The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases
Abstract
1. Introduction
2. Genes Implicated in Microcephaly
| Gene Name a | Protein Product | Protein Function | Refs |
|---|---|---|---|
| CHROMATIN- AND CHROMOSOME-ASSOCIATED PROTEINS | |||
| MCPH1 | Microcephalin, BRCT-repeat inhibitor of TERT expression 1 | Regulates chromosome condensation; acts in intra-S and G2/M checkpoints | [17,18,19] |
| MCPH10 | ZNF335 | Chromatin remodeling complex regulating neuronal gene expression and cell fate | [20] |
| MCPH11 | PHC1 | Component of the Polycomb group (PcG) PRC1-like complex, regulating transcriptional repression and chromatin remodeling of developmental genes, e.g., Hox genes. | [21,22] |
| MCPH21 MCPH22 MCPH23 | NCAPD2 (condensin I, subunit 1); NCAPD3 (condensin II, subunit D3); NCAPH (condensin I, subunit H) | Cooperate in compaction of interphase chromatin into mitotic chromosomes. | [23,24,25,26] |
| CENTROSOME DUPLICATION AND FUNCTION | |||
| MCPH3 | CDK5RAP2/CEP215, regulator of cyclin-dependent kinase 5 | Centriole engagement and microtubule nucleation | [27,28,29] |
| MCPH2 | WD40-Repeat Protein 62 | Spindle organization, centrosome duplication | [30,31] |
| MCPH8 | CEP135 | Centriole biogenesis, duplication and elongation | [32,33] |
| MCPH9 | CEP152 | Centrosome duplication | [34,35] |
| MCPH14 | SAS-6 | Centrosome duplication, procentriole formation | [36,37] |
| MCPH6 | CENPJ | Centrosome- and kinetochore-associated protein | [38,39,40] |
| MCPH7 | STIL, SCL-interrupting locus protein | Centriole assembly and duplication | [41,42] |
| SPINDLE AND MICROTUBULE FUNCTION AND DYNAMICS | |||
| MCPH5 | ASPM | Mitotic spindle regulator | [43,44,45] |
| MCPH17 | CIT, Citron Rho-interacting kinase | Cytokinesis, localizes the kinesin KIF14 to the central spindle and midbody | [46,47] |
| MCPH20 | KIF14 | Kinesin motor protein, acts at microtubules and midbody via interaction with CIT/MCPH17 | [48,49] |
| MCPH25 | MAP11 Microtubule associated protein 11 | Mitotic spindle dynamics | [50] |
| KINETOCHORES | |||
| MCPH4 | CASC5/KNL1 | Kinetochore assembly, microtubule attachments, SAC signaling | [51] |
| MCPH13 | CENP-E | Stabilization of kinetochore-microtubule attachments, chromosome congression | [52,53] |
| CELL CYCLE TRANSITIONS | |||
| MCPH12 | CDK6 | Cell cycle kinase, cell cycle entry | [54,55] |
| MCPH16 | ANKLE2, Ankyrin repeat and LEM domain-containing protein 2 | Regulates nuclear envelope reassembly at mitotic exit, promotes dephosphorylation of BAF/BANF1 possibly via PP2A | [56,57] |
