Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways
Abstract
1. Introduction
- What additional transposons’, viruses’, and genes’ silencing roles might explain the expansion of Piwi genes in culicine mosquitoes but not in anopheline mosquitoes and Drosophilids?
- Can mosquito Piwi pathways cause transcriptional chromatin silencing like in Drosophilids?
- What drives the immense diversity of piRNAs between mosquito species and strains, different transposons in their piRNA cluster loci or different states of virus infections?
- Can we harness or manipulate the mosquito Piwi/piRNA pathways to develop novel antiviral effectors for vector control strategies?
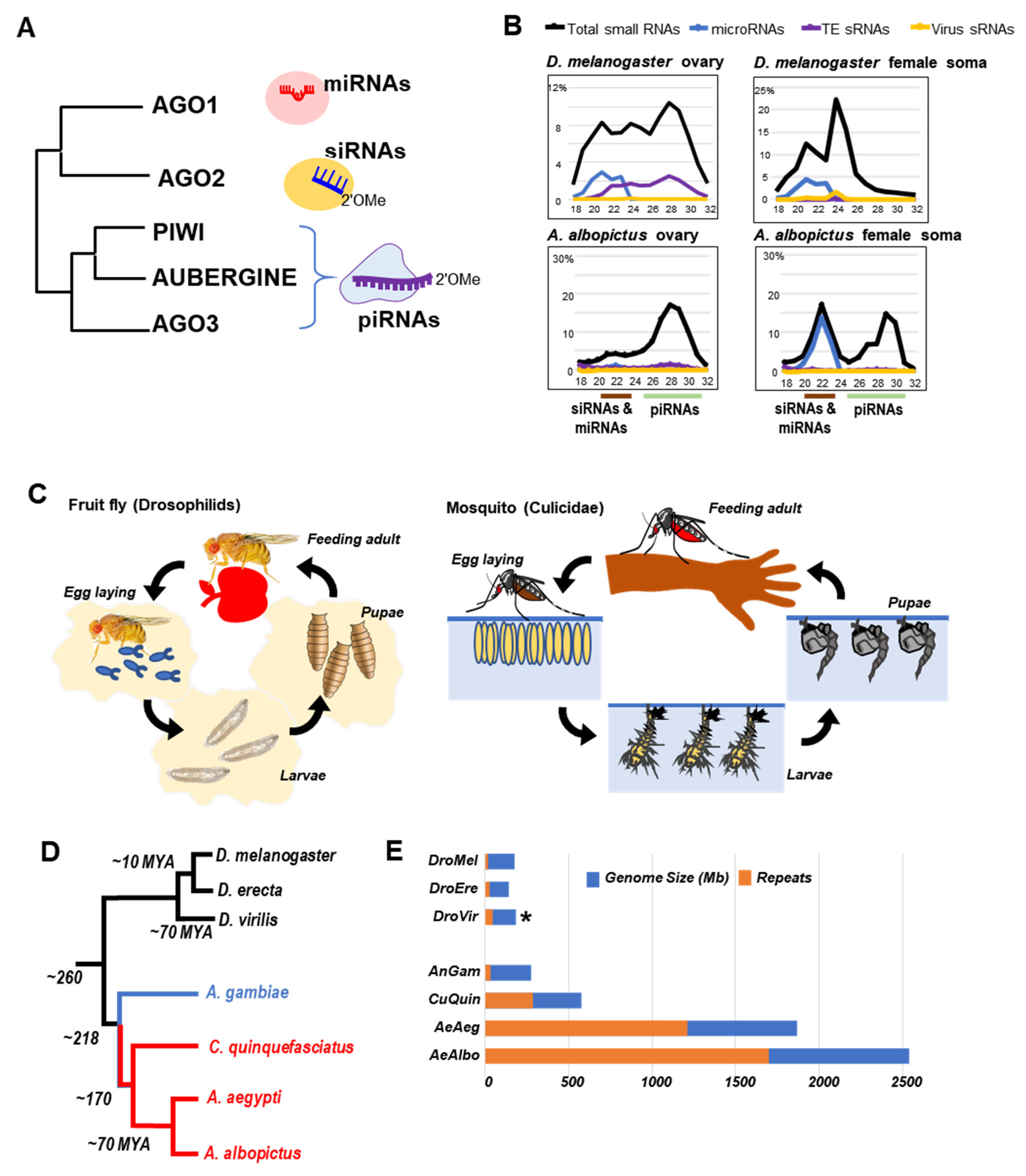
2. Genomic, Physiological and Evolutionary Differences between Drosophilids and Mosquitoes
3. Genome Transposon Composition and Germline Specificity of piRNAs in Drosophilids and Mosquitoes
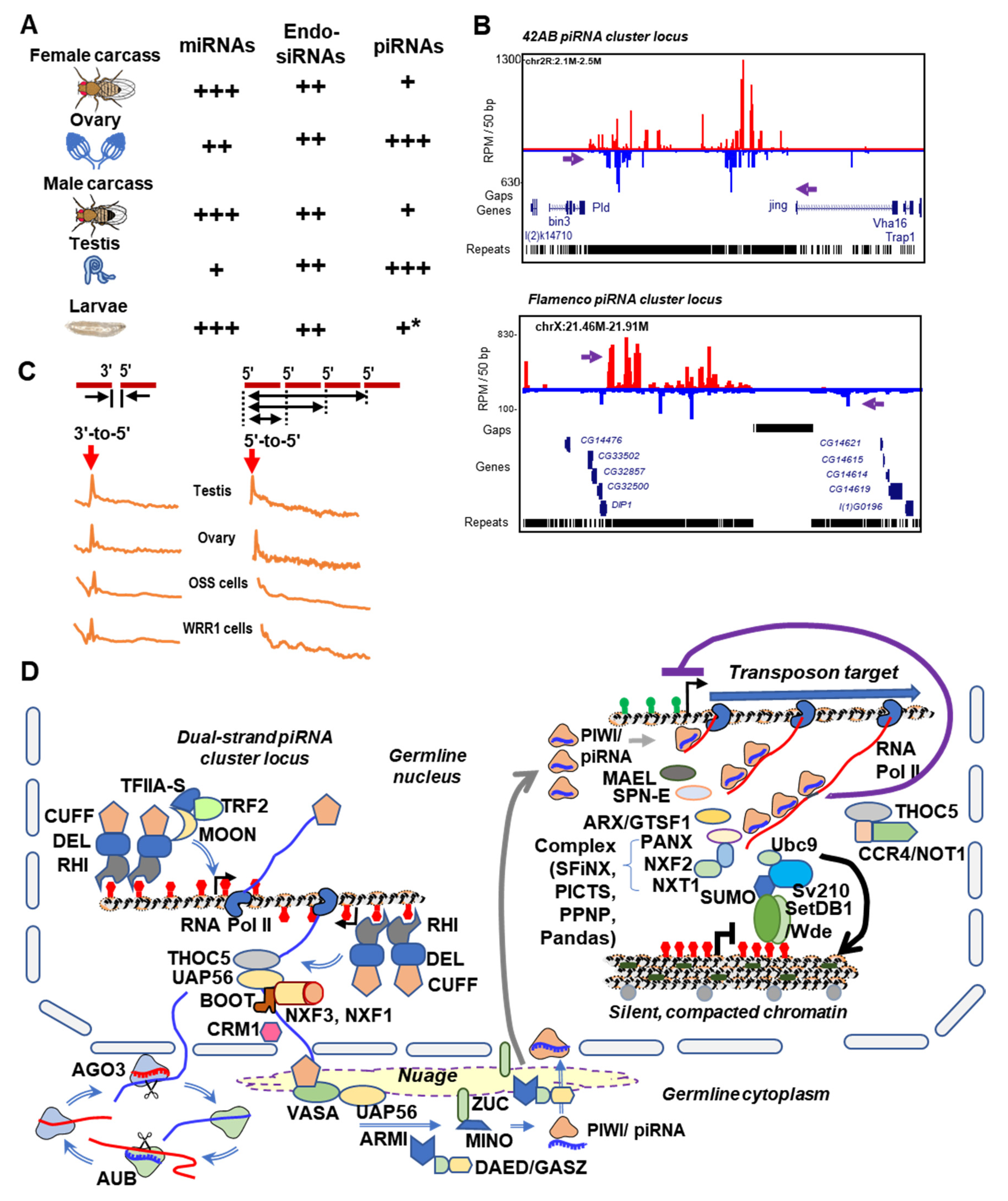
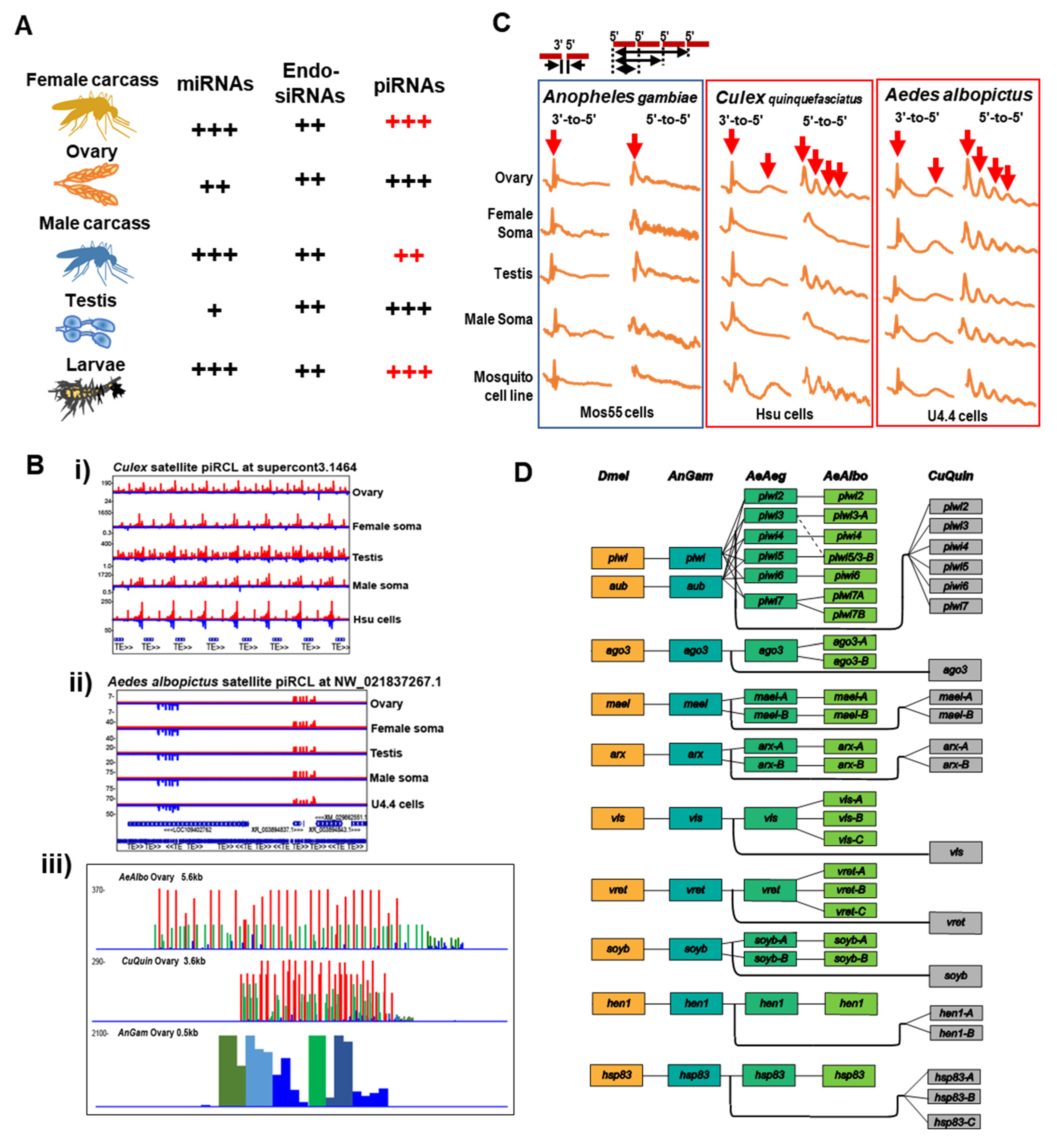
4. The piRNA Clusters of Drosophila and Culicidae—Genomic Memory Bank of Transposon Invasion Versus Unique piRNA Clusters Containing Satellite DNA Repeats
