Phosphorylation-Dependent Differences in CXCR4-LASP1-AKT1 Interaction between Breast Cancer and Chronic Myeloid Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs and Mutagenesis
2.2. Cell Culture and Transfection
2.3. Western Blot
2.4. Generation of GST-LASP1, His-LASP1 and GST-CRKL
2.5. GST-LASP1 Phosphorylation
2.6. Immunoprecipitation and Pull-Down
2.7. Generation of Cxcr4−/− Bone Marrow Derived Dendritic Cells
2.8. Radioactive Kinase Assay
2.9. PCR-Primer for Chemokine Receptors
2.10. Life Cell Imaging
2.11. Immunofluorescence
2.12. SH3 Domain Mapping
2.13. Statistics
3. Results
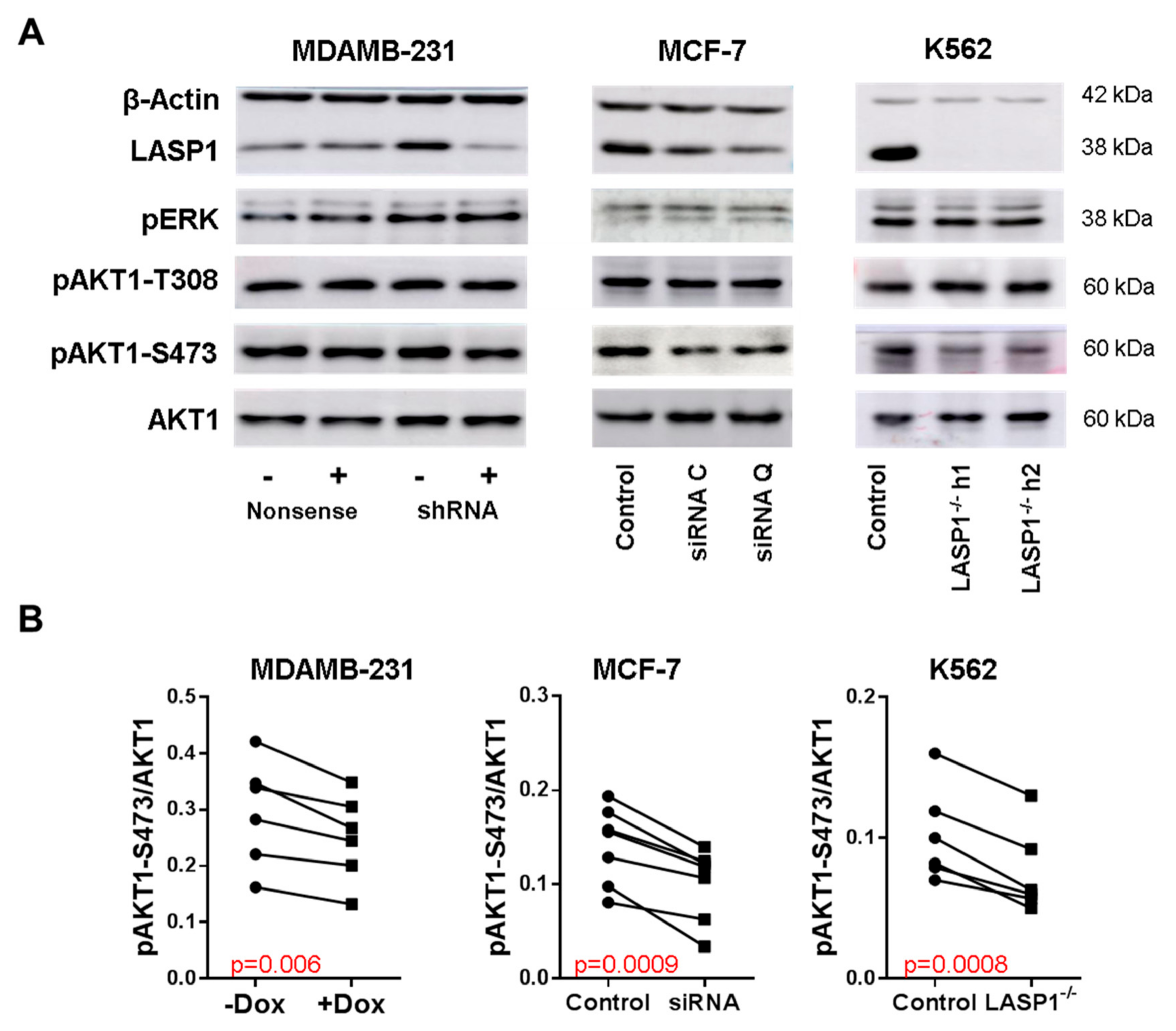
3.1. LASP1 Depletion Negatively Regulates AKT1-S473 Phosphorylation
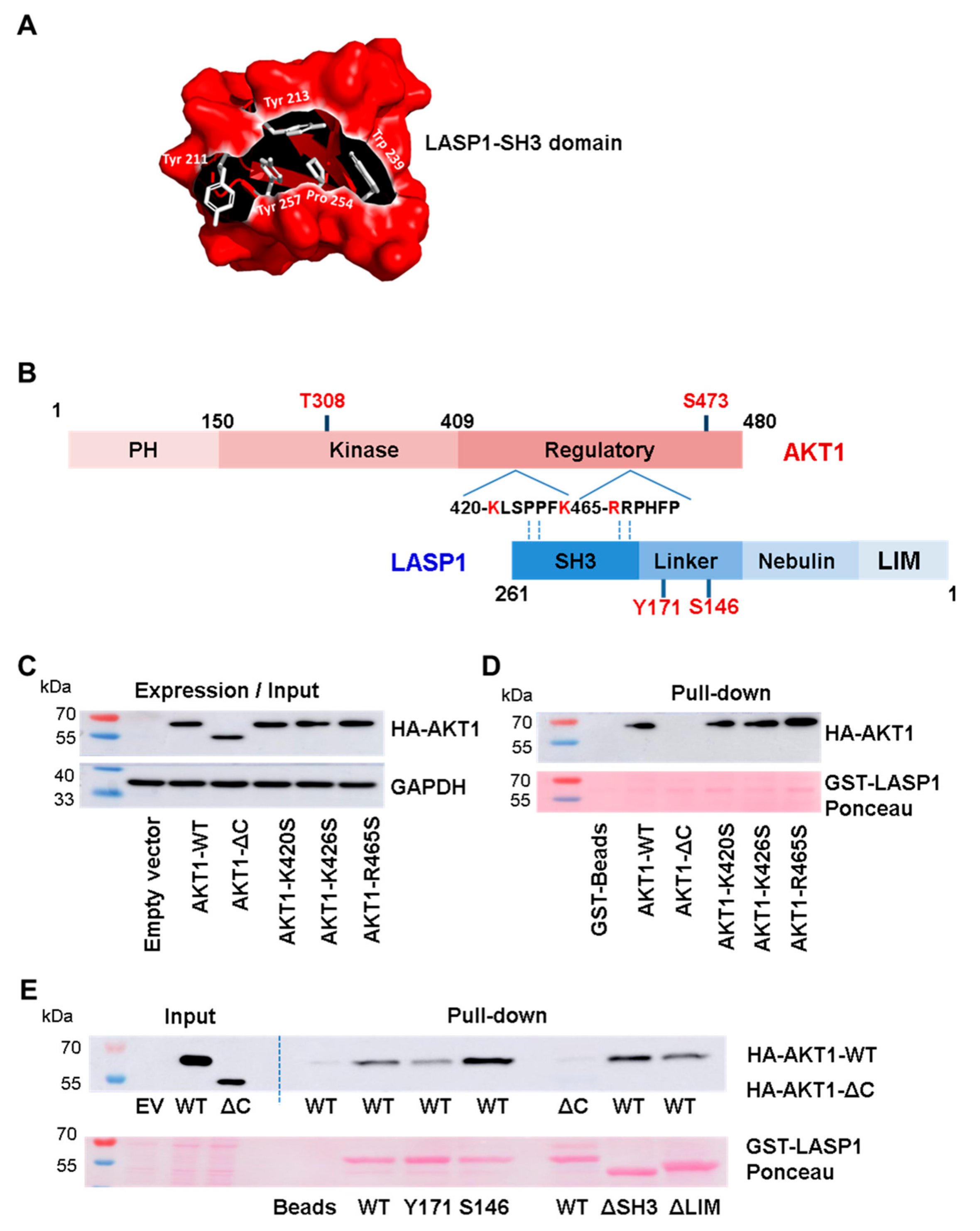
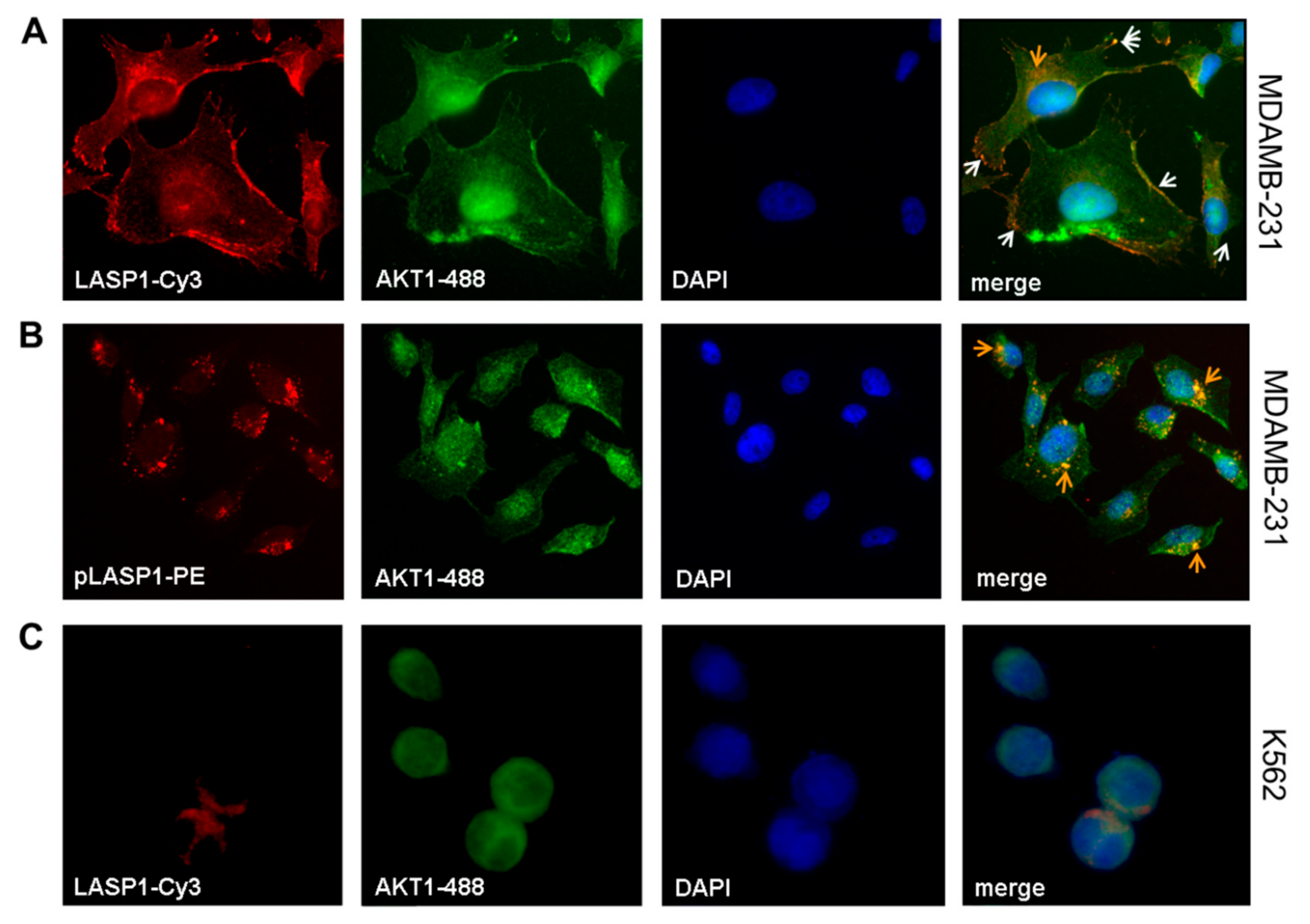
3.2. LASP1-AKT1 Interaction in MDAMB-231 Breast Cancer Cells
3.3. AKT1 Preferentially Binds to LASP1 Phosphorylated at S146
3.4. Interaction between Linker Region of LASP1 and AKT1 C-Terminus
3.5. CXCR4-LASP1 Binding
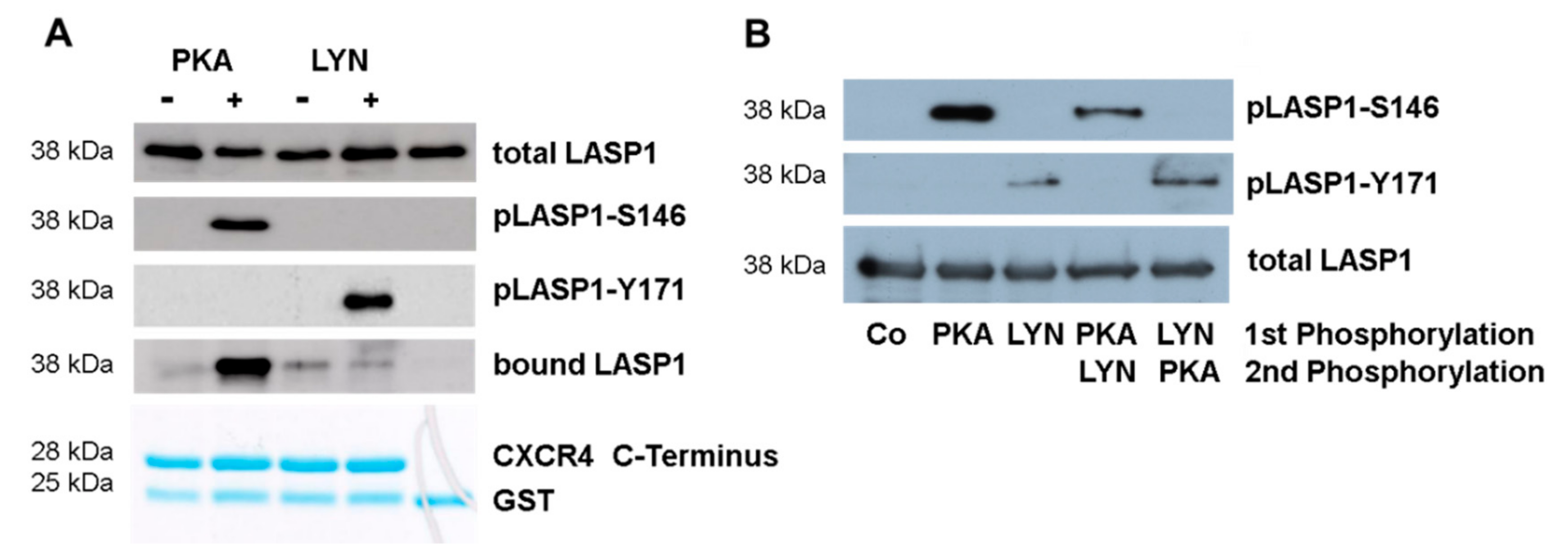
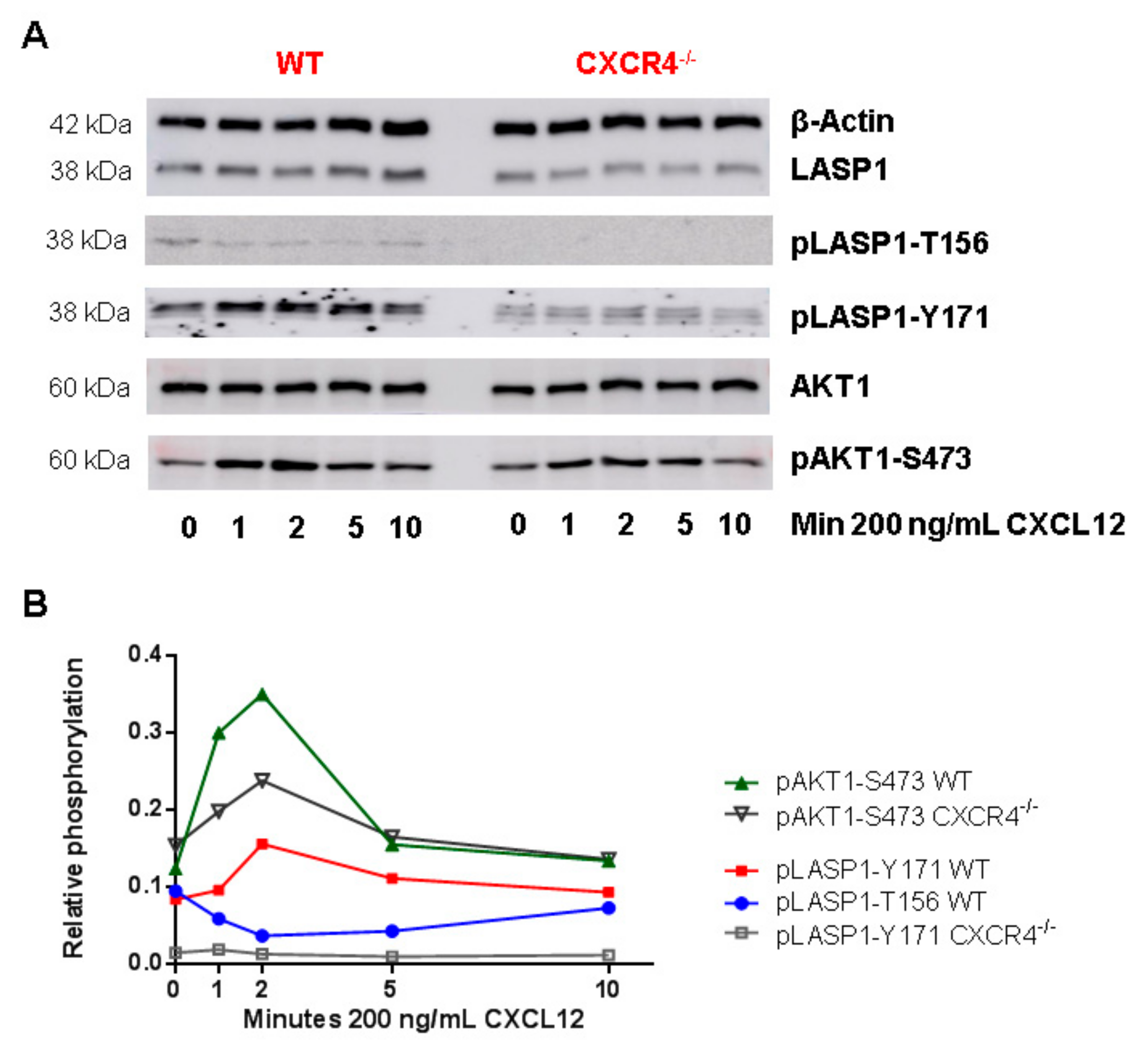
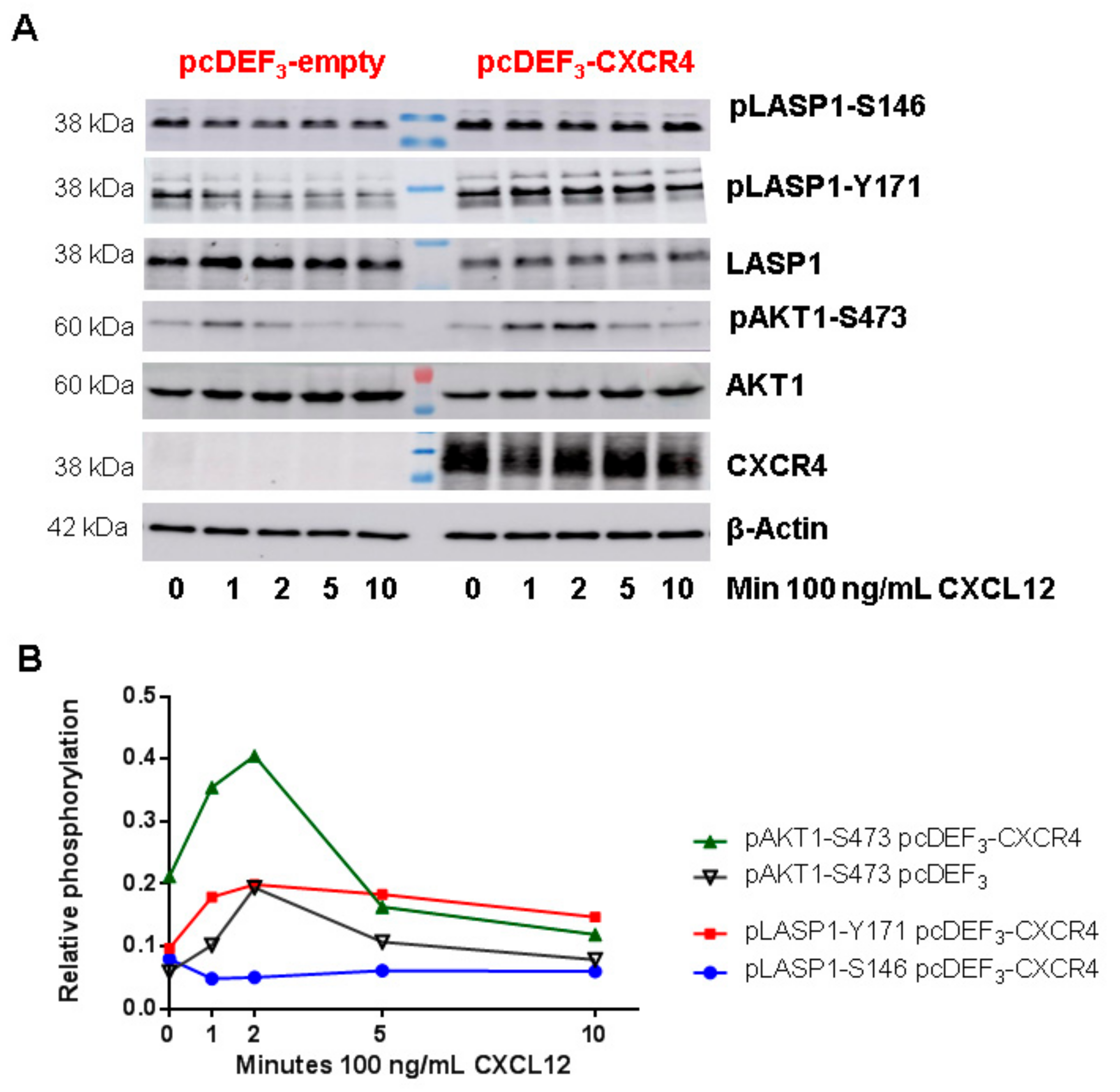
3.6. CXCR4-Induces LASP1 Phosphorylation in Intact Cells
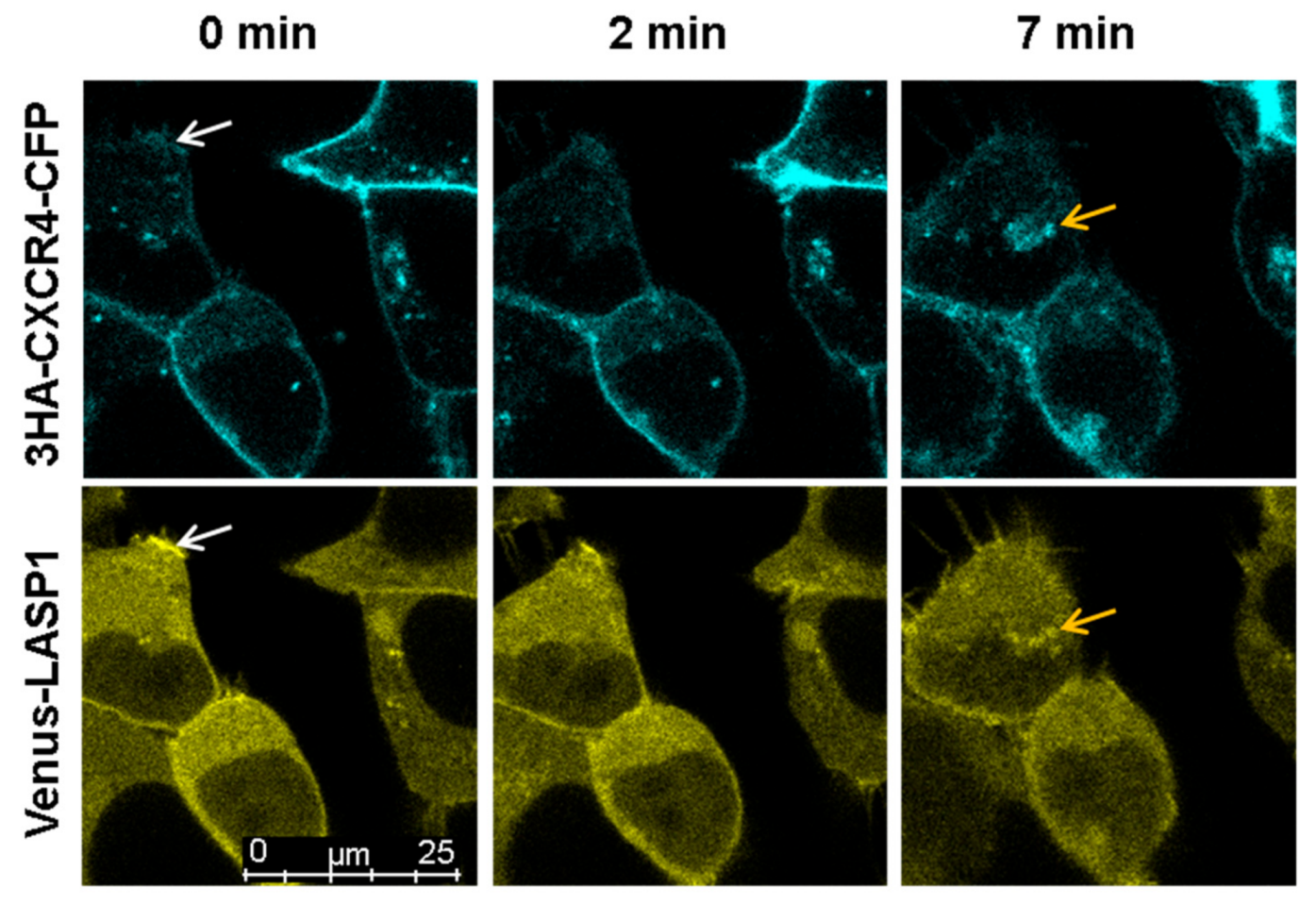
3.7. LASP1-CXCR4 Co-Localization in Intact Cells by Life Cell Imaging
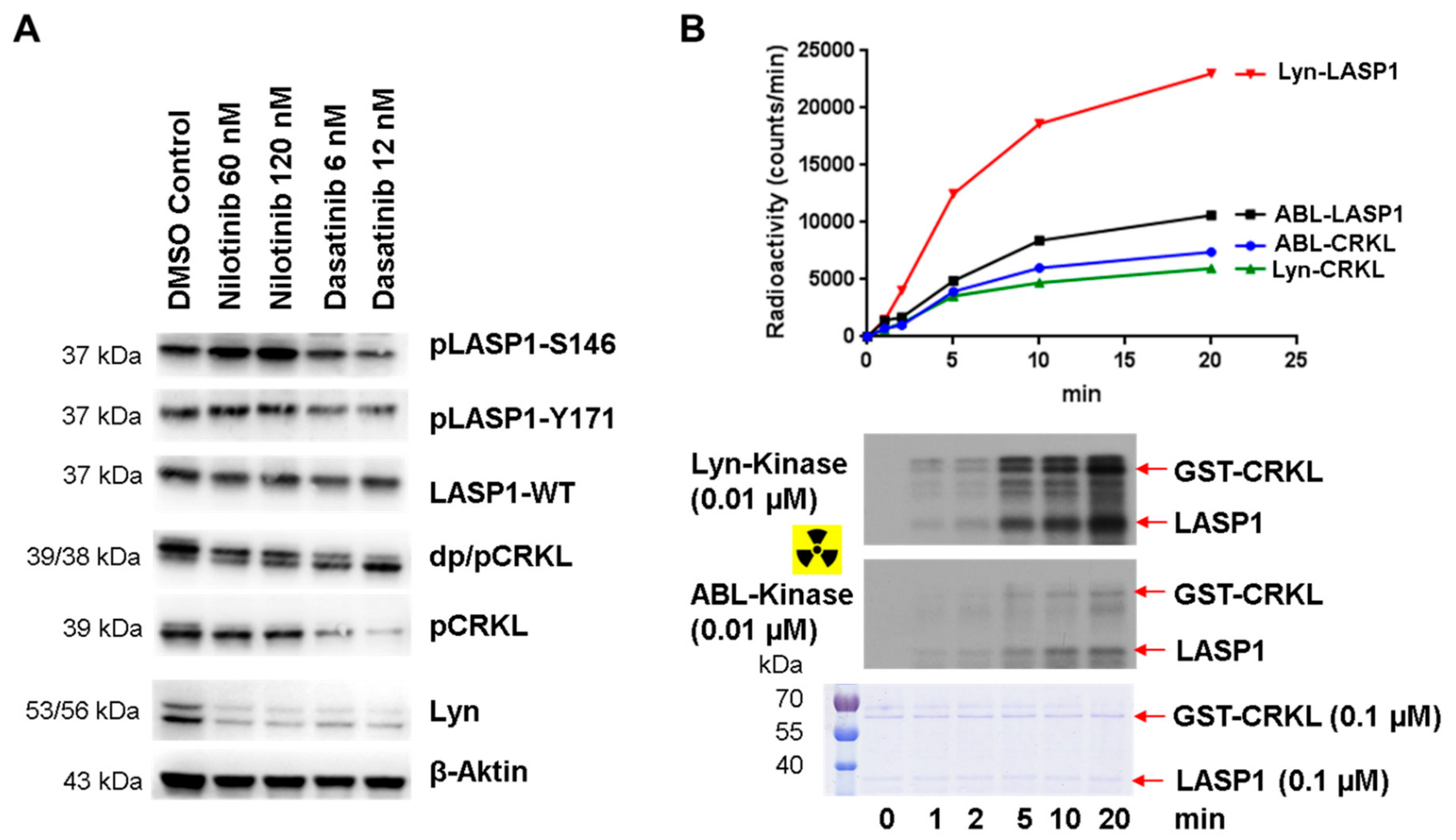
3.8. Enhanced pLASP1-S146 in K562 CML Cells during TKI Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tomasetto, C.; Moog-Lutz, C.; Regnier, C.H.; Schreiber, V.; Basset, P.; Rio, M.C. Lasp-1 (MLN 50) defines a new LIM protein subfamily characterized by the association of LIM and SH3 domains. FEBS Lett. 1995, 373, 245–249. [Google Scholar] [CrossRef]