| GLOBAL CELL ORGANIZATION AND FUNCTION | |||
| MCPH18 | WDFY3, WD repeat and FYVE domain-containing protein 3 | Component of the autophagic machinery | [11,58] |
| MCPH19 | COPB2, Coatomer subunit beta | Component of the Golgi and vesicular trafficking system | [10,59] |
| NEURAL CELL-SPECIFIC FUNCTION | |||
| MCPH15 | MFSD2A, Sodium-dependent lysophosphatidylcholine symporter 1 | Expressed at the blood–brain barrier, transport of fatty acids | [9,60] |
2.1. Centrosome Duplication and Function in Microcephaly
2.2. Spindle and Microtubule Function and Dynamics
3. The Rising Role of Kinetochores in Microcephaly
3.1. Roles of Kinetochores in Chromosome Segregation and Mitotic Fidelity
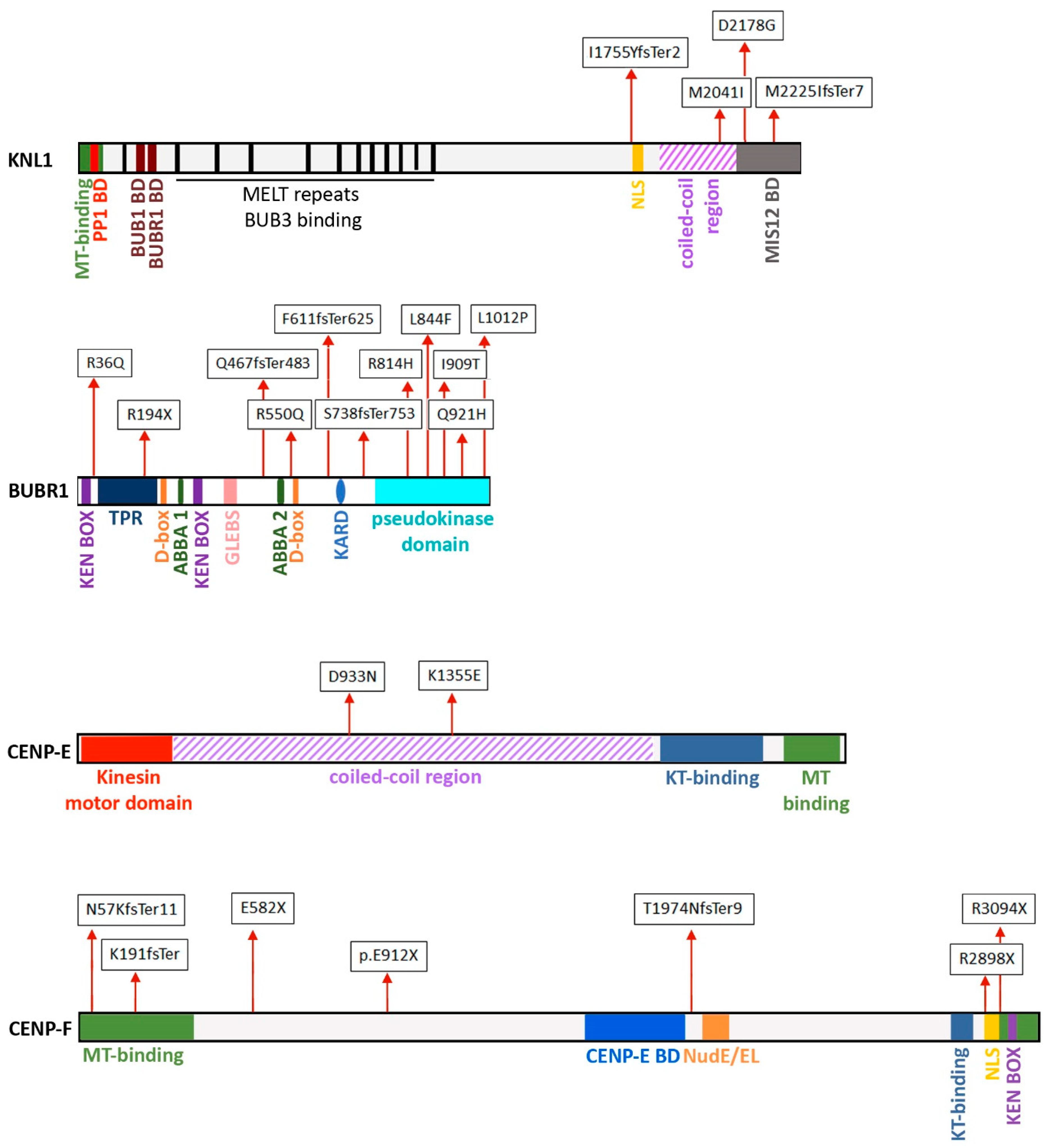
3.2. Kinetochore Gene Mutations Associated with Microcephaly Syndromes
3.2.1. CASC5/KNL
3.2.2. BUBR1 and other SAC Proteins
3.2.3. CENP-E
3.2.4. CENP-F
3.2.5. NUP133 and Other Nucleoporins
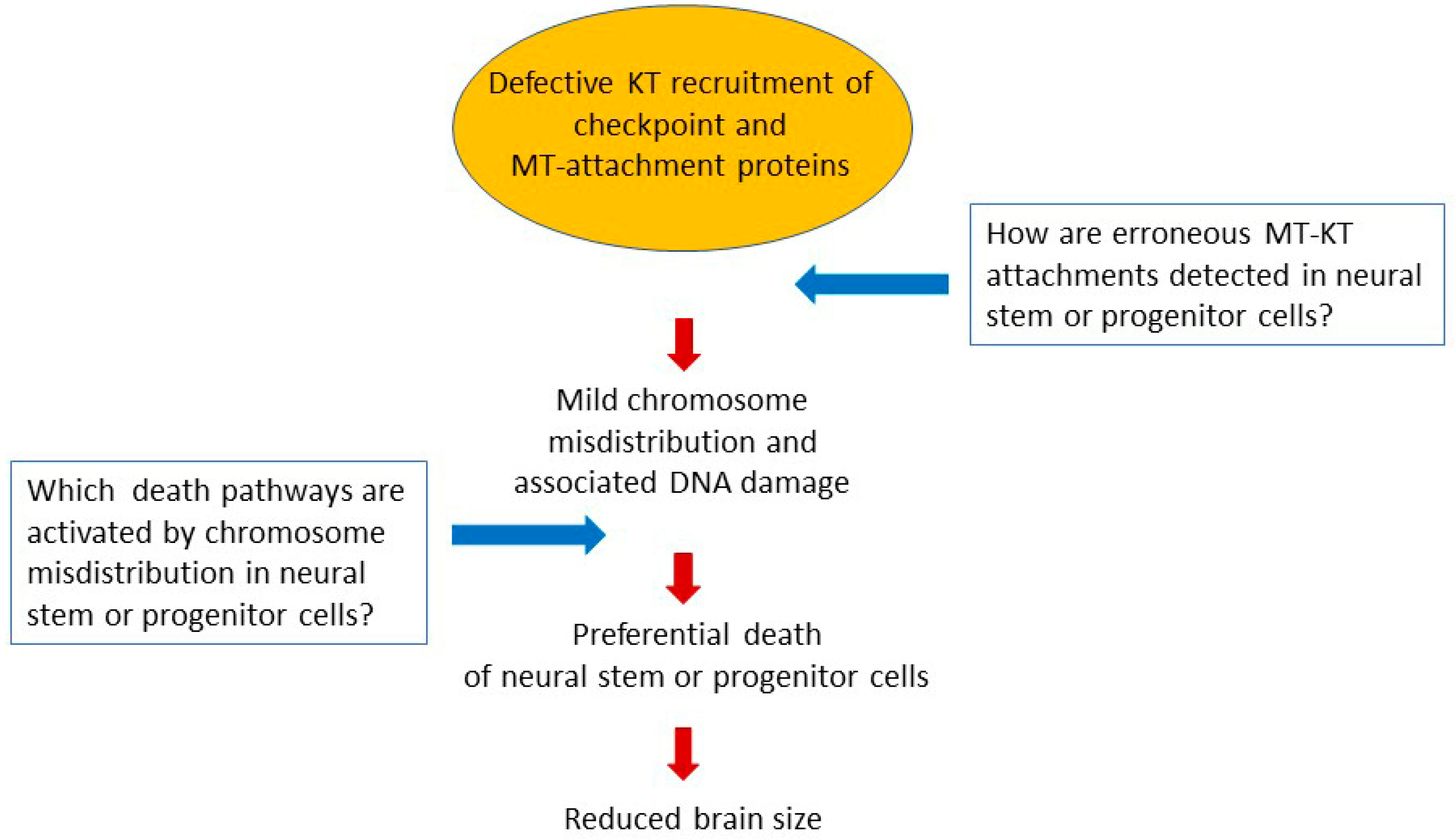
4. Forward Looks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levine, M.S.; Holland, A.J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 2018, 32, 620–638. [Google Scholar] [CrossRef] [PubMed]
- Bakhoum, S.F.; Landau, D.A. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb. Perspect. Med. 2017, 7, a029611. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, D.; Bae, B.-I.; Walsh, C.A. The Genetics of Primary Microcephaly. Annu. Rev. Genomics Hum. Genet. 2018, 19, 177–200. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.; Coleman, K.; Reid, S.; Plaja, A.; Firth, H.; FitzPatrick, D.; Kidd, A.; Méhes, K.; Nash, R.; Robin, N.; et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 2004, 36, 1159–1161. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Asfahani, R.; Carroll, P.; Bicknell, L.; Lescai, F.; Bright, A.; Chanudet, E.; Brooks, A.; Christou-Savina, S.; Osman, G.; et al. The kinetochore protein, CENPF, is mutated in human ciliopathy and microcephaly phenotypes. J. Med. Genet. 2015, 52, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, E.C.; Walsh, C.A. Genetic causes of microcephaly and lessons for neuronal development. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 461–478. [Google Scholar] [CrossRef]
- Dolk, H. The predictive value of microcephaly during the first year of life for mental retardation at seven years. Dev. Med. Child Neurol. 1991, 33, 974–983. [Google Scholar] [CrossRef]
