snR30/U17 Small Nucleolar Ribonucleoprotein: A Critical Player during Ribosome Biogenesis
Abstract
1. Introduction
2. snR30 Biogenesis, Sequence, and Structure
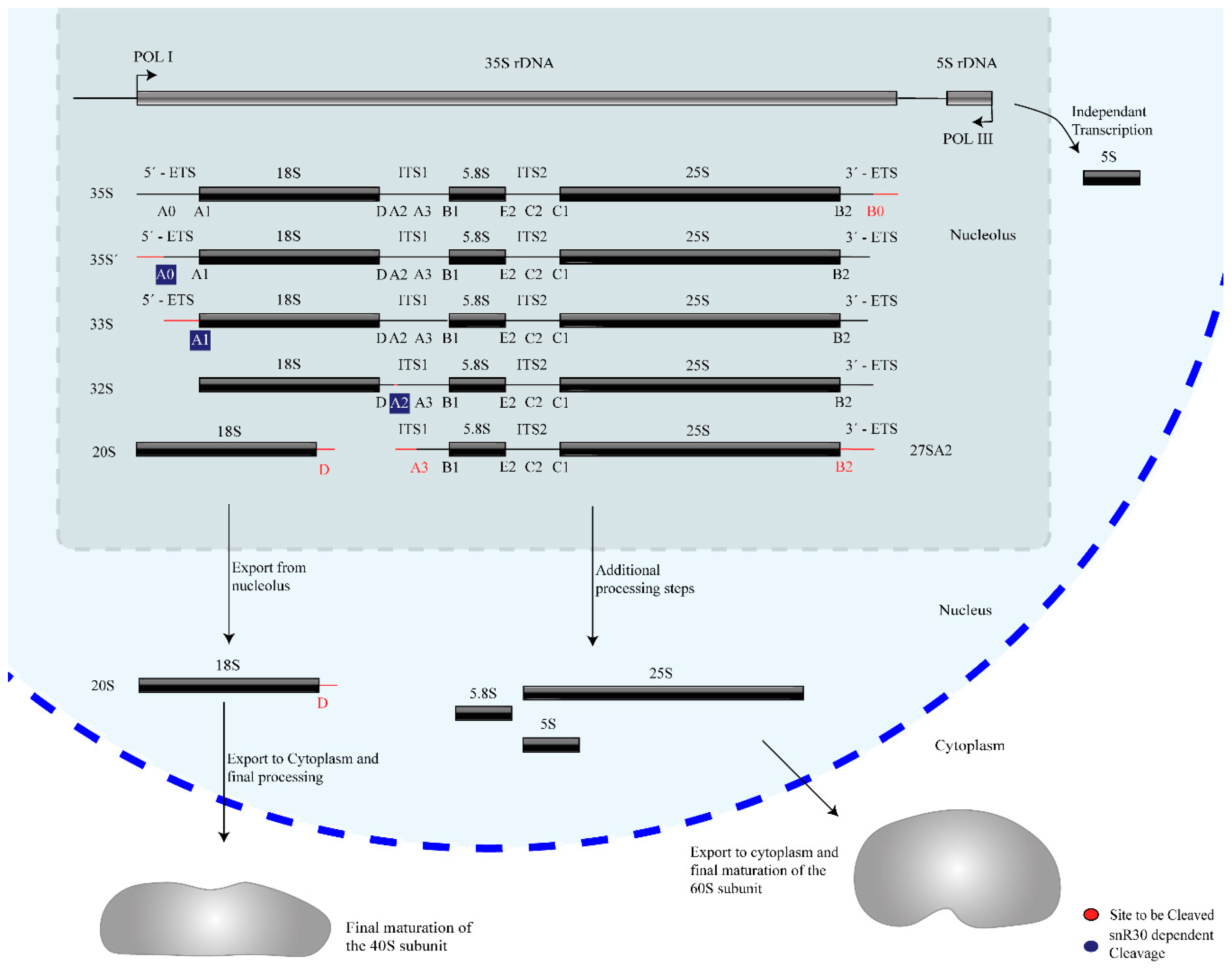
2.1. Transcription and Processing
2.2. snR30 snoRNP Formation and Nucleolar Localization
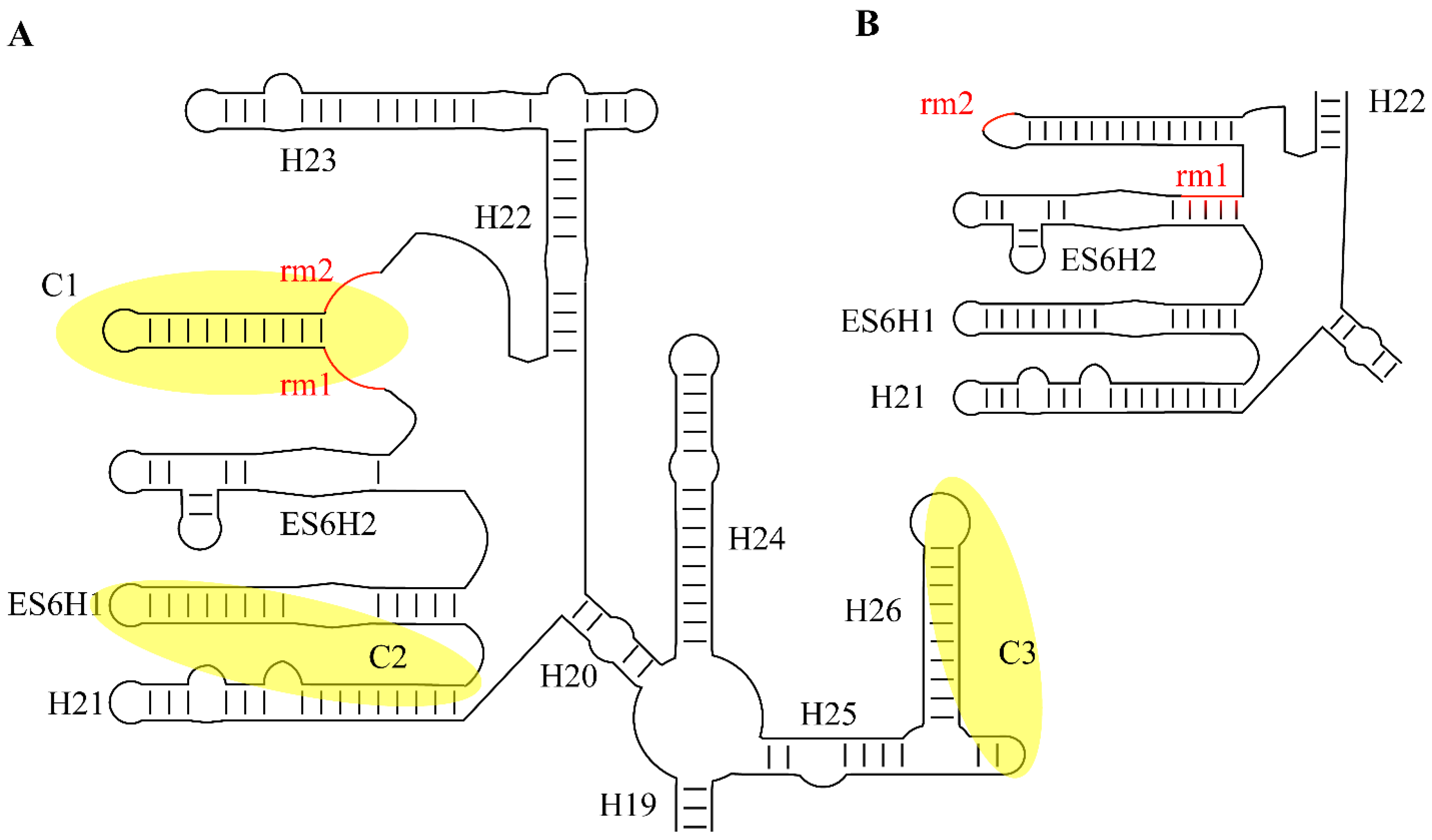
2.3. Conservation of snR30/U17 Structure, Sequence, and Motifs

| Organism | Common Name | Gene Name | Length (nt) | m1 | m2 | Accession Number | Reference |
|---|---|---|---|---|---|---|---|
| Apteryx rowi | Kiwi | E1/U17 | 205 | AUAUUCCUA | AAACCAU | XR_003255265.1 | N/A |
| Caenorhabditis elegans | Round worm | CeN96 | 221 | AUAUUCCUU | AAACCAU | AY948718.1 | [80] |
| Caretta caretta | Turtle | U17 | 213 | AUAUUCCUA | AAACCAU | AJ306558 | [79] |
| Candida glabrata | Yeast | snR30 | 576 | AUAUUCCUG | AAACCAU | URS00006E7B8C | [81] a |
| Danio rerio | Zebra fish | E1/U17 | >140 | AUAUUCCUA | AAACCAU | LR812563.1 | N/A |
| Fugu rubripes | Pufferfish | U17 | 218 | AUAUUCCUA | AAACCAU | X94942 | [82] |
| Homo sapiens | Humans | U17 | 207 | AUAUUCCUA | AAACCAU | L16791 | [74] |
| Kazachstania naganishii | Yeast | snR30 | 589 | AUAUUCCUA | AAACCAU | URS0000BE6F45 | [81] a |
| Mus musculus | Mouse | U17/SNORA73 | 203 | AUAUUCCUA | AAACCAU | XR_003836540.1 | N/A |
| Pelomedusa subrufa | Turtle | U17 | 215 | AUAUUCCUA | AAACCAU c | AJ306565 | [79] |
| Saccharomyces cerevisiae | Yeast | snR30 | 606 | AUAUUCCUA | AAACCAU | NR_132204 | [74] |
| Saccharomyces pombe | Yeast | U17 | 325 | AUAUUCCUA | AAACCAU | AJ544685 | [74] |
| Tetrahymena thermophila | Algae | U17 | ~240 b | AUAUUCCUG | AAACCAU | AJ544686 | [74] |
| Xenopus laevis | Frog | U17 | 222 | AUAUUCCUA | AAACCAU | X71081 | [54] |
3. Role of snR30/U17 in rRNA Processing
3.1. Site-Specific Binding to the 18S rRNA
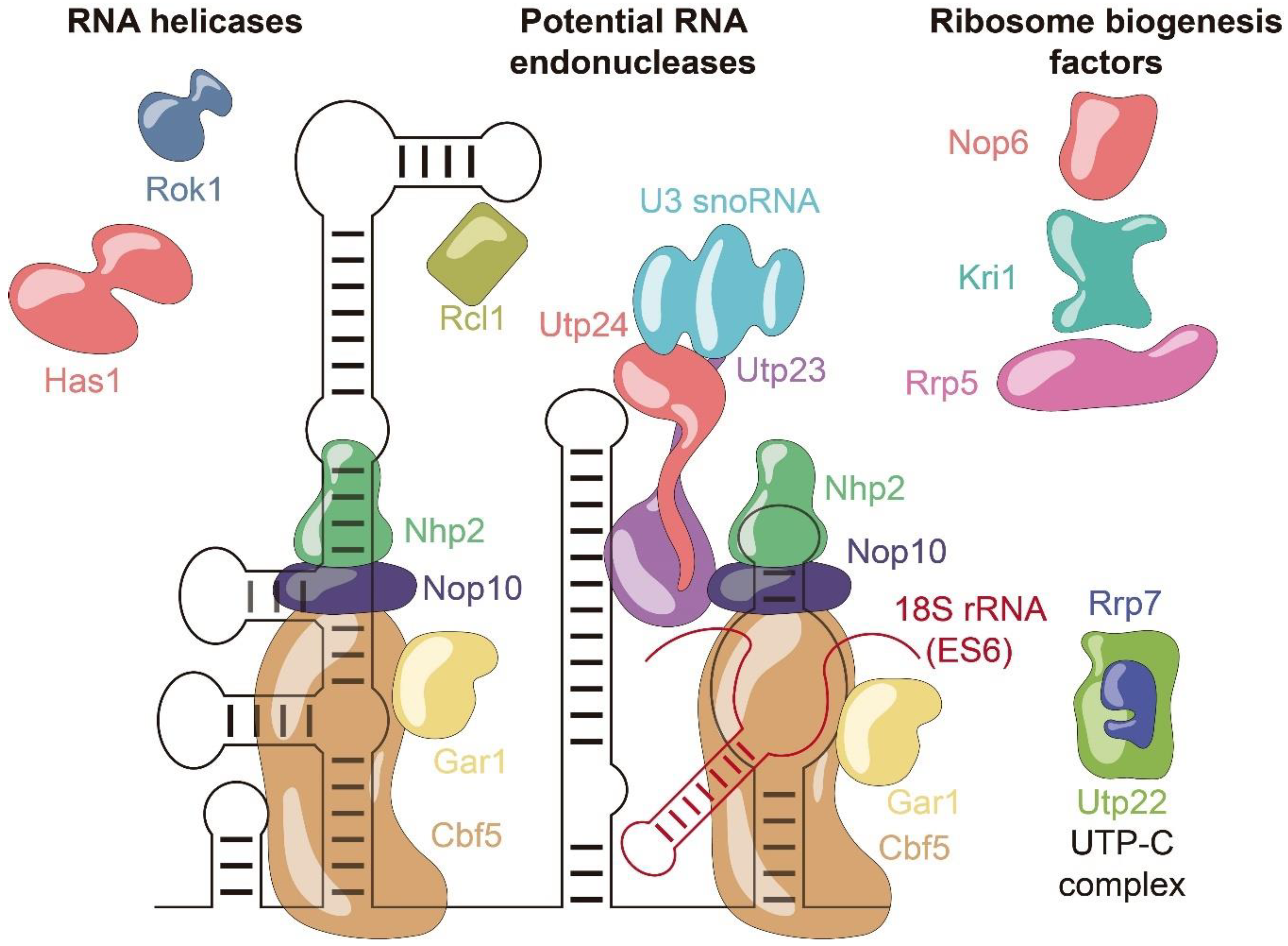
3.2. snR30 Protein Recruitment
3.3. Hypothetical Functions of snR30 during Ribosome Formation
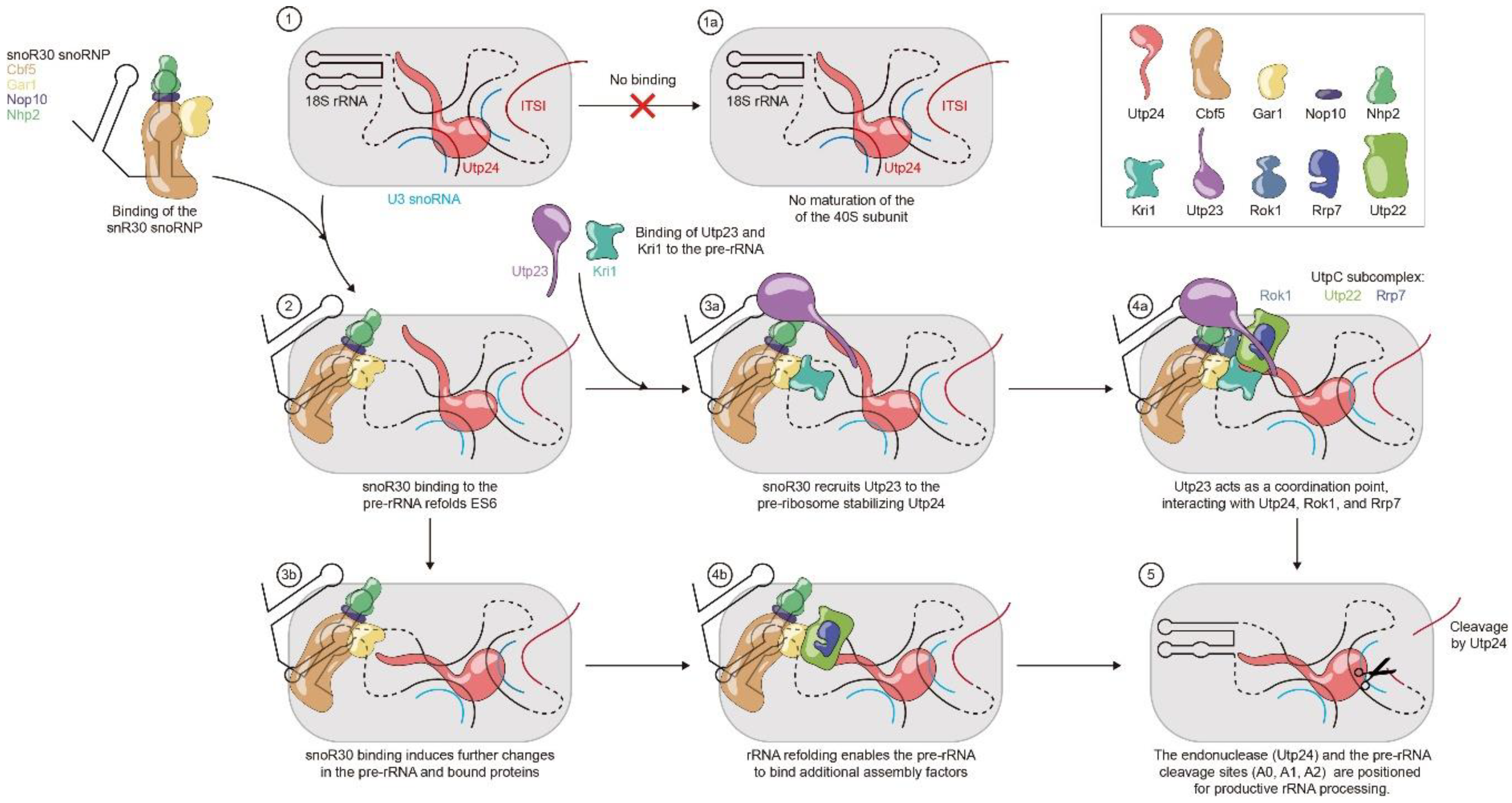
3.3.1. snR30 Mechanism—Hypothesis 1
3.3.2. snR30 Mechanism—Hypothesis 2
4. snR30 Release from the Pre-Ribosomal Particle
4.1. Required Factors for snR30 Release
4.2. Timing of the snR30 snoRNP Release
5. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| cryo-EM | cryo-electron microscopy |
| ES | eukaryotic expansion segment |
| ETS | external transcribed spacer |
| ITS | internal transcribed spacer |
| pre-rRNA | precursor ribosomal RNA |
| RNP | Ribonucleoprotein |
| rRCS | ribosomal RNA complementary sequence |
| rRNA | ribosomal ribonucleic acid |
| scaRNA | small Cajal body RNAs |
| snoRNP | small nucleolar ribonucleoprotein |
| SSU | small subunit of the ribosome |
| Utp | U-three protein |
References