5. Rapid Evolution of Piwi Genes and piRNA Pathways
6. Culicine Mosquitoes Display Extraordinarily Phased piRNA Biogenesis Patterns
7. Viral piRNAs in Drosophila versus Mosquitoes: Minor in the Former and Major in the Latter?
8. Is the Role of Active Transposons Causing Hybrid Fertility in Drosophila Conserved in Mosquitoes?
9. How Conserved is the AGO2 Endo-siRNA Pathway between Drosophila and Mosquitoes?
10. RNAi Pathways as Applications for Mosquito Vector Control
11. Final Thoughts: Strengthening the Fly-Mosquito Partnership for Future Biomedical Progress
Funding
Acknowledgments
Conflicts of Interest
References
- Karess, R.E.; Rubin, G.M. Analysis of P transposable element functions in drosophila. Cell 1984, 38, 135–146. [Google Scholar] [CrossRef]
- Bingham, P.M.; Kidwell, M.G.; Rubin, G.M. The molecular basis of P-M hybrid dysgenesis: The role of the P element, a P-strain-specific transposon family. Cell 1982, 29, 995–1004. [Google Scholar] [CrossRef]
- Kidwell, M.G. Hybrid Dysgenesis in DROSOPHILA MELANOGASTER: Nature and Inheritance of P Element Regulation. Genetics 1985, 111, 337–350. [Google Scholar]
- Conway, M.J.; Colpitts, T.M.; Fikrig, E. Role of the Vector in Arbovirus Transmission. Annu. Rev. Virol. 2014, 1, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Lequime, S.; Lambrechts, L. Vertical transmission of arboviruses in mosquitoes: A historical perspective. Infect. Genet. Evol. 2014, 28, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Akkouche, A.; Rebollo, R.; Burlet, N.; Esnault, C.; Martinez, S.; Viginier, B.; Terzian, C.; Vieira, C.; Fablet, M. Tirant, a newly discovered active endogenous retrovirus in Drosophila simulans. J. Virol. 2012, 86, 3675–3681. [Google Scholar] [CrossRef]
- Dolei, A. Endogenous retroviruses and human disease. Expert Rev. Clin. Immunol. 2006, 2, 149–167. [Google Scholar] [CrossRef]
- Feschotte, C.; Gilbert, C. Endogenous viruses: Insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef]
- Malik, H.S.; Henikoff, S.; Eickbush, T.H. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000, 10, 1307–1318. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Wolf, Y.I.; Krupovic, M. Evolutionary entanglement of mobile genetic elements and host defence systems: Guns for hire. Nat. Rev. Genet. 2020, 21, 119–131. [Google Scholar] [CrossRef]
- Johnson, W.E. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 2019, 17, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Obbard, D.J.; Gordon, K.H.J.; Buck, A.H.; Jiggins, F.M. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007, 318, 761–764. [Google Scholar] [CrossRef]
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Hannon, G.J. Uncovering RNAi mechanisms in plants: Biochemistry enters the foray. FEBS Lett. 2005, 579, 5899–5903. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-L.; Rana, T.M. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell 2002, 10, 549–561. [Google Scholar] [CrossRef]
- Tijsterman, M.; Plasterk, R.H.A. Dicers at RISC; the mechanism of RNAi. Cell 2004, 117, 1–3. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Cheng, Z.; Zhu, Q. AGO2 and its partners: A silencing complex, a chromatin modulator, and new features. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 33–53. [Google Scholar] [CrossRef]
- Chawla, G.; Luhur, A.; Sokol, N. Analysis of MicroRNA Function in Drosophila. Methods Mol. Biol. 2016, 1478, 79–94. [Google Scholar] [CrossRef]
- Asgari, S. Role of microRNAs in arbovirus/vector interactions. Viruses 2014, 6, 3514–3534. [Google Scholar] [CrossRef]
- Dubey, S.K.; Shrinet, J.; Jain, J.; Ali, S.; Sunil, S. Aedes aegypti microRNA miR-2b regulates ubiquitin-related modifier to control chikungunya virus replication. Sci. Rep. 2017, 7, 17666. [Google Scholar] [CrossRef] [PubMed]
- Avila-Bonilla, R.G.; Yocupicio-Monroy, M.; Marchat, L.A.; Pérez-Ishiwara, D.G.; Cerecedo-Mercado, D.A.; Del Ángel, R.M.; Salas-Benito, J.S. miR-927 has pro-viral effects during acute and persistent infection with dengue virus type 2 in C6/36 mosquito cells. J. Gen. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.K.; Shrinet, J.; Sunil, S. Aedes aegypti microRNA, miR-2944b-5p interacts with 3′UTR of chikungunya virus and cellular target vps-13 to regulate viral replication. PLoS Negl. Trop. Dis. 2019, 13, e0007429. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Asgari, S. MicroRNA-like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc. Natl. Acad. Sci. USA 2014, 111, 2746–2751. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Olson, K.E.; Blair, C.D.; Garcia-Blanco, M.A.; Cullen, B.R. A “microRNA-like” small RNA expressed by Dengue virus? Proc. Natl. Acad. Sci. USA 2014, 111, E2359. [Google Scholar] [CrossRef] [PubMed]
- Finol, E. Are viral small RNA regulating Dengue virus replication beyond serotype 2? Proc. Natl. Acad. Sci. USA 2014, 111, E2915–E2916. [Google Scholar] [CrossRef]
- Feng, X.; Zhou, S.; Wang, J.; Hu, W. microRNA profiles and functions in mosquitoes. PLoS Negl. Trop. Dis. 2018, 12, e0006463. [Google Scholar] [CrossRef]
- Kritikou, E. Endo-siRNAs truly endogenous. Nat. Rev. Mol. Cell Boil. 2008, 9, 427. [Google Scholar] [CrossRef]
- Okamura, K.; Lai, E.C. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Boil. 2008, 9, 673–678. [Google Scholar] [CrossRef]
- Golden, D.E.; Gerbasi, V.R.; Sontheimer, E.J. An inside job for siRNAs. Mol. Cell 2008, 31, 309–312. [Google Scholar] [CrossRef]
- Claycomb, J.M. Ancient endo-siRNA pathways reveal new tricks. Curr. Biol. 2014, 24, R703–R715. [Google Scholar] [CrossRef] [PubMed]
- Lucas, K.J.; Myles, K.M.; Raikhel, A.S. Small RNAs: A new frontier in mosquito biology. Trends Parasitol. 2013, 29, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Bamezai, S.; Rawat, V.P.S.; Buske, C. Concise Review: The Piwi-piRNA Axis: Pivotal beyond Transposon Silencing. Stem Cells 2012, 30, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Munafò, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Fejes Tóth, K.; Aravin, A.A. piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 2017, 33, 882–894. [Google Scholar] [CrossRef]
- Czech, B.; Hannon, G.J. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef]
- Varjak, M.; Leggewie, M.; Schnettler, E. The antiviral piRNA response in mosquitoes? J. Gen. Virol. 2018, 99, 1551–1562. [Google Scholar] [CrossRef]
- Schraivogel, D.; Meister, G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem. Sci. 2014, 39, 420–431. [Google Scholar] [CrossRef]
- Okamura, K. Diversity of animal small RNA pathways and their biological utility. Wiley Interdiscip. Rev. RNA 2012, 3, 351–368. [Google Scholar] [CrossRef]
- Axtell, M.J.; Westholm, J.O.; Lai, E.C. Vive la différence: Biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011, 12, 221. [Google Scholar] [CrossRef]
- Clark, J.P.; Lau, N.C. Piwi Proteins and piRNAs step onto the systems biology stage. Adv. Exp. Med. Biol. 2014, 825, 159–197. [Google Scholar] [CrossRef] [PubMed]
- Parhad, S.S.; Theurkauf, W.E. Rapid evolution and conserved function of the piRNA pathway. Open Biol. 2019, 9, 180181. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Srivastav, S.P.; Gamez, S.; Feitosa-Suntheimer, F.; Patterson, E.I.; Johnson, R.M.; Matson, E.R.; Gold, A.S.; Brackney, D.E.; Connor, J.H.; et al. An integrated mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frézal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2018, 2, 174–181. [Google Scholar] [CrossRef]
- Flynn, J.M.; Long, M.; Wing, R.A.; Clark, A.G. Evolutionary Dynamics of Abundant 7-bp Satellites in the Genome of Drosophila virilis. Mol. Biol. Evol. 2020, 37, 1362–1375. [Google Scholar] [CrossRef]
- Stark, A.; Lin, M.F.; Kheradpour, P.; Pedersen, J.S.; Parts, L.; Carlson, J.W.; Crosby, M.A.; Rasmussen, M.D.; Roy, S.; Deoras, A.N.; et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 2007, 450, 219–232. [Google Scholar] [CrossRef]
- Hahn, M.W.; Han, M.V.; Han, S.-G. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007, 3, e197. [Google Scholar] [CrossRef]
- Matthews, B.J.; Dudchenko, O.; Kingan, S.B.; Koren, S.; Antoshechkin, I.; Crawford, J.E.; Glassford, W.J.; Herre, M.; Redmond, S.N.; Rose, N.H.; et al. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 2018, 563, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Dudchenko, O.; Batra, S.S.; Omer, A.D.; Nyquist, S.K.; Hoeger, M.; Durand, N.C.; Shamim, M.S.; Machol, I.; Lander, E.S.; Aiden, A.P.; et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 2017, 356, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Carissimo, G.; Pain, A.; Belda, E.; Vernick, K.D. Highly focused transcriptional response of Anopheles coluzzii to O’nyong nyong arbovirus during the primary midgut infection. BMC Genom. 2018, 19, 526. [Google Scholar] [CrossRef] [PubMed]