- Butt, E.; Raman, D. New Frontiers for the Cytoskeletal Protein LASP1. Front. Oncol. 2018, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Mihlan, S.; Reiss, C.; Thalheimer, P.; Herterich, S.; Gaetzner, S.; Kremerskothen, J.; Pavenstadt, H.J.; Lewandrowski, U.; Sickmann, A.; Butt, E. Nuclear import of LASP-1 is regulated by phosphorylation and dynamic protein-protein interactions. Oncogene 2013, 32, 2107–2113. [Google Scholar] [CrossRef] [PubMed]
- Traenka, J.; Hauck, C.R.; Lewandrowski, U.; Sickmann, A.; Gambaryan, S.; Thalheimer, P.; Butt, E. Integrin-dependent translocation of LASP-1 to the cytoskeleton of activated platelets correlates with LASP-1 phosphorylation at tyrosine 171 by Src-kinase. Thromb. Haemostasis 2009, 102, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Park, Z.Y.; Lin, D.; Brahmbhatt, A.A.; Rio, M.C.; Yates, J.R., 3rd; Klemke, R.L. Regulation of cell migration and survival by focal adhesion targeting of Lasp-1. J. Cell Biol. 2004, 165, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Raman, D.; Sai, J.; Neel, N.F.; Chew, C.S.; Richmond, A. LIM and SH3 protein-1 modulates CXCR2-mediated cell migration. PLoS ONE 2010, 5, e10050. [Google Scholar] [CrossRef]
- Doring, Y.; Pawig, L.; Weber, C.; Noels, H. The CXCL12/CXCR4 chemokine ligand/receptor axis in cardiovascular disease. Front. Physiol. 2014, 5, 212. [Google Scholar] [CrossRef]
- Jin, L.; Tabe, Y.; Konoplev, S.; Xu, Y.; Leysath, C.E.; Lu, H.; Kimura, S.; Ohsaka, A.; Rios, M.B.; Calvert, L.; et al. CXCR4 up-regulation by imatinib induces chronic myelogenous leukemia (CML) cell migration to bone marrow stroma and promotes survival of quiescent CML cells. Mol. Cancer Ther. 2008, 7, 48–58. [Google Scholar] [CrossRef]
- Yeung, K.Y.; Gooley, T.A.; Zhang, A.; Raftery, A.E.; Radich, J.P.; Oehler, V.G. Predicting relapse prior to transplantation in chronic myeloid leukemia by integrating expert knowledge and expression data. Bioinformatics 2012, 28, 823–830. [Google Scholar] [CrossRef][Green Version]