- Morris-Rosendahl, D.J.; Kaindl, A.M. What next-generation sequencing (NGS) technology has enabled us to learn about primary autosomal recessive microcephaly (MCPH). Mol. Cell. Probes 2015, 29, 271–281. [Google Scholar] [CrossRef]
- Guemez-Gamboa, A.; Nguyen, L.N.; Yang, H.; Zaki, M.S.; Kara, M.; Ben-Omran, T.; Akizu, N.; Rosti, R.O.; Rosti, B.; Scott, E.; et al. Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat. Genet. 2015, 47, 809–813. [Google Scholar] [CrossRef]
- DiStasio, A.; Driver, A.; Sund, K.; Donlin, M.; Muraleedharan, R.M.; Pooya, S.; Kline-Fath, B.; Kaufman, K.M.; Prows, C.A.; Schorry, E.; et al. Copb2 is essential for embryogenesis and hypomorphic mutations cause human microcephaly. Hum. Mol. Genet. 2017, 26, 4836–4848. [Google Scholar] [CrossRef]
- Kadir, R.; Harel, T.; Markus, B.; Perez, Y.; Bakhrat, A.; Cohen, I.; Volodarsky, M.; Feintsein-Linial, M.; Chervinski, E.; Zlotogora, J.; et al. ALFY-Controlled DVL3 Autophagy Regulates Wnt Signaling, Determining Human Brain Size. PLoS Genet. 2016, 12, e1005919. [Google Scholar] [CrossRef] [PubMed]
- Heald, R.; Khodjakov, A. Thirty years of search and capture: The complex simplicity of mitotic spindle assembly. J. Cell Biol. 2015, 211, 1103–1111. [Google Scholar] [CrossRef] [PubMed]
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.D. Molecular Mechanisms of Spindle Assembly Checkpoint Activation and Silencing. Prog. Mol. Subcell. Biol. 2017, 56, 429–455. [Google Scholar]
- McIntosh, J.R. Assessing the Contributions of Motor Enzymes and Microtubule Dynamics to Mitotic Chromosome Motions. Annu. Rev. Cell Dev. Biol. 2017, 33, 1–22. [Google Scholar] [CrossRef]
- Jackson, A.P.; Eastwood, H.; Bell, S.M.; Adu, J.; Toomes, C.; Carr, I.M.; Roberts, E.; Hampshire, D.J.; Crow, Y.J.; Mighell, A.J.; et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am. J. Hum. Genet. 2002, 71, 136–142. [Google Scholar] [CrossRef]
- Trimborn, M.; Bell, S.M.; Felix, C.; Rashid, Y.; Jafri, H.; Griffiths, P.D.; Neumann, L.M.; Krebs, A.; Reis, A.; Sperling, K.; et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am. J. Hum. Genet. 2004, 75, 261–266. [Google Scholar] [CrossRef]
- Lin, S.Y.; Rai, R.; Li, K.; Xu, Z.X.; Elledge, S.J. BRIT1/MCPH1 is a DNA damage responsive protein that regulates the Brca1-Chk1 pathway, implicating checkpoint dysfunction in microcephaly. Proc. Natl. Acad. Sci. USA 2005, 102, 15105–15109. [Google Scholar] [CrossRef]