- Brustolin, M.; Pujhari, S.; Henderson, C.A.; Rasgon, J.L. Anopheles mosquitoes may drive invasion and transmission of Mayaro virus across geographically diverse regions. PLoS Negl. Trop. Dis. 2018, 12, e0006895. [Google Scholar] [CrossRef]
- Sardelis, M. Vector Competence of Selected North American Culex and Coquillettidia Mosquitoes for West Nile Virus. Emerg. Infect. Dis. 2001, 7, 1018–1022. [Google Scholar] [CrossRef]
- Bingham, A.M.; Burkett-Cadena, N.D.; Hassan, H.K.; Unnasch, T.R. Vector Competence and Capacity of Culex erraticus (Diptera: Culicidae) for Eastern Equine Encephalitis Virus in the Southeastern United States. J. Med. Entomol. 2016, 53, 473–476. [Google Scholar] [CrossRef]
- Vega-Rúa, A.; Zouache, K.; Girod, R.; Failloux, A.-B.; Lourenço-de-Oliveira, R. High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of Chikungunya virus. J. Virol. 2014, 88, 6294–6306. [Google Scholar] [CrossRef]
- McElroy, K.L.; Tsetsarkin, K.A.; Vanlandingham, D.L.; Higgs, S. Role of the yellow fever virus structural protein genes in viral dissemination from the Aedes aegypti mosquito midgut. J. Gen. Virol. 2006, 87, 2993–3001. [Google Scholar] [CrossRef]
- Kotsakiozi, P.; Gloria-Soria, A.; Caccone, A.; Evans, B.; Schama, R.; Martins, A.J.; Powell, J.R. Tracking the return of Aedes aegypti to Brazil, the major vector of the dengue, chikungunya and Zika viruses. PLoS Negl. Trop. Dis. 2017, 11, e0005653. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; von Mering, C.; Letunic, I.; Torrents, D.; Suyama, M.; Copley, R.R.; Christophides, G.K.; Thomasova, D.; Holt, R.A.; Subramanian, G.M.; et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 2002, 298, 149–159. [Google Scholar] [CrossRef]
- Rahman, R.; Chirn, G.-W.; Kanodia, A.; Sytnikova, Y.A.; Brembs, B.; Bergman, C.M.; Lau, N.C. Unique transposon landscapes are pervasive across Drosophila melanogaster genomes. Nucleic Acids Res. 2015, 43, 10655–10672. [Google Scholar] [CrossRef] [PubMed]
- Hill, T. Transposable element dynamics are consistent across the Drosophila phylogeny, despite drastically differing content. bioRxiv 2020. [Google Scholar] [CrossRef]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Flutre, T.; Duprat, E.; Feuillet, C.; Quesneville, H. Considering transposable element diversification in de novo annotation approaches. PLoS ONE 2011, 6, e16526. [Google Scholar] [CrossRef] [PubMed]
- Kaminker, J.S.; Bergman, C.M.; Kronmiller, B.; Carlson, J.; Svirskas, R.; Patel, S.; Frise, E.; Wheeler, D.A.; Lewis, S.E.; Rubin, G.M.; et al. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 2002, 3, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, E.S.; Barbash, D.A. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol. Biol. Evol. 2013, 30, 1816–1829. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Nene, V.; Wortman, J.R.; Lawson, D.; Haas, B.; Kodira, C.; Tu, Z.J.; Loftus, B.; Xi, Z.; Megy, K.; Grabherr, M.; et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 2007, 316, 1718–1723. [Google Scholar] [CrossRef]
- Neafsey, D.E.; Waterhouse, R.M.; Abai, M.R.; Aganezov, S.S.; Alekseyev, M.A.; Allen, J.E.; Amon, J.; Arcà, B.; Arensburger, P.; Artemov, G.; et al. Mosquito genomics. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 2015, 347, 1258522. [Google Scholar] [CrossRef]
- Arensburger, P.; Megy, K.; Waterhouse, R.M.; Abrudan, J.; Amedeo, P.; Antelo, B.; Bartholomay, L.; Bidwell, S.; Caler, E.; Camara, F.; et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 2010, 330, 86–88. [Google Scholar] [CrossRef]
- Chen, X.-G.; Jiang, X.; Gu, J.; Xu, M.; Wu, Y.; Deng, Y.; Zhang, C.; Bonizzoni, M.; Dermauw, W.; Vontas, J.; et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc. Natl. Acad. Sci. USA 2015, 112, E5907–E5915. [Google Scholar] [CrossRef] [PubMed]
- Marsano, R.M.; Leronni, D.; D’Addabbo, P.; Viggiano, L.; Tarasco, E.; Caizzi, R. Mosquitoes LTR retrotransposons: A deeper view into the genomic sequence of Culex quinquefasciatus. PLoS ONE 2012, 7, e30770. [Google Scholar] [CrossRef] [PubMed]
- Palatini, U.; Masri, R.A.; Cosme, L.V.; Koren, S.; Thibaud-Nissen, F.; Biedler, J.K.; Krsticevic, F.; Johnston, J.S.; Halbach, R.; Crawford, J.E.; et al. Improved reference genome of the arboviral vector Aedes albopictus. Genome Biol. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Lu, J. Silencing of Transposable Elements by piRNAs in Drosophila: An Evolutionary Perspective. Genom. Proteom. Bioinform. 2017, 15, 164–176. [Google Scholar] [CrossRef]
- Biryukova, I.; Ye, T. Endogenous siRNAs and piRNAs derived from transposable elements and genes in the malaria vector mosquito Anopheles gambiae. BMC Genom. 2015, 16, 278. [Google Scholar] [CrossRef]
- Tassetto, M.; Kunitomi, M.; Whitfield, Z.J.; Dolan, P.T.; Sánchez-Vargas, I.; Garcia-Knight, M.; Ribiero, I.; Chen, T.; Olson, K.E.; Andino, R. Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Saint-Leandre, B.; Capy, P.; Hua-Van, A.; Filée, J. piRNA and Transposon Dynamics in Drosophila: A Female Story. Genome Biol. Evol. 2020, 12, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; Richard McCombie, W.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.C.; Wood, J.G.; Chang, C.; Tam, A.D.; Franklin, M.J.; Siegel, E.R.; Helfand, S.L. A somatic piRNA pathway in the Drosophila fat body ensures metabolic homeostasis and normal lifespan. Nat. Commun. 2016, 7, 13856. [Google Scholar] [CrossRef]
- Story, B.; Ma, X.; Ishihara, K.; Li, H.; Hall, K.; Peak, A.; Anoja, P.; Park, J.; Haug, J.; Blanchette, M.; et al. Defining the expression of piRNA and transposable elements in Drosophila ovarian germline stem cells and somatic support cells. Life Sci. Alliance 2019, 2, e201800211. [Google Scholar] [CrossRef]
- Quénerch’Du, E.; Anand, A.; Kai, T. The piRNA pathway is developmentally regulated during spermatogenesis in Drosophila. RNA 2016, 22, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Nagao, A.; Mituyama, T.; Huang, H.; Chen, D.; Siomi, M.C.; Siomi, H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 2010, 16, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Merkle, J.A.; Wittes, J.; Schüpbach, T. Signaling between somatic follicle cells and the germline patterns the egg and embryo of Drosophila. Curr. Top. Dev. Biol. 2020, 140, 55–86. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R. Germ Plasm Biogenesis—An Oskar-Centric Perspective. Curr. Top. Dev. Biol. 2016, 116, 679–707. [Google Scholar] [CrossRef] [PubMed]
- Venkei, Z.G.; Choi, C.P.; Feng, S.; Chen, C.; Jacobsen, S.E.; Kim, J.K.; Yamashita, Y.M. A kinesin Klp10A mediates cell cycle-dependent shuttling of Piwi between nucleus and nuage. PLoS Genet. 2020, 16, e1008648. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, X.; Cao, C.; Wen, Y.; Sakashita, A.; Chen, S.; Zhang, J.; Zhang, Y.; Zhou, L.; Luo, M.; et al. UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat. Commun. 2019, 10, 4705. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T.; Watanabe, T.; Kuramochi-Miyagawa, S.; Takemoto, N.; Shiromoto, Y.; Kudo, A.; Kanai-Azuma, M.; Tashiro, F.; Miyazaki, S.; Katanaya, A.; et al. Mouse GTSF1 is an essential factor for secondary piRNA biogenesis. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Wu, P.-H.; Fu, Y.; Cecchini, K.; Özata, D.M.; Arif, A.; Yu, T.; Colpan, C.; Gainetdinov, I.; Weng, Z.; Zamore, P.D. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat. Genet. 2020, 52, 728–739. [Google Scholar] [CrossRef]
- Shiromoto, Y.; Kuramochi-Miyagawa, S.; Nagamori, I.; Chuma, S.; Arakawa, T.; Nishimura, T.; Hasuwa, H.; Tachibana, T.; Ikawa, M.; Nakano, T. GPAT2 is required for piRNA biogenesis, transposon silencing, and maintenance of spermatogonia in mice. Boil. Reprod. 2019, 101, 248–256. [Google Scholar] [CrossRef]
- Choi, C.P.; Tay, R.J.; Starostik, M.R.; Feng, S.; Moresco, J.J.; Montgomery, B.E.; Xu, E.; Hammonds, M.A.; Schatz, M.C.; Montgomery, T.A.; et al. SNPC-1.3 is a sex-specific transcription factor that drives male piRNA expression in C. elegans. bioRxiv 2020. [Google Scholar] [CrossRef]
- Arensburger, P.; Hice, R.H.; Wright, J.A.; Craig, N.L.; Atkinson, P.W. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genom. 2011, 12, 606. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Dong, Y.; Gu, J.; Puthiyakunnon, S.; Wu, Y.; Chen, X.-G. Developmental piRNA profiles of the invasive vector mosquito Aedes albopictus. Parasit. Vectors 2016, 9, 524. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.Y.; Simakov, O.; Bridge, D.M.; Cartwright, P.; Bellantuono, A.J.; Kuhn, A.; Holstein, T.W.; David, C.N.; Steele, R.E.; Martínez, D.E. Expansion of a single transposable element family is associated with genome-size increase and radiation in the genus Hydra. Proc. Natl. Acad. Sci. USA 2019, 116, 22915–22917. [Google Scholar] [CrossRef] [PubMed]