- Herrmann, A.B.; Muller, M.L.; Orth, M.F.; Muller, J.P.; Zernecke, A.; Hochhaus, A.; Ernst, T.; Butt, E.; Frietsch, J.J. Knockout of LASP1 in CXCR4 expressing CML cells promotes cell persistence, proliferation and TKI resistance. J. Cell. Mol. Med. 2020. [Google Scholar] [CrossRef]
- Frietsch, J.J.; Kastner, C.; Grunewald, T.G.; Schweigel, H.; Nollau, P.; Ziermann, J.; Clement, J.H.; La Rosee, P.; Hochhaus, A.; Butt, E. LASP1 is a novel BCR-ABL substrate and a phosphorylation-dependent binding partner of CRKL in chronic myeloid leukemia. Oncotarget 2014, 5, 5257–5271. [Google Scholar] [CrossRef][Green Version]
- Chew, C.S.; Chen, X.; Parente, J.A., Jr.; Tarrer, S.; Okamoto, C.; Qin, H.Y. Lasp-1 binds to non-muscle F-actin in vitro and is localized within multiple sites of dynamic actin assembly in vivo. J. Cell Sci. 2002, 115, 4787–4799. [Google Scholar] [CrossRef][Green Version]
- Orth, M.F.; Cazes, A.; Butt, E.; Grunewald, T.G. An update on the LIM and SH3 domain protein 1 (LASP1): A versatile structural, signaling, and biomarker protein. Oncotarget 2015, 6, 26–42. [Google Scholar] [CrossRef]
- Duvall-Noelle, N.; Karwandyar, A.; Richmond, A.; Raman, D. LASP-1: A nuclear hub for the UHRF1-DNMT1-G9a-Snail1 complex. Oncogene 2016, 35, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Endres, M.; Kneitz, S.; Orth, M.F.; Perera, R.K.; Zernecke, A.; Butt, E. Regulation of matrix metalloproteinases (MMPs) expression and secretion in MDA-MB-231 breast cancer cells by LIM and SH3 protein 1 (LASP1). Oncotarget 2016, 7, 64244–64259. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhao, H.; Chen, H.; Yao, Q. CXCR4 in breast cancer: Oncogenic role and therapeutic targeting. Drug Des. Dev. Ther. 2015, 9, 4953–4964. [Google Scholar] [CrossRef]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef]
- Tian, Y.; Yin, H.; Deng, X.; Tang, B.; Ren, X.; Jiang, T. CXCL12 induces migration of oligodendrocyte precursor cells through the CXCR4activated MEK/ERK and PI3K/AKT pathways. Mol. Med. Rep. 2018, 18, 4374–4380. [Google Scholar] [CrossRef]