- Yang, Y.J.; Baltus, A.E.; Mathew, R.S.; Murphy, E.A.; Evrony, G.D.; Gonzalez, D.M.; Wang, E.P.; Marshall-Walker, C.A.; Barry, B.J.; Murn, J.; et al. Microcephaly gene links trithorax and REST/NRSF to control neural stem cell proliferation and differentiation. Cell 2012, 151, 1097–1112. [Google Scholar] [CrossRef]
- Gunster, M.J.; Satijn, D.P.; Hamer, K.M.; den Blaauwen, J.L.; de Bruijn, D.; Alkema, M.J.; van Lohuizen, M.; van Driel, R.; Otte, A.P. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol. Cell. Biol. 1997, 17, 2326–2335. [Google Scholar] [CrossRef] [PubMed]
- Awad, S.; Al-Dosari, M.S.; Al-Yacoub, N.; Colak, D.; Salih, M.A.; Alkuraya, F.S.; Poizat, C. Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum. Mol. Genet. 2013, 22, 2200–2213. [Google Scholar] [CrossRef] [PubMed]
- Schmiesing, J.A.; Gregson, H.C.; Zhou, S.; Yokomori, K. A Human Condensin Complex Containing hCAP-C-hCAP-E and CNAP1, a Homolog of Xenopus XCAP-D2, Colocalizes with Phosphorylated Histone H3 during the Early Stage of Mitotic Chromosome Condensation. Mol. Cell. Biol. 2000, 20, 6996–7006. [Google Scholar] [CrossRef] [PubMed]
- Aono, N.; Sutani, T.; Tomonaga, T.; Mochida, S.; Yanagida, M. Cnd2 has dual roles in mitotic condensation and interphase. Nature 2002, 417, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Losada, A.; Hirano, M.; Myers, M.P.; Neuwald, A.F.; Hirano, T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 2003, 115, 109–121. [Google Scholar] [CrossRef]
- Martin, C.A.; Murray, J.E.; Carroll, P.; Leitch, A.; Mackenzie, K.J.; Halachev, M.; Fetit, A.E.; Keith, C.; Bicknell, L.S.; Fluteau, A.; et al. Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 2016, 30, 2158–2172. [Google Scholar] [CrossRef]
- Moynihan, L.; Jackson, A.P.; Roberts, E.; Karbani, G.; Lewis, I.; Corry, P.; Turner, G.; Mueller, R.F.; Lench, N.J.; Woods, C.G. A third novel locus for primary autosomal recessive microcephaly maps to chromosome 9q34. Am. J. Hum. Genet. 2000, 66, 724–727. [Google Scholar] [CrossRef]
- Graser, S.; Stierhof, Y.D.; Nigg, E.A. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 2007, 120, 4321–4331. [Google Scholar] [CrossRef]
- Lizarraga, S.B.; Margossian, S.P.; Harris, M.H.; Campagna, D.R.; Han, A.P.; Blevins, S.; Mudbhary, R.; Barker, J.E.; Walsh, C.A.; Fleming, M.D. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development 2010, 137, 1907–1917. [Google Scholar] [CrossRef]
- Bilgüvar, K.; Öztürk, A.K.; Louvi, A.; Kwan, K.Y.; Choi, M.; Tatli, B.; Yalnizoǧlu, D.; Tüysüz, B.; Çaǧlayan, A.O.; Gökben, S.; et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 2010, 467, 207–210. [Google Scholar] [CrossRef]