- Talla, V.; Suh, A.; Kalsoom, F.; Dincă, V.; Vila, R.; Friberg, M.; Wiklund, C.; Backström, N. Rapid Increase in Genome Size as a Consequence of Transposable Element Hyperactivity in Wood-White (Leptidea) Butterflies. Genome Boil. Evol. 2017, 9, 2491–2505. [Google Scholar] [CrossRef]
- Akbari, O.S.; Antoshechkin, I.; Amrhein, H.; Williams, B.; Diloreto, R.; Sandler, J.; Hay, B.A. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 2013, 3, 1493–1509. [Google Scholar] [CrossRef]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.F.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L.; et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.-C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef]
- Vourekas, A.; Alexiou, P.; Vrettos, N.; Maragkakis, M.; Mourelatos, Z. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 2016, 531, 390–394. [Google Scholar] [CrossRef]
- Barckmann, B.; Pierson, S.; Dufourt, J.; Papin, C.; Armenise, C.; Port, F.; Grentzinger, T.; Chambeyron, S.; Baronian, G.; Desvignes, J.-P.; et al. Aubergine iCLIP Reveals piRNA-Dependent Decay of mRNAs Involved in Germ Cell Development in the Early Embryo. Cell Rep. 2015, 12, 1205–1216. [Google Scholar] [CrossRef]
- Girardi, E.; Miesen, P.; Pennings, B.; Frangeul, L.; Saleh, M.-C.; van Rij, R.P. Histone-derived piRNA biogenesis depends on the ping-pong partners Piwi5 and Ago3 in Aedes aegypti. Nucleic Acids Res. 2017, 45, 4881–4892. [Google Scholar] [CrossRef]
- Betting, V.; Joosten, J.; Halbach, R.; Thaler, M.; Miesen, P.; Van Rij, R.P. A regulatory network of a piRNA and lncRNA initiates responder and trailer piRNA formation during embryonic development of Aedes mosquitoes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lau, N.C.; Robine, N.; Martin, R.; Chung, W.J.; Niki, Y.; Berezikov, E.; Lai, E.C. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009, 19, 1776–1785. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Inagaki, S.; Mituyama, T.; Kawamura, Y.; Ono, Y.; Sakota, E.; Kotani, H.; Asai, K.; Siomi, H.; Siomi, M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009, 461, 1296–1299. [Google Scholar] [CrossRef]
- Fagegaltier, D.; Falciatori, I.; Czech, B.; Castel, S.; Perrimon, N.; Simcox, A.; Hannon, G.J. Oncogenic transformation of Drosophila somatic cells induces a functional piRNA pathway. Genes Dev. 2016, 30, 1623–1635. [Google Scholar] [CrossRef]
- Vrettos, N.; Maragkakis, M.; Alexiou, P.; Mourelatos, Z. Kc167, a widely used Drosophila cell line, contains an active primary piRNA pathway. RNA 2017, 23, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Mohammed, J.; Bortolamiol-Becet, D.; Tsai, H.; Robine, N.; Westholm, J.O.; Ladewig, E.; Dai, Q.; Okamura, K.; Flynt, A.S.; et al. Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res. 2014, 24, 1236–1250. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, Y.; Zhang, H.; Farley, G.; Wang, J.; Quarles, K.A.; Weng, Z.; Zamore, P.D. The genome of the Hi5 germ cell line from, an agricultural pest and novel model for small RNA biology. eLife 2018, 7. [Google Scholar] [CrossRef]
- Kawaoka, S.; Hayashi, N.; Suzuki, Y.; Abe, H.; Sugano, S.; Tomari, Y.; Shimada, T.; Katsuma, S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 2009, 15, 1258–1264. [Google Scholar] [CrossRef]
- Kawaoka, S.; Izumi, N.; Katsuma, S.; Tomari, Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell 2011, 43, 1015–1022. [Google Scholar] [CrossRef]
- Honda, S.; Kirino, Y.; Maragkakis, M.; Alexiou, P.; Ohtaki, A.; Murali, R.; Mourelatos, Z.; Kirino, Y. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 2013, 19, 1405–1418. [Google Scholar] [CrossRef]
- Goriaux, C.; Théron, E.; Brasset, E.; Vaury, C. History of the discovery of a master locus producing piRNAs: The flamenco/COM locus in Drosophila melanogaster. Front. Genet. 2014, 5, 257. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. A cytological demonstration of the location of an interchange between two non-homologous chromosomes of zea mays. Proc. Natl. Acad. Sci. USA 1930, 16, 791–796. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Kneuss, E.; Munafò, M.; Eastwood, E.L.; Deumer, U.-S.; Preall, J.B.; Hannon, G.J.; Czech, B. Specialization of the Drosophila nuclear export family protein Nxf3 for piRNA precursor export. Genes Dev. 2019, 33, 1208–1220. [Google Scholar] [CrossRef]
- Ohtani, H.; Iwasaki, Y.W.; Shibuya, A.; Siomi, H.; Siomi, M.C.; Saito, K. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013, 27, 1656–1661. [Google Scholar] [CrossRef]
- Li, C.; Vagin, V.V.; Lee, S.; Xu, J.; Ma, S.; Xi, H.; Seitz, H.; Horwich, M.D.; Syrzycka, M.; Honda, B.M.; et al. Collapse of Germline piRNAs in the Absence of Argonaute3 Reveals Somatic piRNAs in Flies. Cell 2009, 137, 509–521. [Google Scholar] [CrossRef]
- Sato, K.; Siomi, M.C. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 32–42. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, J.; Koppetsch, B.S.; Wang, J.; Tipping, C.; Ma, S.; Weng, Z.; Theurkauf, W.E.; Zamore, P.D. Heterotypic piRNA Ping-Pong Requires Qin, a Protein with Both E3 Ligase and Tudor Domains. Mol. Cell 2011, 44, 1005. [Google Scholar] [CrossRef][Green Version]
- Saito, K.; Nishida, K.M.; Mori, T.; Kawamura, Y.; Miyoshi, K.; Nagami, T.; Siomi, H.; Siomi, M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006, 20, 2214–2222. [Google Scholar] [CrossRef]
- Guzzardo, P.M.; Muerdter, F.; Hannon, G.J. The piRNA pathway in flies: Highlights and future directions. Curr. Opin. Genet. Dev. 2013, 23, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 Homolog Rhino Is Required for Transposon Silencing and piRNA Production by Dual-Strand Clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.A.; Stuwe, E.; Luo, Y.; Ninova, M.; Le Thomas, A.; Rozhavskaya, E.; Li, S.; Vempati, S.; Laver, J.D.; Patel, D.J.; et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol. Cell 2016, 63, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Stuwe, E.; Li, S.; Du, J.; Marinov, G.; Rozhkov, N.; Chen, Y.-C.A.; Luo, Y.; Sachidanandam, R.; Toth, K.F.; et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014, 28, 1667–1680. [Google Scholar] [CrossRef]
- Pane, A.; Jiang, P.; Zhao, D.Y.; Singh, M.; Schüpbach, T. The Cutoff protein regulates piRNA cluster expression and piRNA production in theDrosophilagermline. EMBO J. 2011, 30, 4601–4615. [Google Scholar] [CrossRef]
- Czech, B.; Preall, J.B.; McGinn, J.; Hannon, G.J. A Transcriptome-wide RNAi Screen in the Drosophila Ovary Reveals Factors of the Germline piRNA Pathway. Mol. Cell 2013, 50, 749–761. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Schultz, N.; Zhang, F.; Parhad, S.S.; Tu, S.; Vreven, T.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 2014, 157, 1353–1363. [Google Scholar] [CrossRef]
- Dönertas, D.; Sienski, G.; Brennecke, J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013, 27, 1693–1705. [Google Scholar] [CrossRef]
- Goriaux, C.; Desset, S.; Renaud, Y.; Vaury, C.; Brasset, E. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 2014, 15, 411–418. [Google Scholar] [CrossRef]
- Brasset, E.; Taddei, A.R.; Arnaud, F.; Faye, B.; Fausto, A.M.; Mazzini, M.; Giorgi, F.; Vaury, C. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology 2006, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Chalvet, F.; Teysset, L.; Terzian, C.; Prud’Homme, N.; Santamaria, P.; Bucheton, A.; Pélisson, A. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999, 18, 2659–2669. [Google Scholar] [CrossRef] [PubMed]
- Pélisson, A.; Song, S.U.; Prud’Homme, N.; Smith, P.A.; Bucheton, A.; Corces, V.G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994, 13, 4401–4411. [Google Scholar] [CrossRef] [PubMed]
- Pelisson, A.; Mejlumian, L.; Robert, V.; Terzian, C.; Bucheton, A. Drosophila germline invasion by the endogenous retrovirus gypsy: Involvement of the viral env gene. Insect Biochem. Mol. Biol. 2002, 32, 1249–1256. [Google Scholar] [CrossRef]
- Sokolova, O.A.; Mikhaleva, E.A.; Kharitonov, S.L.; Abramov, Y.A.; Gvozdev, V.A.; Klenov, M.S. Special vulnerability of somatic niche cells to transposable element activation in Drosophila larval ovaries. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- George, P.; Jensen, S.; Pogorelcnik, R.; Lee, J.; Xing, Y.; Brasset, E.; Vaury, C.; Sharakhov, I.V. Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenetics Chromatin 2015, 8, 50. [Google Scholar] [CrossRef]