- Eliceiri, G.L. The vertebrate E1/U17 small nucleolar ribonucleoprotein particle. J. Cell. Biochem. 2006, 98, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.; Ferreira-Cerca, S.; Hurt, E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013, 126, 4815–4821. [Google Scholar] [CrossRef] [PubMed]
- Klinge, S.; Woolford, J.L., Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Phipps, K.R.; Charette, J.; Baserga, S.J. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev. RNA 2011, 2, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kos, M.; Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Kister, K.P.; Muller, B.; Eckert, W.A. Complex endonucleolytic cleavage pattern during early events in the processing of pre-rRNA in the lower eukaryote, Tetrahymena thermophila. Nucleic Acids Res. 1983, 11, 3487–3502. [Google Scholar] [CrossRef] [PubMed]
- Hadjiolova, K.V.; Nicoloso, M.; Mazan, S.; Hadjiolov, A.A.; Bachellerie, J.P. Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem. 1993, 212, 211–215. [Google Scholar] [CrossRef]
- Kiss-Laszlo, Z.; Henry, Y.; Bachellerie, J.P.; Caizergues-Ferrer, M.; Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 1996, 85, 1077–1088. [Google Scholar] [CrossRef]
- Ni, J.; Tien, A.L.; Fournier, M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997, 89, 565–573. [Google Scholar] [CrossRef]
- Tollervey, D.; Kiss, T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997, 9, 337–342. [Google Scholar] [CrossRef]
- Lafontaine, D.L.; Tollervey, D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 1999, 5, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.; Tollervey, D. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell Biol. 2000, 20, 2650–2659. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.J.; Segault, V.; Charpentier, B.; Nottrott, S.; Fabrizio, P.; Bachi, A.; Wilm, M.; Rosbash, M.; Branlant, C.; Luhrmann, R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 2000, 103, 457–466. [Google Scholar] [CrossRef]
- Girard, J.P.; Lehtonen, H.; Caizerguesferrer, M.; Amalric, F.; Tollervey, D.; Lapeyre, B. Gar1 is an essential small nucleolar rnp protein required for pre-ribosomal-rna processing in yeast. EMBO J. 1992, 11, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.J.; Bousquet-Antonelli, C.; Henry, Y.; Caizergues-Ferrer, M.; Tollervey, D. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Gene Dev. 1998, 12, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Henras, A.; Henry, Y.; Bousquet-Antonelli, C.; Noaillac-Depeyre, J.; Gelugne, J.P.; Caizergues-Ferrer, M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998, 17, 7078–7090. [Google Scholar] [CrossRef]
- Zebarjadian, Y.; King, T.; Fournier, M.J.; Clarke, L.; Carbon, J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell Biol. 1999, 19, 7461–7472. [Google Scholar] [CrossRef]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef]