- Shohayeb, B.; Lim, N.R.; Ho, U.; Xu, Z.; Dottori, M.; Quinn, L.; Ng, D.C.H. The Role of WD40-Repeat Protein 62 (MCPH2) in Brain Growth: Diverse Molecular and Cellular Mechanisms Required for Cortical Development. Mol. Neurobiol. 2018, 55, 5409–5424. [Google Scholar] [CrossRef] [PubMed]
- Kleylein-Sohn, J.; Westendorf, J.; Le Clech, M.; Habedanck, R.; Stierhof, Y.D.; Nigg, E.A. Plk4-Induced Centriole Biogenesis in Human Cells. Dev. Cell 2007, 13, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.S.; Baig, S.M.; Neumann, S.; Nürnberg, G.; Farooq, M.; Ahmad, I.; Alef, T.; Hennies, H.C.; Technau, M.; Altmüller, J.; et al. A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am. J. Hum. Genet. 2012, 90, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.S.; Wilkinson, C.J.; Mayor, T.; Mortensen, P.; Nigg, E.A.; Mann, M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003, 426, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Guernsey, D.L.; Jiang, H.; Hussin, J.; Arnold, M.; Bouyakdan, K.; Perry, S.; Babineau-Sturk, T.; Beis, J.; Dumas, N.; Evans, S.C.; et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am. J. Hum. Genet. 2010, 87, 40–51. [Google Scholar] [CrossRef]
- Leidel, S.; Delattre, M.; Cerutti, L.; Baumer, K.; Gönczy, P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005, 7, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Rupp, V.M.; Orpinell, M.; Hussain, M.S.; Altmüller, J.; Steinmetz, M.O.; Enzinger, C.; Thiele, H.; Höhne, W.; Nürnberg, G.; et al. A missense mutation in the PISA domain of HsSAS-6 causes autosomal recessive primary microcephaly in a large consanguineous pakistani family. Hum. Mol. Genet. 2014, 23, 5940–5949. [Google Scholar] [CrossRef]
- Hung, L.-Y.; Tang, C.-J.C.; Tang, T.K. Protein 4.1 R-135 Interacts with a Novel Centrosomal Protein (CPAP) Which Is Associated with the gamma -Tubulin Complex. Mol. Cell. Biol. 2000, 20, 7813–7825. [Google Scholar] [CrossRef]
- Leal, G.F.; Roberts, E.; Silva, E.O.; Costa, S.M.R.; Hampshire, D.J.; Woods, C.G. A novel locus for autosomal recessive primary microcephaly (MCPH6) maps to 13q12.2. J. Med. Genet. 2003, 40, 540–542. [Google Scholar] [CrossRef][Green Version]
- Bond, J.; Roberts, E.; Springell, K.; Lizarraga, S.; Scott, S.; Higgins, J.; Hampshire, D.J.; Morrison, E.E.; Leal, G.F.; Silva, E.O.; et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005, 37, 353–355. [Google Scholar] [CrossRef]