- Aguiar, E.R.G.R.; de Almeida, J.P.P.; Queiroz, L.R.; Oliveira, L.S.; Olmo, R.P.; de Faria, I.J.d.S.; Imler, J.-L.; Gruber, A.; Matthews, B.J.; Marques, J.T. A single unidirectional piRNA cluster similar to the locus is the major source of EVE-derived transcription and small RNAs in mosquitoes. RNA 2020, 26, 581–594. [Google Scholar] [CrossRef]
- Ishizu, H.; Kinoshita, T.; Hirakata, S.; Komatsuzaki, C.; Siomi, M.C. Distinct and Collaborative Functions of Yb and Armitage in Transposon-Targeting piRNA Biogenesis. Cell Rep. 2019, 27, 1822–1835.e8. [Google Scholar] [CrossRef]
- Duc, C.; Yoth, M.; Jensen, S.; Mouniee, N.; Bergman, C.M.; Vaury, C.; Brasset, E. Trapping a somatic endogenous retrovirus into a germline piRNA cluster immunizes the germline against further invasion. Genome Biol. 2019, 20, 127. [Google Scholar] [CrossRef]
- Yu, B.; Lin, Y.A.; Parhad, S.S.; Jin, Z.; Ma, J.; Theurkauf, W.E.; Zhang, Z.Z.Z.; Huang, Y. Structural insights into Rhino-Deadlock complex for germline piRNA cluster specification. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Parhad, S.S.; Tu, S.; Weng, Z.; Theurkauf, W.E. Adaptive Evolution Leads to Cross-Species Incompatibility in the piRNA Transposon Silencing Machinery. Dev. Cell 2017, 43, 60–70.e5. [Google Scholar] [CrossRef] [PubMed]
- Vermaak, D.; Henikoff, S.; Malik, H.S. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005, 1, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Rogers, A.K.; Webster, A.; Marinov, G.K.; Liao, S.E.; Perkins, E.M.; Hur, J.K.; Aravin, A.A.; Tóth, K.F. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Sienski, G.; Dönertas, D.; Brennecke, J. Transcriptional Silencing of Transposons by Piwi and Maelstrom and Its Impact on Chromatin State and Gene Expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, S.; Miao, N.; Xu, P.; Lu, X.; Zhang, Y.; Wang, M.; Ouyang, X.; Yuan, X.; Liu, W.; et al. A Pandas complex adapted for piRNA-guided transcriptional silencing and heterochromatin formation. Nat. Cell Biol. 2019, 21, 1261–1272. [Google Scholar] [CrossRef]
- Ninova, M.; Chen, Y.-C.A.; Godneeva, B.; Rogers, A.K.; Luo, Y.; Fejes Tóth, K.; Aravin, A.A. Su(var)2-10 and the SUMO Pathway Link piRNA-Guided Target Recognition to Chromatin Silencing. Mol. Cell 2020, 77, 556–570.e6. [Google Scholar] [CrossRef]
- ElMaghraby, M.F.; Andersen, P.R.; Puhringer, F.; Hohmann, U.; Meixner, K.; Lendl, T.; Tirian, L.; Brennecke, J. A Heterochromatin-Specific RNA Export Pathway Facilitates piRNA Production. Cell 2019, 178, 964–979.e20. [Google Scholar] [CrossRef]
- Batki, J.; Schnabl, J.; Wang, J.; Handler, D.; Andreev, V.I.; Stieger, C.E.; Novatchkova, M.; Lampersberger, L.; Kauneckaite, K.; Xie, W.; et al. The nascent RNA binding complex SFiNX licenses piRNA-guided heterochromatin formation. Nat. Struct. Mol. Biol. 2019, 26, 720–731. [Google Scholar] [CrossRef]
- Murano, K.; Iwasaki, Y.W.; Ishizu, H.; Mashiko, A.; Shibuya, A.; Kondo, S.; Adachi, S.; Suzuki, S.; Saito, K.; Natsume, T.; et al. Nuclear RNA export factor variant initiates piRNA-guided co-transcriptional silencing. EMBO J. 2019, 38, e102870. [Google Scholar] [CrossRef]
- Fabry, M.H.; Ciabrelli, F.; Munafo, M.; Eastwood, E.L.; Kneuss, E.; Falciatori, I.; Falconio, F.A.; Hannon, G.J.; Czech, B. piRNA-guided co-transcriptional silencing coopts nuclear export factors. eLife 2019, 8. [Google Scholar] [CrossRef]
- Mugat, B.; Nicot, S.; Varela-Chavez, C.; Jourdan, C.; Sato, K.; Basyuk, E.; Juge, F.; Siomi, M.C.; Pelisson, A.; Chambeyron, S. The Mi-2 nucleosome remodeler and the Rpd3 histone deacetylase are involved in piRNA-guided heterochromatin formation. Nat. Commun. 2020, 11, 2818. [Google Scholar] [CrossRef]
- Kordyukova, M.; Sokolova, O.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Glukhov, S.; Kalmykova, A. Nuclear Ccr4-Not mediates the degradation of telomeric and transposon transcripts at chromatin in the Drosophila germline. Nucleic Acids Res. 2020, 48, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, B.; Liu, P.; Li, J.; Chen, X.; Gu, J. piRNA Profiling of Dengue Virus Type 2-Infected Asian Tiger Mosquito and Midgut Tissues. Viruses 2018, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.H.; Salmela, H.; Obbard, D.J. Duplication and Diversification of Dipteran Argonaute Genes, and the Evolutionary Divergence of Piwi and Aubergine. Genome Biol. Evol. 2016, 8, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Miesen, P.; Girardi, E.; van Rij, R.P. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015, 43, 6545–6556. [Google Scholar] [CrossRef]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC Protein Family: United by Structure, Divergent in Function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef]
- Milewska, A.; Kindler, E.; Vkovski, P.; Zeglen, S.; Ochman, M.; Thiel, V.; Rajfur, Z.; Pyrc, K. APOBEC3-mediated restriction of RNA virus replication. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Bruno, M.; Mahgoub, M.; Macfarlan, T.S. The Arms Race between KRAB-Zinc Finger Proteins and Endogenous Retroelements and Its Impact on Mammals. Annu. Rev. Genet. 2019, 53, 393–416. [Google Scholar] [CrossRef]
- Jacobs, F.M.; Greenberg, D.; Nguyen, N.; Haeussler, M.; Ewing, A.D.; Katzman, S.; Paten, B.; Salama, S.R.; Haussler, D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 2014, 516, 242–245. [Google Scholar] [CrossRef]
- Soleimani, S.; Valizadeh Arshad, Z.; Moradi, S.; Ahmadi, A.; Davarpanah, S.J.; Azimzadeh Jamalkandi, S. Small regulatory noncoding RNAs in Drosophila melanogaster: Biogenesis and biological functions. Brief. Funct. Genom. 2020, 19, 309–323. [Google Scholar] [CrossRef]
- Lau, N.C. Small RNAs in the animal gonad: Guarding genomes and guiding development. Int. J. Biochem. Cell Biol. 2010, 42, 1334–1347. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Narry Kim, V.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Boil. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790.e5. [Google Scholar] [CrossRef]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Hoa, N.T.; Keene, K.M.; Olson, K.E.; Zheng, L. Characterization of RNA interference in an Anopheles gambiae cell line. Insect Biochem. Mol. Boil. 2003, 33, 949–957. [Google Scholar] [CrossRef]
- Joosten, J.; Miesen, P.; Taskopru, E.; Pennings, B.; Jansen, P.; Huynen, M.A.; Vermeulen, M.; Van Rij, R.P. The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 2019, 47, 2546–2559. [Google Scholar] [CrossRef]
- Mohn, F.; Handler, D.; Brennecke, J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef]
- Feltzin, V.L.; Khaladkar, M.; Abe, M.; Parisi, M.; Hendriks, G.-J.; Kim, J.; Bonini, N.M. The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell 2015, 14, 443–452. [Google Scholar] [CrossRef]
- Hayashi, R.; Schnabl, J.; Handler, D.; Mohn, F.; Ameres, S.L.; Brennecke, J. Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 2016, 539, 588–592. [Google Scholar] [CrossRef]
- Ge, D.T.; Wang, W.; Tipping, C.; Gainetdinov, I.; Weng, Z.; Zamore, P.D. The RNA-Binding ATPase, Armitage, Couples piRNA Amplification in Nuage to Phased piRNA Production on Mitochondria. Mol. Cell 2019, 74, 982–995.e6. [Google Scholar] [CrossRef] [PubMed]
- Xiol, J.; Spinelli, P.; Laussmann, M.A.; Homolka, D.; Yang, Z.; Cora, E.; Coute, Y.; Conn, S.; Kadlec, J.; Sachidanandam, R.; et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 2014, 157, 1698–1711. [Google Scholar] [CrossRef] [PubMed]
- Wenda, J.M.; Homolka, D.; Yang, Z.; Spinelli, P.; Sachidanandam, R.; Pandey, R.R.; Pillai, R.S. Distinct Roles of RNA Helicases MVH and TDRD9 in PIWI Slicing-Triggered Mammalian piRNA Biogenesis and Function. Dev. Cell 2017, 41, 623–637.e9. [Google Scholar] [CrossRef] [PubMed]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef]
- Yamashiro, H.; Siomi, M.C. PIWI-Interacting RNA in Drosophila: Biogenesis, Transposon Regulation, and Beyond. Chem. Rev. 2018, 118, 4404–4421. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.-X.; Ding, S.-W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef]
- Chotkowski, H.L.; Ciota, A.T.; Jia, Y.; Puig-Basagoiti, F.; Kramer, L.D.; Shi, P.-Y.; Glaser, R.L. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology 2008, 377, 197–206. [Google Scholar] [CrossRef]
- Zambon, R.A.; Vakharia, V.N.; Wu, L.P. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell. Microbiol. 2006, 8, 880–889. [Google Scholar] [CrossRef]
- Petit, M.; Mongelli, V.; Frangeul, L.; Blanc, H.; Jiggins, F.; Saleh, M.-C. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2016, 113, E4218–E4227. [Google Scholar] [CrossRef]