- Liang, X.H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef]
- Baudin-Baillieu, A.; Fabret, C.; Liang, X.H.; Piekna-Przybylska, D.; Fournier, M.J.; Rousset, J.P. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009, 37, 7665–7677. [Google Scholar] [CrossRef]
- Piekna-Przybylska, D.; Przybylski, P.; Baudin-Baillieu, A.; Rousset, J.P.; Fournier, M.J. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem. 2008, 283, 26026–26036. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Liu, Q.; Fournier, M.J. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 2007, 28, 965–977. [Google Scholar] [CrossRef] [PubMed]
- King, T.H.; Liu, B.; McCully, R.R.; Fournier, M.J. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 2003, 11, 425–435. [Google Scholar] [CrossRef]
- Tollervey, D.; Guthrie, C. Deletion of a yeast small nuclear RNA gene impairs growth. EMBO J. 1985, 4, 3873–3878. [Google Scholar] [CrossRef] [PubMed]
- Bally, M.; Hughes, J.; Cesareni, G. Snr30—A new, essential small nuclear-rna from saccharomyces-cerevisiae. Nucleic Acids Res. 1988, 16, 5291–5303. [Google Scholar] [CrossRef]
- Morrissey, J.P.; Tollervey, D. Yeast Snr30 is a small nucleolar rna required for 18s ribosomal-rna synthesis. Mol. Cell. Biol. 1993, 13, 2469–2477. [Google Scholar] [CrossRef]
- Jinn, S.; Brandis, K.A.; Ren, A.; Chacko, A.; Dudley-Rucker, N.; Gale, S.E.; Sidhu, R.; Fujiwara, H.; Jiang, H.; Olsen, B.N.; et al. snoRNA U17 regulates cellular cholesterol trafficking. Cell Metab. 2015, 21, 855–867. [Google Scholar] [CrossRef]
- Dutca, L.M.; Gallagher, J.E.; Baserga, S.J. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011, 39, 5164–5180. [Google Scholar] [CrossRef]
- Brink, M.F.; Verbeet, M.P.; de Boer, H.A. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993, 12, 3987–3996. [Google Scholar] [CrossRef]
- Hughes, J.M.; Ares, M., Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5’ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991, 10, 4231–4239. [Google Scholar] [CrossRef]
- Li, H.D.; Zagorski, J.; Fournier, M.J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1990, 10, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.Q.; Fournier, M.J. U14 base-pairs with 18S rRNA: A novel snoRNA interaction required for rRNA processing. Genes Dev. 1995, 9, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.P.; Tollervey, D. U14 small nucleolar RNA makes multiple contacts with the pre-ribosomal RNA. Chromosoma 1997, 105, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Hackert, P.; Ruprecht, M.; Simm, S.; Bruning, L.; Mirus, O.; Sloan, K.E.; Kudla, G.; Schleiff, E.; Bohnsack, M.T. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA 2014, 20, 1173–1182. [Google Scholar] [CrossRef]
- Tollervey, D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987, 6, 4169–4175. [Google Scholar] [CrossRef]