- Martini, M.; De Santis, M.C.; Braccini, L.; Gulluni, F.; Hirsch, E. PI3K/AKT signaling pathway and cancer: An updated review. Ann. Med. 2014, 46, 372–383. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB signaling: Navigating the network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Zhong, C.; Chen, Y.; Tao, B.; Peng, L.; Peng, T.; Yang, X.; Xia, X.; Chen, L. LIM and SH3 protein 1 regulates cell growth and chemosensitivity of human glioblastoma via the PI3K/AKT pathway. BMC Cancer 2018, 18, 722. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Y.; Wang, X.; Zhang, H.; Zhang, Y.; Gao, Y.; Weng, M.; Wang, L.; Liang, H.; Li, M.; et al. LASP-1 induces proliferation, metastasis and cell cycle arrest at the G2/M phase in gallbladder cancer by down-regulating S100P via the PI3K/AKT pathway. Cancer Lett. 2016, 372, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Tang, L.; Wu, L.; Li, K.; Wang, H.; Li, W.; Wu, J.; Li, M.; Wang, S.; Zhao, L. LASP1 promotes nasopharyngeal carcinoma progression through negatively regulation of the tumor suppressor PTEN. Cell Death Dis. 2018, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Cai, Y.; Xu, L.; Yao, X.; Shi, J.; Zhang, F.; Luo, Y.; Zheng, K.; Liu, J.; Deng, F.; et al. Loss of the 14-3-3sigma is essential for LASP1-mediated colorectal cancer progression via activating PI3K/AKT signaling pathway. Sci. Rep. 2016, 6, 25631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Fan, C.; Wang, L.; Li, A.; Zhou, H.; Cai, L.; Miao, Y.; Li, Q.; Qiu, X.; et al. Lasp1 promotes malignant phenotype of non-small-cell lung cancer via inducing phosphorylation of FAK-AKT pathway. Oncotarget 2017, 8, 75102–75113. [Google Scholar] [CrossRef]
- Zeng, M.; van der Donk, W.A.; Chen, J. Lanthionine synthetase C-like protein 2 (LanCL2) is a novel regulator of Akt. Mol. Biol. Cell 2014, 25, 3954–3961. [Google Scholar] [CrossRef]