- Kumar, A.; Girimaji, S.C.; Duvvari, M.R.; Blanton, S.H. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 2008, 84, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Vulprecht, J.; David, A.; Tibelius, A.; Castiel, A.; Konotop, G.; Liu, F.; Bestvater, F.; Raab, M.S.; Zentgraf, H.; Izraeli, S.; et al. STIL is required for centriole duplication in human cells. J. Cell Sci. 2012, 125, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Bond, J.; Roberts, E.; Mochida, G.H.; Hampshire, D.J.; Scott, S.; Askham, J.M.; Springell, K.; Mahadevan, M.; Crow, Y.J.; Markham, A.F.; et al. ASPM is a major determinant of cerebral cortical size. Nat. Genet. 2002, 32, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.L.; Kosodo, Y.; Enard, W.; Pääbo, S.; Huttner, W.B. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA 2006, 103, 10438–10443. [Google Scholar] [CrossRef] [PubMed]
- Létard, P.; Drunat, S.; Vial, Y.; Duerinckx, S.; Ernault, A.; Amram, D.; Arpin, S.; Bertoli, M.; Busa, T.; Ceulemans, B.; et al. Autosomal recessive primary microcephaly due to ASPM mutations: An update. Hum. Mutat. 2018, 39, 319–332. [Google Scholar] [CrossRef]
- Di Cunto, F.; Calautti, E.; Hsiao, J.; Ong, L.; Topley, G.; Turco, E.; Dotto, G.P. Citron Rho-interacting kinase, a novel tissue-specific Ser/Thr kinase encompassing the Rho-Rac-binding protein citron. J. Biol. Chem. 1998, 273, 29706–29711. [Google Scholar] [CrossRef]
- Li, H.; Bielas, S.L.; Zaki, M.S.; Ismail, S.; Farfara, D.; Um, K.; Rosti, R.O.; Scott, E.C.; Tu, S.; Chi, N.C.; et al. Biallelic Mutations in Citron Kinase Link Mitotic Cytokinesis to Human Primary Microcephaly. Am. J. Hum. Genet. 2016, 99, 501–510. [Google Scholar] [CrossRef]
- Gruneberg, U.; Neef, R.; Li, X.; Chan, E.H.Y.; Chalamalasetty, R.B.; Nigg, E.A.; Barr, F.A. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 2006, 172, 363–372. [Google Scholar] [CrossRef]
- Moawia, A.; Shaheen, R.; Rasool, S.; Waseem, S.S.; Ewida, N.; Budde, B.; Kawalia, A.; Motameny, S.; Khan, K.; Fatima, A.; et al. Mutations of KIF14 cause primary microcephaly by impairing cytokinesis. Ann. Neurol. 2017, 82, 562–577. [Google Scholar] [CrossRef]
- Perez, Y.; Bar-Yaacov, R.; Kadir, R.; Wormser, O.; Shelef, I.; Birk, O.S.; Flusser, H.; Birnbaum, R.Y. Mutations in the microtubule-associated protein MAP11 (C7orf43) cause microcephaly in humans and zebrafish. Brain 2019, 142, 574–585. [Google Scholar] [CrossRef]