- Saito, K.; Siomi, M.C. Small RNA-Mediated Quiescence of Transposable Elements in Animals. Dev. Cell 2010, 19, 687–697. [Google Scholar] [CrossRef]
- Rozhkov, N.V.; Aravin, A.A.; Zelentsova, E.S.; Schostak, N.G.; Sachidanandam, R.; McCombie, W.R.; Hannon, G.J.; Evgen’Ev, M.B. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA 2010, 16, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Shpiz, S.; Ryazansky, S.; Olovnikov, I.; Abramov, Y.; Kalmykova, A. Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the drosophila germline. PLoS Genet. 2014, 10, e1004138. [Google Scholar] [CrossRef]
- Li, H. Induction and Suppression of RNA Silencing by an Animal Virus. Science 2002, 296, 1319–1321. [Google Scholar] [CrossRef]
- Galiana-Arnoux, D.; Dostert, C.; Schneemann, A.; Hoffmann, J.A.; Imler, J.-L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat. Immunol. 2006, 7, 590–597. [Google Scholar] [CrossRef]
- van Rij, R.P.; Saleh, M.-C.; Berry, B.; Foo, C.; Houk, A.; Antoniewski, C.; Andino, R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006, 20, 2985–2995. [Google Scholar] [CrossRef]
- Wang, X.-H.; Aliyari, R.; Li, W.-X.; Li, H.-W.; Kim, K.; Carthew, R.; Atkinson, P.; Ding, S.-W. RNA interference directs innate immunity against viruses in adult Drosophila. Science 2006, 312, 452–454. [Google Scholar] [CrossRef]
- Sabin, L.R.; Zhou, R.; Gruber, J.J.; Lukinova, N.; Bambina, S.; Berman, A.; Lau, C.-K.; Thompson, C.B.; Cherry, S. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 2009, 138, 340–351. [Google Scholar] [CrossRef]
- Tassetto, M.; Kunitomi, M.; Andino, R. Circulating Immune Cells Mediate a Systemic RNAi-Based Adaptive Antiviral Response in Drosophila. Cell 2017, 169, 314–325.e13. [Google Scholar] [CrossRef]
- Harsh, S.; Ozakman, Y.; Kitchen, S.M.; Paquin-Proulx, D.; Nixon, D.F.; Eleftherianos, I. Dicer-2 Regulates Resistance and Maintains Homeostasis against Zika Virus Infection in Drosophila. J. Immunol. 2018, 201, 3058–3072. [Google Scholar] [CrossRef]
- Nayak, A.; Berry, B.; Tassetto, M.; Kunitomi, M.; Acevedo, A.; Deng, C.; Krutchinsky, A.; Gross, J.; Antoniewski, C.; Andino, R. Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 2010, 17, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Martins, N.; Lemoine, A.; Santiago, E.; Paro, S.; Imler, J.-L.; Meignin, C. A Transgenic Flock House Virus Replicon Reveals an RNAi Independent Antiviral Mechanism Acting in Drosophila Follicular Somatic Cells. G3 2019, 9, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Soares, Z.G.; Gonçalves, A.N.A.; de Oliveira, K.P.V.; Marques, J.T. Viral RNA recognition by the Drosophila small interfering RNA pathway. Microbes Infect. 2014, 16, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Viginier, B.; Saint-Michel, É.; Arnaud, F.; Ratinier, M.; Fablet, M. Viral infection impacts transposable element transcript amounts in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 12249–12257. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Liu, Y.; Wang, P.; Xiao, X. Mosquito Defense Strategies against Viral Infection. Trends Parasitol. 2016, 32, 177–186. [Google Scholar] [CrossRef]
- Goic, B.; Stapleford, K.A.; Frangeul, L.; Doucet, A.J.; Gausson, V.; Blanc, H.; Schemmel-Jofre, N.; Cristofari, G.; Lambrechts, L.; Vignuzzi, M.; et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Miesen, P.; Ivens, A.; Buck, A.H.; van Rij, R.P. Small RNA Profiling in Dengue Virus 2-Infected Aedes Mosquito Cells Reveals Viral piRNAs and Novel Host miRNAs. PLoS Negl. Trop. Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef]
- Göertz, G.; Miesen, P.; Overheul, G.; van Rij, R.; van Oers, M.; Pijlman, G. Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells. Viruses 2019, 11, 271. [Google Scholar] [CrossRef]
- Belda, E.; Nanfack-Minkeu, F.; Eiglmeier, K.; Carissimo, G.; Holm, I.; Diallo, M.; Diallo, D.; Vantaux, A.; Kim, S.; Sharakhov, I.V.; et al. De novo profiling of RNA viruses in Anopheles malaria vector mosquitoes from forest ecological zones in Senegal and Cambodia. BMC Genom. 2019, 20, 664. [Google Scholar] [CrossRef]
- Siu, R.W.C.; Fragkoudis, R.; Simmonds, P.; Donald, C.L.; Chase-Topping, M.E.; Barry, G.; Attarzadeh-Yazdi, G.; Rodriguez-Andres, J.; Nash, A.A.; Merits, A.; et al. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: Characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J. Virol. 2011, 85, 2907–2917. [Google Scholar] [CrossRef]
- Paradkar, P.N.; Trinidad, L.; Voysey, R.; Duchemin, J.-B.; Walker, P.J. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 18915–18920. [Google Scholar] [CrossRef] [PubMed]
- Rückert, C.; Prasad, A.N.; Garcia-Luna, S.M.; Robison, A.; Grubaugh, N.D.; Weger-Lucarelli, J.; Ebel, G.D. Small RNA responses of Culex mosquitoes and cell lines during acute and persistent virus infection. Insect Biochem. Mol. Biol. 2019, 109, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Weger-Lucarelli, J.; Rückert, C.; Grubaugh, N.D.; Misencik, M.J.; Armstrong, P.M.; Stenglein, M.D.; Ebel, G.D.; Brackney, D.E. Adventitious viruses persistently infect three commonly used mosquito cell lines. Virology 2018, 521, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Barreto-Vieira, D.F.; Jácome, F.C.; da Silva, M.A.N.; Caldas, G.C.; de Filippis, A.M.B.; de Sequeira, P.C.; de Souza, E.M.; Andrade, A.A.; Manso, P.P.d.A.; Trindade, G.F.; et al. Structural investigation of C6/36 and Vero cell cultures infected with a Brazilian Zika virus. PLoS ONE 2017, 12, e0184397. [Google Scholar] [CrossRef] [PubMed]
- Buchman, A.; Gamez, S.; Li, M.; Antoshechkin, I.; Li, H.-H.; Wang, H.-W.; Chen, C.-H.; Klein, M.J.; Duchemin, J.-B.; Paradkar, P.N.; et al. Engineered resistance to Zika virus in transgenic Aedes aegypti expressing a polycistronic cluster of synthetic small RNAs. Proc. Natl. Acad. Sci. USA 2019, 116, 3656–3661. [Google Scholar] [CrossRef]
- Wu, X.; Hong, H.; Yue, J.; Wu, Y.; Li, X.; Jiang, L.; Li, L.; Li, Q.; Gao, G.; Yang, X. Inhibitory effect of small interfering RNA on dengue virus replication in mosquito cells. Virol. J. 2010, 7, 270. [Google Scholar] [CrossRef]
- Saldaña, M.A.; Etebari, K.; Hart, C.E.; Widen, S.G.; Wood, T.G.; Thangamani, S.; Asgari, S.; Hughes, G.L. Zika virus alters the microRNA expression profile and elicits an RNAi response in Aedes aegypti mosquitoes. PLoS Negl. Trop. Dis. 2017, 11, e0005760. [Google Scholar] [CrossRef]
- Léger, P.; Lara, E.; Jagla, B.; Sismeiro, O.; Mansuroglu, Z.; Coppée, J.Y.; Bonnefoy, E.; Bouloy, M. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J. Virol. 2013, 87, 1631–1648. [Google Scholar] [CrossRef]
- Holmes, E.C. The Evolution of Endogenous Viral Elements. Cell Host Microbe 2011, 10, 368–377. [Google Scholar] [CrossRef]
- Katzourakis, A.; Gifford, R.J. Endogenous viral elements in animal genomes. PLoS Genet. 2010, 6, e1001191. [Google Scholar] [CrossRef]
- Palatini, U.; Miesen, P.; Carballar-Lejarazu, R.; Ometto, L.; Rizzo, E.; Tu, Z.; van Rij, R.P.; Bonizzoni, M. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genom. 2017, 18, 512. [Google Scholar] [CrossRef] [PubMed]
- Lequime, S.; Lambrechts, L. Discovery of flavivirus-derived endogenous viral elements in two Anopheles mosquito genomes supports the existence of Anopheles-associated insect-specific flaviviruses. Virus Evol. 2017, 3, vew035. [Google Scholar] [CrossRef] [PubMed]
- Houé, V.; Bonizzoni, M.; Failloux, A.-B. Endogenous non-retroviral elements in genomes of Aedes mosquitoes and vector competence. Emerg. Microbes Infect. 2019, 8, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Lourenço-de-Oliveira, R.; Marques, J.T.; Sreenu, V.B.; Nten, C.A.; Guimarães, E.R.; Varjak, M.; Kohl, A.; Failloux, A.-B. Culex quinquefasciatus mosquitoes do not support replication of Zika virus. J. Gen. Virol. 2018, 99, 258–264. [Google Scholar] [CrossRef]
- Ter Horst, A.M.; Nigg, J.C.; Dekker, F.M.; Falk, B.W. Endogenous Viral Elements Are Widespread in Arthropod Genomes and Commonly Give Rise to PIWI-Interacting RNAs. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Suzuki, Y.; Baidaliuk, A.; Miesen, P.; Frangeul, L.; Crist, A.B.; Merkling, S.H.; Fontaine, A.; Lequime, S.; Moltini-Conclois, I.; Blanc, H.; et al. Non-retroviral Endogenous Viral Element Limits Cognate Virus Replication in Aedes aegypti Ovaries. Curr. Biol. 2020. [Google Scholar] [CrossRef]
- Dobzhansky, T. On the Sterility of the Interracial Hybrids in Drosophila Pseudoobscura. Proc. Natl. Acad. Sci. USA 1933, 19, 397–403. [Google Scholar] [CrossRef]
- Dobzhansky, T. Role of the Autosomes in the Drosophila Pseudoobscura Hybrids. Proc. Natl. Acad. Sci. USA 1933, 19, 950–953. [Google Scholar] [CrossRef]