- Stolting, M.; Wiesner, C.; van Vliet, V.; Butt, E.; Pavenstadt, H.; Linder, S.; Kremerskothen, J. Lasp-1 regulates podosome function. PLoS ONE 2012, 7, e35340. [Google Scholar] [CrossRef]
- Butt, E.; Gambaryan, S.; Gottfert, N.; Galler, A.; Marcus, K.; Meyer, H.E. Actin binding of human LIM and SH3 protein is regulated by cGMP- and cAMP-dependent protein kinase phosphorylation on serine 146. J. Biol. Chem. 2003, 278, 15601–15607. [Google Scholar] [CrossRef]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Traenka, C.; Remke, M.; Korshunov, A.; Bender, S.; Hielscher, T.; Northcott, P.A.; Witt, H.; Ryzhova, M.; Felsberg, J.; Benner, A.; et al. Role of LIM and SH3 protein 1 (LASP1) in the metastatic dissemination of medulloblastoma. Cancer Res. 2010, 70, 8003–8014. [Google Scholar] [CrossRef]
- Walter, U.; Miller, P.; Wilson, F.; Menkes, D.; Greengard, P. Immunological distinction between guanosine 3’:5’-monophosphate-dependent and adenosine 3’:5’-monophosphate-dependent protein kinases. J. Biol. Chem. 1980, 255, 3757–3762. [Google Scholar] [PubMed]
- Nie, Y.; Waite, J.; Brewer, F.; Sunshine, M.J.; Littman, D.R.; Zou, Y.R. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 2004, 200, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Vorlova, S.; Koch, M.; Manthey, H.D.; Cochain, C.; Busch, M.; Chaudhari, S.M.; Stegner, D.; Yepes, M.; Lorenz, K.; Nolte, M.W.; et al. Coagulation factor XII induces pro-inflammatory cytokine responses in macrophages and promotes atherosclerosis in mice. Thromb. Haemostasis 2017, 117, 176–187. [Google Scholar] [CrossRef]