- Genin, A.; Desir, J.; Lambert, N.; Biervliet, M.; Van der Aa, N.; Pierquin, G.; Killian, A.; Tosi, M.; Urbina, M.; Lefort, A.; et al. Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Hum. Mol. Genet. 2012, 21, 5306–5317. [Google Scholar] [CrossRef] [PubMed]
- Yen, T.J.; Li, G.; Schaar, B.T.; Szilak, I.; Cleveland, D.W. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 1992, 359, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Mirzaa, G.M.; Vitre, B.; Carpenter, G.; Abramowicz, I.; Gleeson, J.G.; Paciorkowski, A.R.; Cleveland, D.W.; Dobyns, W.B.; O’Driscoll, M. Mutations in CENPE define a novel kinetochore-centromeric mechanism for microcephalic primordial dwarfism. Hum. Genet. 2014, 133, 1023–1039. [Google Scholar] [CrossRef] [PubMed]
- Meyerson, M.; Harlow, E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Biol. 1994, 14, 2077–2086. [Google Scholar] [CrossRef]
- Hussain, M.S.; Baig, S.M.; Neumann, S.; Peche, V.S.; Szczepanski, S.; Nürnberg, G.; Tariq, M.; Jameel, M.; Khan, T.N.; Fatima, A.; et al. CDK6 associates with the centrosome during mitosis and is mutated in a large pakistani family with primary microcephaly. Hum. Mol. Genet. 2013, 22, 5199–5214. [Google Scholar] [CrossRef]
- Asencio, C.; Davidson, I.F.; Santarella-Mellwig, R.; Ly-Hartig, T.B.N.; Mall, M.; Wallenfang, M.R.; Mattaj, I.W.; Gorjánácz, M. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell 2012, 150, 122–135. [Google Scholar] [CrossRef]
- Shaheen, R.; Maddirevula, S.; Ewida, N.; Alsahli, S.; Abdel-Salam, G.M.H.; Zaki, M.S.; Al Tala, S.; Alhashem, A.; Softah, A.; Al-Owain, M.; et al. Genomic and phenotypic delineation of congenital microcephaly. Genet. Med. 2019, 21, 545–552. [Google Scholar] [CrossRef]
- Clausen, T.H.; Lamark, T.; Isakson, P.; Finley, K.; Larsen, K.B.; Brech, A.; Øvervatn, A.; Stenmark, H.; Bjørkøy, G.; Simonsen, A.; et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010, 6, 330–344. [Google Scholar] [CrossRef]
- Harrison-Lavoie, K.J.; Lewis, V.A.; Hynes, G.M.; Collison, K.S.; Nutland, E.; Willison, K.R. A 102 kDa subunit of a Golgi-associated particle has homology to beta subunits of trimeric G proteins. EMBO J. 1993, 12, 2847–2853. [Google Scholar] [CrossRef]
- Angers, M.; Uldry, M.; Kong, D.; Gimble, J.M.; Jetten, A.M. Mfsd2a encodes a novel major facilitator superfamily domain-containing protein highly induced in brown adipose tissue during fasting and adaptive thermogenesis. Biochem. J. 2008, 416, 347–355. [Google Scholar] [CrossRef]