- Cheng, J.; Kellner, N.; Berninghausen, O.; Hurt, E.; Beckmann, R. 3.2-A-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat. Struct. Mol. Biol. 2017, 24, 954–964. [Google Scholar] [CrossRef]
- Barandun, J.; Chaker-Margot, M.; Hunziker, M.; Molloy, K.R.; Chait, B.T.; Klinge, S. The complete structure of the small-subunit processome. Nat. Struct. Mol. Biol. 2017, 24, 944–953. [Google Scholar] [CrossRef]
- Chaker-Margot, M.; Barandun, J.; Hunziker, M.; Klinge, S. Architecture of the yeast small subunit processome. Science 2017, 355. [Google Scholar] [CrossRef]
- Dragon, F.; Gallagher, J.E.; Compagnone-Post, P.A.; Mitchell, B.M.; Porwancher, K.A.; Wehner, K.A.; Wormsley, S.; Settlage, R.E.; Shabanowitz, J.; Osheim, Y.; et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002, 417, 967–970. [Google Scholar] [CrossRef]
- Grandi, P.; Rybin, V.; Bassler, J.; Petfalski, E.; Strauss, D.; Marzioch, M.; Schafer, T.; Kuster, B.; Tschochner, H.; Tollervey, D.; et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 2002, 10, 105–115. [Google Scholar] [CrossRef]
- Zhang, L.M.; Wu, C.; Cai, G.H.; Chen, S.; Ye, K.Q. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Gene Dev. 2016, 30, 718–732. [Google Scholar] [CrossRef]
- Chaker-Margot, M.; Hunziker, M.; Barandun, J.; Dill, B.D.; Klinge, S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat. Struct. Mol. Biol. 2015, 22, 920–923. [Google Scholar] [CrossRef] [PubMed]
- Chaker-Margot, M.; Klinge, S. Assembly and early maturation of large subunit precursors. RNA 2019, 25, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xie, Z.; Yang, F.; Ye, K. Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res. 2017, 45, 6837–6847. [Google Scholar] [CrossRef] [PubMed]
- Perez-Fernandez, J.; Roman, A.; De Las Rivas, J.; Bustelo, X.R.; Dosil, M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007, 27, 5414–5429. [Google Scholar] [CrossRef] [PubMed]
- Kornprobst, M.; Turk, M.; Kellner, N.; Cheng, J.; Flemming, D.; Kos-Braun, I.; Kos, M.; Thoms, M.; Berninghausen, O.; Beckmann, R.; et al. Architecture of the 90S pre-ribosome: A structural view on the birth of the eukaryotic ribosome. Cell 2016, 166, 380–393. [Google Scholar] [CrossRef]
- Cheng, J.; Bassler, J.; Fischer, P.; Lau, B.; Kellner, N.; Kunze, R.; Griesel, S.; Kallas, M.; Berninghausen, O.; Strauss, D.; et al. Thermophile 90S pre-ribosome structures reveal the reverse order of co-transcriptional 18S rRNA subdomain integration. Mol. Cell 2019, 75, 1256–1269.e1257. [Google Scholar] [CrossRef]
- Ursic, D.; Himmel, K.L.; Gurley, K.A.; Webb, F.; Culbertson, M.R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997, 25, 4778–4785. [Google Scholar] [CrossRef]
- Steinmetz, E.J.; Brow, D.A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell Biol. 1996, 16, 6993–7003. [Google Scholar] [CrossRef]