- Zhou, R.; Shao, Z.; Liu, J.; Zhan, W.; Gao, Q.; Pan, Z.; Wu, L.; Xu, L.; Ding, Y.; Zhao, L. COPS5 and LASP1 synergistically interact to downregulate 14-3-3sigma expression and promote colorectal cancer progression via activating PI3K/AKT pathway. Int. J. Cancer 2018, 142, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, T.G.; Kammerer, U.; Kapp, M.; Eck, M.; Dietl, J.; Butt, E.; Honig, A. Nuclear localization and cytosolic overexpression of LASP-1 correlates with tumor size and nodal-positivity of human breast carcinoma. BMC Cancer 2007, 7, 198. [Google Scholar] [CrossRef]
- Saksela, K.; Permi, P. SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett. 2012, 586, 2609–2614. [Google Scholar] [CrossRef]
- Nishi, H.; Shaytan, A.; Panchenko, A.R. Physicochemical mechanisms of protein regulation by phosphorylation. Front. Genet. 2014, 5, 270. [Google Scholar] [CrossRef]
- Pinna, L.A.; Ruzzene, M. How do protein kinases recognize their substrates? Biochim. Biophys. Acta 1996, 1314, 191–225. [Google Scholar] [CrossRef]
- Lane, S.W.; Gilliland, D.G. Leukemia stem cells. Semin. Cancer Biol. 2010, 20, 71–76. [Google Scholar] [CrossRef]
- Keicher, C.; Gambaryan, S.; Schulze, E.; Marcus, K.; Meyer, H.E.; Butt, E. Phosphorylation of mouse LASP-1 on threonine 156 by cAMP- and cGMP-dependent protein kinase. Biochem. Biophys. Res. Commun. 2004, 324, 308–316. [Google Scholar] [CrossRef]
- Xu, D.; Li, R.; Wu, J.; Jiang, L.; Zhong, H.A. Drug design targeting the CXCR4/CXCR7/CXCL12 pathway. Curr. Top. Med. Chem. 2016, 16, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- La Rosee, P.; Holm-Eriksen, S.; Konig, H.; Hartel, N.; Ernst, T.; Debatin, J.; Mueller, M.C.; Erben, P.; Binckebanck, A.; Wunderle, L.; et al. Phospho-CRKL monitoring for the assessment of BCR-ABL activity in imatinib-resistant chronic myeloid leukemia or Ph+ acute lymphoblastic leukemia patients treated with nilotinib. Haematologica 2008, 93, 765–769. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gioia, R.; Leroy, C.; Drullion, C.; Lagarde, V.; Etienne, G.; Dulucq, S.; Lippert, E.; Roche, S.; Mahon, F.X.; Pasquet, J.M. Quantitative phosphoproteomics revealed interplay between Syk and Lyn in the resistance to nilotinib in chronic myeloid leukemia cells. Blood 2011, 118, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Tabe, Y.; Jin, L.; Iwabuchi, K.; Wang, R.Y.; Ichikawa, N.; Miida, T.; Cortes, J.; Andreeff, M.; Konopleva, M. Role of stromal microenvironment in nonpharmacological resistance of CML to imatinib through Lyn/CXCR4 interactions in lipid rafts. Leukemia 2012, 26, 883–892. [Google Scholar] [CrossRef]
- Mahon, F.X.; Hayette, S.; Lagarde, V.; Belloc, F.; Turcq, B.; Nicolini, F.; Belanger, C.; Manley, P.W.; Leroy, C.; Etienne, G.; et al. Evidence that resistance to nilotinib may be due to BCR-ABL, Pgp, or Src kinase overexpression. Cancer Res. 2008, 68, 9809–9816. [Google Scholar] [CrossRef]
- Sobolik, T.; Su, Y.J.; Wells, S.; Ayers, G.D.; Cook, R.S.; Richmond, A. CXCR4 drives the metastatic phenotype in breast cancer through induction of CXCR2 and activation of MEK and PI3K pathways. Mol. Biol. Cell 2014, 25, 566–582. [Google Scholar] [CrossRef]
- Busillo, J.M.; Benovic, J.L. Regulation of CXCR4 signaling. Biochim. Biophys. Acta 2007, 1768, 952–963. [Google Scholar] [CrossRef]
- Domanska, U.M.; Kruizinga, R.C.; Nagengast, W.B.; Timmer-Bosscha, H.; Huls, G.; de Vries, E.G.; Walenkamp, A.M. A review on CXCR4/CXCL12 axis in oncology: No place to hide. Eur. J. Cancer 2013, 49, 219–230. [Google Scholar] [CrossRef]
- Houshmand, M.; Simonetti, G.; Circosta, P.; Gaidano, V.; Cignetti, A.; Martinelli, G.; Saglio, G.; Gale, R.P. Chronic myeloid leukemia stem cells. Leukemia 2019, 33, 1543–1556. [Google Scholar] [CrossRef]
- Xiao, L.Y.; Kan, W.M. Cyclic AMP (cAMP) confers drug resistance against DNA damaging agents via PKAIA in CML cells. Eur. J. Pharmacol. 2017, 794, 201–208. [Google Scholar] [CrossRef]