- Dobzhansky, T. Studies on Hybrid Sterility. II. Localization of Sterility Factors in Drosophila Pseudoobscura Hybrids. Genetics 1936, 21, 113–135. [Google Scholar]
- Palopoli, M.F.; Wu, C.I. Genetics of hybrid male sterility between Drosophila sibling species: A complex web of epistasis is revealed in interspecific studies. Trends Genet. 1995, 11, 10. [Google Scholar] [CrossRef]
- Di Giacomo, M.; Comazzetto, S.; Saini, H.; De Fazio, S.; Carrieri, C.; Morgan, M.; Vasiliauskaite, L.; Benes, V.; Enright, A.J.; O’Carroll, D. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol. Cell 2013, 50, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, E.S.; Edelman, N.B.; Barbash, D.A. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012, 10, e1001428. [Google Scholar] [CrossRef] [PubMed]
- Rozhkov, N.V.; Hammell, M.; Hannon, G.J. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013, 27, 400–412. [Google Scholar] [CrossRef]
- Erwin, A.A.; Galdos, M.A.; Wickersheim, M.L.; Harrison, C.C.; Marr, K.D.; Colicchio, J.M.; Blumenstiel, J.P. piRNAs Are Associated with Diverse Transgenerational Effects on Gene and Transposon Expression in a Hybrid Dysgenic Syndrome of D. virilis. PLoS Genet. 2015, 11, e1005332. [Google Scholar] [CrossRef]
- Serrato-Capuchina, A.; Wang, J.; Earley, E.; Peede, D.; Isbell, K.; Matute, D.R. Paternally Inherited P-Element Copy Number Affects the Magnitude of Hybrid Dysgenesis in Drosophila simulans and D. melanogaster. Genome Biol. Evol. 2020, 12, 808–826. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.; Rosales-Stephens, H.-L.; Unckless, R.L. Rapid divergence of the copulation proteins in the Drosophila dunni group is associated with hybrid post-mating-prezygotic incompatibilities. bioRxiv 2020. [Google Scholar] [CrossRef]
- Brennecke, J.; Malone, C.D.; Aravin, A.A.; Sachidanandam, R.; Stark, A.; Hannon, G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008, 322, 1387–1392. [Google Scholar] [CrossRef]
- Kidwell, M.G.; Kidwell, J.F.; Sved, J.A. Hybrid Dysgenesis in drosophila melanogaster: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics 1977, 86, 813–833. [Google Scholar]
- Ota, R.; Kobayashi, S. Myc plays an important role in Drosophila P-M hybrid dysgenesis to eliminate germline cells with genetic damage. Commun. Biol. 2020, 3, 185. [Google Scholar] [CrossRef]
- Kotov, A.A.; Adashev, V.E.; Godneeva, B.K.; Ninova, M.; Shatskikh, A.S.; Bazylev, S.S.; Aravin, A.A.; Olenina, L.V. piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster. Nucleic Acids Res. 2019, 47, 4255–4271. [Google Scholar] [CrossRef]
- Desset, S.; Buchon, N.; Meignin, C.; Coiffet, M.; Vaury, C. In Drosophila melanogaster the COM Locus Directs the Somatic Silencing of Two Retrotransposons through both Piwi-Dependent and -Independent Pathways. PLoS ONE 2008, 3, e1526. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brookfield, J.F.Y. Genetic evidence for repression of somatic P element movements in Drosophila melanogaster consistent with a role for the KP element. Heredity 1996, 76, 384–391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Srivastav, S.P.; Kelleher, E.S. Paternal Induction of Hybrid Dysgenesis in Drosophila melanogaster Is Weakly Correlated with Both P-Element and hobo Element Dosage. G3 2017, 7, 1487–1497. [Google Scholar] [CrossRef] [PubMed]
- Srivastav, S.P.; Rahman, R.; Ma, Q.; Pierre, J.; Bandyopadhyay, S.; Lau, N.C. Har-P, a short P-element variant, weaponizes P-transposase to severely impair Drosophila development. eLife 2019, 8. [Google Scholar] [CrossRef]
- Itoh, M.; Boussy, I.A. Full-size P and KP elements predominate in wild Drosophila melanogaster. Genes Genet. Syst. 2002, 77, 259–267. [Google Scholar] [CrossRef][Green Version]
- Rasmusson, K.E.; Simmons, M.J.; Raymond, J.D.; McLarnon, C.F. Quantitative effects of P elements on hybrid dysgenesis in Drosophila melanogaster. Genetics 1990, 124, 647–662. [Google Scholar]
- Kofler, R. Dynamics of Transposable Element Invasions with piRNA Clusters. Mol. Biol. Evol. 2019, 36, 1457–1472. [Google Scholar] [CrossRef]
- Kelleher, E.S.; Azevedo, R.B.R.; Zheng, Y. The Evolution of Small-RNA-Mediated Silencing of an Invading Transposable Element. Genome Biol. Evol. 2018, 10, 3038–3057. [Google Scholar] [CrossRef]
- Kofler, R. piRNA clusters need a minimum size to control transposable element invasions. Genome Biol. Evol. 2020, 12, 736–749. [Google Scholar] [CrossRef]
- Bonnet, D.D. The hybridization of Aedes aegypti and Aedes albopictus in Hawaii. Proc. Hawaii Entomol. Soc. 1950, 14, 35–39. [Google Scholar]
- Harper, J.P.; Paulson, S.L. Reproductive isolation between Florida strains of Aedes aegypti and Aedes albopictus. J. Am. Mosq. Control Assoc. 1994, 10, 88–92. [Google Scholar] [PubMed]
- Lanzaro, G.C.; Narang, S.K.; Mitchell, S.E.; Kaiser, P.E.; Seawright, J.A. Hybrid Male Sterility in Crosses Between Field and Laboratory Strains of Anopheles quadrimaculatus (Say) (Diptera: Culicidae). J. Med. Entomol. 1988, 25, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Sharakhov, I.V. Premeiotic and meiotic failures lead to hybrid male sterility in the Anopheles gambiae complex. Proc. Biol. Sci. 2019, 286, 20191080. [Google Scholar] [CrossRef] [PubMed]
- Slotman, M.; della Torre, A.; Powell, J.R. The Genetics of Inviability and Male Sterility in Hybrids between Anopheles gambiae and An. arabiensis. Genetics 2004, 167, 275–287. [Google Scholar] [CrossRef]
- Slotman, M.; della Torre, A.; Powell, J.R. Female sterility in hybrids between Anopheles gambiae and A. arabiensis, and the causes of Haldane’s rule. Evolution 2005, 59, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Mathiopoulos, K.D.; della Torre, A.; Predazzi, V.; Petrarca, V.; Coluzzi, M. Cloning of inversion breakpoints in the Anopheles gambiae complex traces a transposable element at the inversion junction. Proc. Natl. Acad. Sci. USA 1998, 95, 12444–12449. [Google Scholar] [CrossRef] [PubMed]
- White, B.J.; Collins, F.H.; Besansky, N.J. Evolution of Anopheles gambiae in Relation to Humans and Malaria. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 111–132. [Google Scholar] [CrossRef]
- Aravin, A.A.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Inhibition of Gene Expression by Homologous Double-Stranded RNA in a Drosophila melanogasterCell Culture. Russ. J. Genet. 2001, 37, 639–642. [Google Scholar] [CrossRef]
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef]
- Aravin, A.A.; Klenov, M.S.; Vagin, V.V.; Rozovskii, Y.M.; Gvozdev, V.A. Role of double-stranded RNA in eukaryotic gene silencing. Mol. Biol. 2002, 36, 180–188. [Google Scholar] [CrossRef]
- Aravin, A.A.; Klenov, M.S.; Vagin, V.V.; Bantignies, F.; Cavalli, G.; Gvozdev, V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004, 24, 6742–6750. [Google Scholar] [CrossRef] [PubMed]
- Klenov, M.S.; Lavrov, S.A.; Stolyarenko, A.D.; Ryazansky, S.S.; Aravin, A.A.; Tuschl, T.; Gvozdev, V.A. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007, 35, 5430–5438. [Google Scholar] [CrossRef] [PubMed]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Mirkovic-Hösle, M.; Förstemann, K. Transposon Defense by Endo-siRNAs, piRNAs and Somatic pilRNAs in Drosophila: Contributions of Loqs-PD and R2D2. PLoS ONE 2014, 9, e84994. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.W.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef]
- Lim, D.-H.; Oh, C.-T.; Lee, L.; Hong, J.-S.; Noh, S.-H.; Hwang, S.; Kim, S.; Han, S.-J.; Lee, Y.S. The endogenous siRNA pathway in Drosophila impacts stress resistance and lifespan by regulating metabolic homeostasis. FEBS Lett. 2011, 585, 3079–3085. [Google Scholar] [CrossRef]
- Hartig, J.V.; Förstemann, K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011, 39, 3836–3851. [Google Scholar] [CrossRef]
- Kawamura, Y.; Saito, K.; Kin, T.; Ono, Y.; Asai, K.; Sunohara, T.; Okada, T.N.; Siomi, M.C.; Siomi, H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 2008, 453, 793–797. [Google Scholar] [CrossRef]
- Okamura, K.; Chung, W.-J.; Ruby, J.G.; Guo, H.; Bartel, D.P.; Lai, E.C. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 2008, 453, 803–806. [Google Scholar] [CrossRef]
- Adelman, Z.N.; Anderson, M.A.E.; Liu, M.; Zhang, L.; Myles, K.M. Sindbis virus induces the production of a novel class of endogenous siRNAs in Aedes aegypti mosquitoes. Insect Mol. Biol. 2012, 21, 357–368. [Google Scholar] [CrossRef]
- Castellano, L.; Rizzi, E.; Krell, J.; Di Cristina, M.; Galizi, R.; Mori, A.; Tam, J.; De Bellis, G.; Stebbing, J.; Crisanti, A.; et al. The germline of the malaria mosquito produces abundant miRNAs, endo-siRNAs, piRNAs and 29-nt small RNAs. BMC Genom. 2015, 16, 100. [Google Scholar] [CrossRef]