- Chanfreau, G.; Legrain, P.; Jacquier, A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 1998, 284, 975–988. [Google Scholar] [CrossRef]
- Mouaikel, J.; Verheggen, C.; Bertrand, E.; Tazi, J.; Bordonne, R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 2002, 9, 891–901. [Google Scholar] [CrossRef]
- Kiss, T.; Filipowicz, W. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 1993, 12, 2913–2920. [Google Scholar] [CrossRef] [PubMed]
- Pelczar, P.; Filipowicz, W. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5’-terminal oligopyrimidine gene family. Mol. Cell Biol. 1998, 18, 4509–4518. [Google Scholar] [CrossRef]
- Cecconi, F.; Mariottini, P.; Loreni, F.; Pierandrei-Amaldi, P.; Campioni, N.; Amaldi, F. U17XS8, a small nucleolar RNA with a 12 nt complementarity to 18S rRNA and coded by a sequence repeated in the six introns of Xenopus laevis ribosomal protein S8 gene. Nucleic Acids Res. 1994, 22, 732–741. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Filipowicz, W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995, 9, 1411–1424. [Google Scholar] [CrossRef]
- Cecconi, F.; Mariottini, P.; Amaldi, F. The Xenopus intron-encoded U17 snoRNA is produced by exonucleolytic processing of its precursor in oocytes. Nucleic Acids Res. 1995, 23, 4670–4676. [Google Scholar] [CrossRef]
- Szczepinska, T.; Kalisiak, K.; Tomecki, R.; Labno, A.; Borowski, L.S.; Kulinski, T.M.; Adamska, D.; Kosinska, J.; Dziembowski, A. DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Res. 2015, 25, 1622–1633. [Google Scholar] [CrossRef]
- Caffarelli, E.; Fatica, A.; Prislei, S.; De Gregorio, E.; Fragapane, P.; Bozzoni, I. Processing of the intron-encoded U16 and U18 snoRNAs: The conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996, 15, 1121–1131. [Google Scholar] [CrossRef]
- Kufel, J.; Grzechnik, P. Small nucleolar RNAs Tell a different tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef]
- Yang, P.K.; Rotondo, G.; Porras, T.; Legrain, P.; Chanfreau, G. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J. Biol. Chem. 2002, 277, 45235–45242. [Google Scholar] [CrossRef]
- Walbott, H.; Machado-Pinilla, R.; Liger, D.; Blaud, M.; Rety, S.; Grozdanov, P.N.; Godin, K.; van Tilbeurgh, H.; Varani, G.; Meier, U.T.; et al. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev. 2011, 25, 2398–2408. [Google Scholar] [CrossRef] [PubMed]
- Caton, E.A.; Kelly, E.K.; Kamalampeta, R.; Kothe, U. Efficient RNA pseudouridylation by eukaryotic H/ACA ribonucleoproteins requires high affinity binding and correct positioning of guide RNA. Nucleic Acids Res. 2018, 46, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Machado-Pinilla, R.; Liger, D.; Leulliot, N.; Meier, U.T. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA 2012, 18, 1833–1845. [Google Scholar] [CrossRef]