- Bernusso, V.A.; Machado-Neto, J.A.; Pericole, F.V.; Vieira, K.P.; Duarte, A.S.; Traina, F.; Hansen, M.D.; Olalla Saad, S.T.; Barcellos, K.S. Imatinib restores VASP activity and its interaction with Zyxin in BCR-ABL leukemic cells. Biochim. Biophys. Acta 2015, 1853, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Maruoka, M.; Sato, M.; Yuan, Y.; Ichiba, M.; Fujii, R.; Ogawa, T.; Ishida-Kitagawa, N.; Takeya, T.; Watanabe, N. Abl-1-bridged tyrosine phosphorylation of VASP by Abelson kinase impairs association of VASP to focal adhesions and regulates leukaemic cell adhesion. Biochem. J. 2012, 441, 889–899. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butt, E.; Stempfle, K.; Lister, L.; Wolf, F.; Kraft, M.; Herrmann, A.B.; Viciano, C.P.; Weber, C.; Hochhaus, A.; Ernst, T.; et al. Phosphorylation-Dependent Differences in CXCR4-LASP1-AKT1 Interaction between Breast Cancer and Chronic Myeloid Leukemia. Cells 2020, 9, 444. https://doi.org/10.3390/cells9020444
Butt E, Stempfle K, Lister L, Wolf F, Kraft M, Herrmann AB, Viciano CP, Weber C, Hochhaus A, Ernst T, et al. Phosphorylation-Dependent Differences in CXCR4-LASP1-AKT1 Interaction between Breast Cancer and Chronic Myeloid Leukemia. Cells. 2020; 9(2):444. https://doi.org/10.3390/cells9020444
Chicago/Turabian StyleButt, Elke, Katrin Stempfle, Lorenz Lister, Felix Wolf, Marcella Kraft, Andreas B. Herrmann, Cristina Perpina Viciano, Christian Weber, Andreas Hochhaus, Thomas Ernst, and et al. 2020. "Phosphorylation-Dependent Differences in CXCR4-LASP1-AKT1 Interaction between Breast Cancer and Chronic Myeloid Leukemia" Cells 9, no. 2: 444. https://doi.org/10.3390/cells9020444
APA StyleButt, E., Stempfle, K., Lister, L., Wolf, F., Kraft, M., Herrmann, A. B., Viciano, C. P., Weber, C., Hochhaus, A., Ernst, T., Hoffmann, C., Zernecke, A., & Frietsch, J. J. (2020). Phosphorylation-Dependent Differences in CXCR4-LASP1-AKT1 Interaction between Breast Cancer and Chronic Myeloid Leukemia. Cells, 9(2), 444. https://doi.org/10.3390/cells9020444





