ |
| F9 | Forward | 5′-CCCATCGTCACTCAAGCCCCAG-3′ |
| F11 | Forward | 5′-AGCAGCCAACCAGCACCACCT-3′ |
| F12 | Forward | 5′-AGGAGGAACAGAAACACGCC-3′ |
| F13 | Forward | 5′-CCTGAAAACCCAGGCCAGCGA-3′ |
| R14 | Reverse | 5′-CGCTGCTGCCGGTAGGTGATG-3′ |
| R13i-a | Reverse | 5′-GCCTGCAGTCACTGGGCACTT-3′ |
| R13i-b | Reverse | 5′-CCTGTCCTGGCCCCAAAT-3′ |
| R8 | Reverse | 5′-GGTCAGGCCGGGCACAAATCT-3′ |
| qPCR primers | ||
| HIPK2-S | Forward | 5’-AGGAGGAACAGAAACACGCC-3’ |
| HIPK2-S | Reverse | 5’-CCTGTCCTGGCCCCAAAT-3’ |
| HIPK2-FL | Forward | 5’-AGGAAGAGTAAGCAGCACCAG-3’ |
| HIPK2-FL | Reverse | 5’-TGCTGATGGTGATGACACTGA-3’ |
| si-S1 | 5′-AUCCUGUAAUCAAUACCUCTT-3′ | siRNA/MWG |
| si-S2 | 5′-GGACAUUGUAUAAGCAGCGTT-3′ | siRNA/MWG |
| si-FL1 | 5′-UCACUCUUCAGGGAGCUCAUCUGGA-3′ | siRNA/MWG |
| siFL2 | 5′-CCAAGGUCAACCAGUACCCUUACAU-3′ | siRNA/MWG |
| GL2§ | 5′-CGUACGCGGAAUACUUCGAUU-3′ | siRNA/MWG |
| si-S3 | 5′-UCUUUAAUCCUGUAAUCAAUACCUC-3′ | stealth siRNA/Invitrogen |
| si-S4 | 5′-GCUUCUGCUGAACAGCACAUUGUAU-3′ | stealth siRNA/Invitrogen |
| si-FL3 | 5′-UCACUCUUCAGGGAGCUCAUCUGGA-3′ | stealth siRNA/Invitrogen |
| si-FL4 | 5′-CCAAGGUCAACCAGUACCCUUACAU-3′ | stealth siRNA/Invitrogen |
| UNC* | Negative Medium GC Duplexes | stealth siRNA/Invitrogen |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatti, V.; Ferrara, M.; Virdia, I.; Matteoni, S.; Monteonofrio, L.; di Martino, S.; Diodoro, M.G.; Di Rocco, G.; Rinaldo, C.; Soddu, S. An Alternative Splice Variant of HIPK2 with Intron Retention Contributes to Cytokinesis. Cells 2020, 9, 484. https://doi.org/10.3390/cells9020484
Gatti V, Ferrara M, Virdia I, Matteoni S, Monteonofrio L, di Martino S, Diodoro MG, Di Rocco G, Rinaldo C, Soddu S. An Alternative Splice Variant of HIPK2 with Intron Retention Contributes to Cytokinesis. Cells. 2020; 9(2):484. https://doi.org/10.3390/cells9020484
Chicago/Turabian StyleGatti, Veronica, Manuela Ferrara, Ilaria Virdia, Silvia Matteoni, Laura Monteonofrio, Simona di Martino, Maria Grazia Diodoro, Giuliana Di Rocco, Cinzia Rinaldo, and Silvia Soddu. 2020. "An Alternative Splice Variant of HIPK2 with Intron Retention Contributes to Cytokinesis" Cells 9, no. 2: 484. https://doi.org/10.3390/cells9020484
APA StyleGatti, V., Ferrara, M., Virdia, I., Matteoni, S., Monteonofrio, L., di Martino, S., Diodoro, M. G., Di Rocco, G., Rinaldo, C., & Soddu, S. (2020). An Alternative Splice Variant of HIPK2 with Intron Retention Contributes to Cytokinesis. Cells, 9(2), 484. https://doi.org/10.3390/cells9020484