- Darzacq, X.; Kittur, N.; Roy, S.; Shav-Tal, Y.; Singer, R.H.; Meier, U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006, 173, 207–218. [Google Scholar] [CrossRef]
- Grozdanov, P.N.; Roy, S.; Kittur, N.; Meier, U.T. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA 2009, 15, 1188–1197. [Google Scholar] [CrossRef]
- Narayanan, A.; Lukowiak, A.; Jady, B.E.; Dragon, F.; Kiss, T.; Terns, R.M.; Terns, M.P. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 1999, 18, 5120–5130. [Google Scholar] [CrossRef]
- Lange, T.S.; Ezrokhi, M.; Amaldi, F.; Gerbi, S.A. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell 1999, 10, 3877–3890. [Google Scholar] [CrossRef][Green Version]
- Dragon, F.; Pogacic, V.; Filipowicz, W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell Biol. 2000, 20, 3037–3048. [Google Scholar] [CrossRef][Green Version]
- Samarsky, D.A.; Fournier, M.J.; Singer, R.H.; Bertrand, E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998, 17, 3747–3757. [Google Scholar] [CrossRef]
- Narayanan, A.; Speckmann, W.; Terns, R.; Terns, M.P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 1999, 10, 2131–2147. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Fayet, E.; Jady, B.E.; Richard, P.; Weber, M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Meier, U.T. RNA modification in Cajal bodies. RNA Biol. 2017, 14, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Atzorn, V.; Fragapane, P.; Kiss, T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell Biol. 2004, 24, 1769–1778. [Google Scholar] [CrossRef]
- Ganot, P.; Bortolin, M.L.; Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997, 89, 799–809. [Google Scholar] [CrossRef]
- Cervelli, M.; Oliverio, M.; Bellini, A.; Bologna, M.; Cecconi, F.; Mariottini, P. Structural and sequence evolution of U17 small nucleolar RNA (snoRNA) and its phylogenetic congruence in chelonians. J. Mol. Evol. 2003, 57, 73–84. [Google Scholar] [CrossRef]
- Hughes, J.M.; Konings, D.A.; Cesareni, G. The yeast homologue of U3 snRNA. EMBO J. 1987, 6, 2145–2155. [Google Scholar] [CrossRef]
- Ruhl, D.D.; Pusateri, M.E.; Eliceiri, G.L. Multiple conserved segments of E1 small nucleolar RNA are involved in the formation of a ribonucleoprotein particle in frog oocytes. Biochem. J. 2000, 348, 517–524. [Google Scholar] [CrossRef]
- Cervelli, M.; Cecconi, F.; Giorgi, M.; Annesi, F.; Oliverio, M.; Mariottini, P. Comparative structure analysis of vertebrate U17 small nucleolar RNA (snoRNA). J. Mol. Evol. 2002, 54, 166–179. [Google Scholar] [CrossRef]