- Myles, K.M.; Wiley, M.R.; Morazzani, E.M.; Adelman, Z.N. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc. Natl. Acad. Sci. USA 2008, 105, 19938–19943. [Google Scholar] [CrossRef]
- Scott, J.C.; Brackney, D.E.; Campbell, C.L.; Bondu-Hawkins, V.; Hjelle, B.; Ebel, G.D.; Olson, K.E.; Blair, C.D. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl. Trop. Dis. 2010, 4, e848. [Google Scholar] [CrossRef]
- Cirimotich, C.M.; Scott, J.C.; Phillips, A.T.; Geiss, B.J.; Olson, K.E. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 2009, 9, 49. [Google Scholar] [CrossRef]
- Brackney, D.E.; Scott, J.C.; Sagawa, F.; Woodward, J.E.; Miller, N.A.; Schilkey, F.D.; Mudge, J.; Wilusz, J.; Olson, K.E.; Blair, C.D.; et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl. Trop. Dis. 2010, 4, e856. [Google Scholar] [CrossRef]
- Keene, K.M.; Foy, B.D.; Sanchez-Vargas, I.; Beaty, B.J.; Blair, C.D.; Olson, K.E. RNA interference acts as a natural antiviral response to O’nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 17240–17245. [Google Scholar] [CrossRef]
- Sánchez-Vargas, I.; Scott, J.C.; Katherine Poole-Smith, B.; Franz, A.W.E.; Barbosa-Solomieu, V.; Wilusz, J.; Olson, K.E.; Blair, C.D. Dengue Virus Type 2 Infections of Aedes aegypti Are Modulated by the Mosquito’s RNA Interference Pathway. PLoS Pathog. 2009, 5, e1000299. [Google Scholar] [CrossRef]
- Bernhardt, S.A.; Simmons, M.P.; Olson, K.E.; Beaty, B.J.; Blair, C.D.; Black, W.C. Rapid intraspecific evolution of miRNA and siRNA genes in the mosquito Aedes aegypti. PLoS ONE 2012, 7, e44198. [Google Scholar] [CrossRef]
- Obbard, D.J.; Jiggins, F.M.; Halligan, D.L.; Little, T.J. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006, 16, 580–585. [Google Scholar] [CrossRef]
- van Mierlo, J.T.; Overheul, G.J.; Obadia, B.; van Cleef, K.W.R.; Webster, C.L.; Saleh, M.-C.; Obbard, D.J.; van Rij, R.P. Novel Drosophila viruses encode host-specific suppressors of RNAi. PLoS Pathog. 2014, 10, e1004256. [Google Scholar] [CrossRef]
- Yen, P.-S.; James, A.; Li, J.-C.; Chen, C.-H.; Failloux, A.-B. Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito Aedes aegypti. Commun. Biol. 2018, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Franz, A.W.E.; Sanchez-Vargas, I.; Adelman, Z.N.; Blair, C.D.; Beaty, B.J.; James, A.A.; Olson, K.E. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc. Natl. Acad. Sci. USA 2006, 103, 4198–4203. [Google Scholar] [CrossRef] [PubMed]
- Moyes, C.L.; Athinya, D.K.; Seethaler, T.; Battle, K.E.; Sinka, M.; Hadi, M.P.; Hemingway, J.; Coleman, M.; Hancock, P.A. Evaluating insecticide resistance across African districts to aid malaria control decisions. Proc. Natl. Acad. Sci. USA 2020, 117, 22042–22050. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Spradling, A.C. Germline Stem Cell Division and Egg Chamber Development in Transplanted Drosophila Germaria. Dev. Biol. 1993, 159, 140–152. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef]
- Cox, D.N.; Chao, A.; Lin, H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 2000, 127, 503–514. [Google Scholar]
- Matthews, B.J.; Vosshall, L.B. How to turn an organism into a model organism in 10 ‘easy’ steps. J. Exp. Biol. 2020, 223, jeb218198. [Google Scholar] [CrossRef]
- Xu, J.; Ren, X.; Sun, J.; Wang, X.; Qiao, H.-H.; Xu, B.-W.; Liu, L.-P.; Ni, J.-Q. A Toolkit of CRISPR-Based Genome Editing Systems in Drosophila. J. Genet. Genom. 2015, 42, 141–149. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Yoshida, H. Drosophila as a Model Organism. Adv. Exp. Med. Biol. 2018, 1076, 1–10. [Google Scholar] [CrossRef]
- Buchman, A.B.; Brogan, D.J.; Sun, R.; Yang, T.; Hsu, P.D.; Akbari, O.S. Programmable RNA Targeting Using CasRx in Flies. CRISPR J. 2020, 3, 164–176. [Google Scholar] [CrossRef]
- Kandul, N.P.; Liu, J.; Hsu, A.D.; Hay, B.A.; Akbari, O.S. A novel drug-inducible sex-separation technique for insects. Nat. Commun. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kandul, N.P.; Liu, J.; Buchman, A.; Gantz, V.M.; Bier, E.; Akbari, O.S. Assessment of a Split Homing Based Gene Drive for Efficient Knockout of Multiple Genes. G3 2020, 10, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Kozeretska, I.A.; Shulha, V.I.; Serga, S.V.; Rozhok, A.I.; Protsenko, O.V.; Lau, N.C. A rapid change in P-element-induced hybrid dysgenesis status in Ukrainian populations of Drosophila melanogaster. Biol. Lett. 2018, 14, 20180184. [Google Scholar] [CrossRef] [PubMed]
- Greppi, C.; Laursen, W.J.; Budelli, G.; Chang, E.C.; Daniels, A.M.; van Giesen, L.; Smidler, A.L.; Catteruccia, F.; Garrity, P.A. Mosquito heat seeking is driven by an ancestral cooling receptor. Science 2020, 367, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Jové, V.; Gong, Z.; Hol, F.J.H.; Zhao, Z.; Sorrells, T.R.; Carroll, T.S.; Prakash, M.; McBride, C.S.; Vosshall, L.B. The Taste of Blood in Mosquitoes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Denlinger, D.L.; Armbruster, P.A. Mosquito Diapause. Annu. Rev. Entomol. 2014, 59, 73–93. [Google Scholar] [CrossRef]
- Li, M.; Yang, T.; Kandul, N.P.; Bui, M.; Gamez, S.; Raban, R.; Bennett, J.; Lanzaro, G.C.; Schmidt, H.; Lee, Y.; et al. Development of a confinable gene drive system in the human disease vector. eLife 2020, 9. [Google Scholar] [CrossRef]
- Li, M.; Bui, M.; Yang, T.; Bowman, C.S.; White, B.J.; Akbari, O.S. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc. Natl. Acad. Sci. USA 2017, 114, E10540–E10549. [Google Scholar] [CrossRef]
- Li, M.; Akbari, O.S.; White, B.J. Highly Efficient Site-Specific Mutagenesis in Malaria Mosquitoes Using CRISPR. G3 2018, 8, 653–658. [Google Scholar] [CrossRef]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 2015, 112, E6736–E6743. [Google Scholar] [CrossRef]
- Gamez, S.; Antoshechkin, I.; Mendez-Sanchez, S.C.; Akbari, O.S. The Developmental Transcriptome of Aedes albopictus, a Major Worldwide Human Disease Vector. G3 2020, 10, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Ramaiah, A.; Adolfi, A.; Halas, P.; Kaduskar, B.; Ngo, L.T.; Jayaprasad, S.; Paul, K.; Whadgar, S.; Srinivasan, S.; et al. Hidden features of the malaria vector mosquito, Anopheles stephensi, revealed by a high-quality reference genome. bioRxiv 2020. [Google Scholar] [CrossRef]
- Casier, K.; Delmarre, V.; Gueguen, N.; Hermant, C.; Viode, E.; Vaury, C.; Ronsseray, S.; Brasset, E.; Teysset, L.; Boivin, A. Environmentally-induced epigenetic conversion of a piRNA cluster. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Chaverra-Rodriguez, D.; Macias, V.M.; Hughes, G.L.; Pujhari, S.; Suzuki, Y.; Peterson, D.R.; Kim, D.; McKeand, S.; Rasgon, J.L. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 2018, 9, 3008. [Google Scholar] [CrossRef] [PubMed]
- Macias, V.M.; McKeand, S.; Chaverra-Rodriguez, D.; Hughes, G.L.; Fazekas, A.; Pujhari, S.; Jasinskiene, N.; James, A.A.; Rasgon, J.L. Cas9-Mediated Gene-Editing in the Malaria Mosquito Anopheles stephensi by ReMOT Control. G3 2020, 10, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Chaverra-Rodriguez, D.; Dalla Benetta, E.; Heu, C.C.; Rasgon, J.L.; Ferree, P.M.; Akbari, O.S. Germline mutagenesis of Nasonia vitripennis through ovarian delivery of CRISPR-Cas9 ribonucleoprotein. Insect Mol. Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Donald, C.L.; Stacey, H.; Watson, M.; Siu, R.W.C.; McFarlane, M.; Fazakerley, J.K.; Kohl, A.; Fragkoudis, R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 2013, 94, 1680–1689. [Google Scholar] [CrossRef]
- Varjak, M.; Dietrich, I.; Sreenu, V.B.; Till, B.E.; Merits, A.; Kohl, A.; Schnettler, E. Spindle-E Acts Antivirally Against Alphaviruses in Mosquito Cells. Viruses 2018, 10, 88. [Google Scholar] [CrossRef]
- Varjak, M.; Donald, C.L.; Mottram, T.J.; Sreenu, V.B.; Merits, A.; Maringer, K.; Schnettler, E.; Kohl, A. Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PLoS Negl. Trop. Dis. 2017, 11, e0006010. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gamez, S.; Srivastav, S.; Akbari, O.S.; Lau, N.C. Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Cells 2020, 9, 2180. https://doi.org/10.3390/cells9102180
Gamez S, Srivastav S, Akbari OS, Lau NC. Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Cells. 2020; 9(10):2180. https://doi.org/10.3390/cells9102180
Chicago/Turabian StyleGamez, Stephanie, Satyam Srivastav, Omar S. Akbari, and Nelson C. Lau. 2020. "Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways" Cells 9, no. 10: 2180. https://doi.org/10.3390/cells9102180
APA StyleGamez, S., Srivastav, S., Akbari, O. S., & Lau, N. C. (2020). Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Cells, 9(10), 2180. https://doi.org/10.3390/cells9102180