- Deng, W.; Zhu, X.; Skogerbo, G.; Zhao, Y.; Fu, Z.; Wang, Y.; He, H.; Cai, L.; Sun, H.; Liu, C.; et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: Genomic features, biogenesis, and expression. Genome Res. 2006, 16, 20–29. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.Y.; Eddy, S.R.; Floden, E.W.; Gardner, P.P.; Jones, T.A.; Tate, J.; et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015, 43, D130–D137. [Google Scholar] [CrossRef] [PubMed]
- Cecconi, F.; Crosio, C.; Mariottini, P.; Cesareni, G.; Giorgi, M.; Brenner, S.; Amaldi, F. A functional role for some Fugu introns larger than the typical short ones: The example of the gene coding for ribosomal protein S7 and snoRNA U17. Nucleic Acids Res. 1996, 24, 3167–3172. [Google Scholar] [CrossRef] [PubMed]
- Lemay, V.; Hossain, A.; Osheim, Y.N.; Beyer, A.L.; Dragon, F. Identification of novel proteins associated with yeast snR30 small nucleolar RNA. Nucleic Acids Res. 2011, 39, 9659–9670. [Google Scholar] [CrossRef]
- Fayet-Lebaron, E.; Atzorn, V.; Henry, Y.; Kiss, T. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 2009, 28, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Lau, B.; La Venuta, G.; Ameismeier, M.; Berninghausen, O.; Hurt, E.; Beckmann, R. 90S pre-ribosome transformation into the primordial 40S subunit. Science 2020, 369, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Spahn, C.M.; Beckmann, R.; Eswar, N.; Penczek, P.A.; Sali, A.; Blobel, G.; Frank, J. Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell 2001, 107, 373–386. [Google Scholar] [CrossRef]
- Wells, G.R.; Weichmann, F.; Sloan, K.E.; Colvin, D.; Watkins, N.J.; Schneider, C. The ribosome biogenesis factor yUtp23/hUTP23 coordinates key interactions in the yeast and human pre-40S particle and hUTP23 contains an essential PIN domain. Nucleic Acids Res. 2017, 45, 4796–4809. [Google Scholar] [CrossRef]
- Wells, G.R.; Weichmann, F.; Colvin, D.; Sloan, K.E.; Kudla, G.; Tollervey, D.; Watkins, N.J.; Schneider, C. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res. 2016, 44, 5399–5409. [Google Scholar] [CrossRef]
- Horn, D.M.; Mason, S.L.; Karbstein, K. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J. Biol. Chem. 2011, 286, 34082–34087. [Google Scholar] [CrossRef]
- Lubben, B.; Fabrizio, P.; Kastner, B.; Luhrmann, R. Isolation and characterization of the small nucleolar ribonucleoprotein particle snr30 from saccharomyces-cerevisiae. J. Biol. Chem. 1995, 270, 11549–11554. [Google Scholar] [CrossRef]
- Liang, X.H.; Fournier, M.J. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol. Cell. Biol. 2006, 26, 7437–7450. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Kos, M.; Tollervey, D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008, 9, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Hoareau-Aveilla, C.; Fayet-Lebaron, E.; Jady, B.E.; Henras, A.K.; Kiss, T. Utp23p is required for dissociation of snR30 small nucleolar RNP from preribosomal particles. Nucleic Acids Res. 2012, 40, 3641–3652. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, M.Y.; Ye, K.Q. Structural and functional analysis of Utp23, a yeast ribosome synthesis factor with degenerate PIN domain. Rna-a Publ. Rna Soc. 2013, 19, 1815–1824. [Google Scholar] [CrossRef]
- Barandun, J.; Hunziker, M.; Klinge, S. Assembly and structure of the SSU processome-a nucleolar precursor of the small ribosomal subunit. Curr. Opin. Struct. Biol. 2018, 49, 85–93. [Google Scholar] [CrossRef]
- Duan, J.; Li, L.; Lu, J.; Wang, W.; Ye, K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol. Cell 2009, 34, 427–439. [Google Scholar] [CrossRef]
- Lebaron, S.; Segerstolpe, A.; French, S.L.; Dudnakova, T.; Alves, F.D.; Granneman, S.; Rappsilber, J.; Beyer, A.L.; Wieslander, L.; Tollervey, D. Rrp5 binding at multiple sites coordinates pre-rRNA processing and assembly. Mol. Cell 2013, 52, 707–719. [Google Scholar] [CrossRef]
- Venema, J.; Tollervey, D. RRP5 is required for formation of both 18S and 5.85 rRNA in yeast. EMBO J. 1996, 15, 5701–5714. [Google Scholar] [CrossRef]
- Khoshnevis, S.; Liu, X.; Dattolo, M.D.; Karbstein, K. Rrp5 establishes a checkpoint for 60S assembly during 40S maturation. RNA 2019, 25, 1164–1176. [Google Scholar] [CrossRef]
- Khoshnevis, S.; Askenasy, I.; Johnson, M.C.; Dattolo, M.D.; Young-Erdos, C.L.; Stroupe, M.E.; Karbstein, K. The DEAD-box protein rok1 orchestrates 40S and 60S ribosome assembly by promoting the release of Rrp5 from Pre-40S ribosomes to allow for 60S maturation. PLoS Biol. 2016, 14. [Google Scholar] [CrossRef]
- Watkins, N.J.; Gottschalk, A.; Neubauer, G.; Kastner, B.; Fabrizio, P.; Mann, M.; Luhrmann, R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 1998, 4, 1549–1568. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.; Tollervey, D. Birth of the snoRNPs: The evolution of the modification-guide snoRNAs. Trends Biochem. Sci. 1998, 23, 383–388. [Google Scholar] [CrossRef]
- Li, S.; Duan, J.; Li, D.; Yang, B.; Dong, M.; Ye, K. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev. 2011, 25, 2409–2421. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vos, T.J.; Kothe, U. snR30/U17 Small Nucleolar Ribonucleoprotein: A Critical Player during Ribosome Biogenesis. Cells 2020, 9, 2195. https://doi.org/10.3390/cells9102195
Vos TJ, Kothe U. snR30/U17 Small Nucleolar Ribonucleoprotein: A Critical Player during Ribosome Biogenesis. Cells. 2020; 9(10):2195. https://doi.org/10.3390/cells9102195
Chicago/Turabian StyleVos, Timothy John, and Ute Kothe. 2020. "snR30/U17 Small Nucleolar Ribonucleoprotein: A Critical Player during Ribosome Biogenesis" Cells 9, no. 10: 2195. https://doi.org/10.3390/cells9102195
APA StyleVos, T. J., & Kothe, U. (2020). snR30/U17 Small Nucleolar Ribonucleoprotein: A Critical Player during Ribosome Biogenesis. Cells, 9(10), 2195. https://doi.org/10.3390/cells9102195