- Marthiens, V.; Rujano, M.A.; Pennetier, C.; Tessier, S.; Paul-Gilloteaux, P.; Basto, R. Centrosome amplification causes microcephaly. Nat. Cell Biol. 2013, 15, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Nano, M.; Basto, R. Consequences of centrosome dysfunction during brain development. In Advances in Experimental Medicine and Biology; Springer New York LLC: New York, NY, USA, 2017; Volume 1002, pp. 19–45. [Google Scholar]
- Maiato, H.; Logarinho, E. Mitotic spindle multipolarity without centrosome amplification. Nat. Cell Biol. 2014, 16, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Paronett, E.M.; Meechan, D.W.; Karpinski, B.A.; LaMantia, A.-S.; Maynard, T.M. Ranbp1, Deleted in DiGeorge/22q11.2 Deletion Syndrome, is a Microcephaly Gene That Selectively Disrupts Layer 2/3 Cortical Projection Neuron Generation. Cereb. Cortex 2015, 25, 3977–3993. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, B.; Ciciarello, M.; Mangiacasale, R.; Palena, A.; Tassin, A.-M.; Cundari, E.; Lavia, P. Mammalian RanBP1 regulates centrosome cohesion during mitosis. J. Cell Sci. 2003, 116, 3399–3411. [Google Scholar] [CrossRef]
- Lavia, P. The GTPase RAN regulates multiple steps of the centrosome life cycle. Chromosome Res. 2016, 24, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Saade, M.; Blanco-Ameijeiras, J.; Gonzalez-Gobartt, E.; Martí, E. A centrosomal view of CNS growth. Development 2018, 145, dev170613. [Google Scholar] [CrossRef]
- Basit, S.; Al-Harbi, K.M.; Alhijji, S.A.M.; Albalawi, A.M.; Alharby, E.; Eldardear, A.; Samman, M.I. CIT, a gene involved in neurogenic cytokinesis, is mutated in human primary microcephaly. Hum. Genet. 2016, 135, 1199–1207. [Google Scholar] [CrossRef]
- Harding, B.N.; Moccia, A.; Drunat, S.; Soukarieh, O.; Tubeuf, H.; Chitty, L.S.; Verloes, A.; Gressens, P.; El Ghouzzi, V.; Joriot, S.; et al. Mutations in Citron Kinase Cause Recessive Microlissencephaly with Multinucleated Neurons. Am. J. Hum. Genet. 2016, 99, 511–520. [Google Scholar] [CrossRef]
- Shaheen, R.; Hashem, A.; Abdel-Salam, G.M.H.; Al-Fadhli, F.; Ewida, N.; Alkuraya, F.S. Mutations in CIT, encoding citron rho-interacting serine/threonine kinase, cause severe primary microcephaly in humans. Hum. Genet. 2016, 135, 1191–1197. [Google Scholar] [CrossRef]
- Bianchi, F.T.; Tocco, C.; Pallavicini, G.; Liu, Y.; Vernì, F.; Merigliano, C.; Bonaccorsi, S.; El-Assawy, N.; Priano, L.; Gai, M.; et al. Citron Kinase Deficiency Leads to Chromosomal Instability and TP53-Sensitive Microcephaly. Cell Rep. 2017, 18, 1674–1686. [Google Scholar] [CrossRef]
- Gai, M.; Bianchi, F.T.; Vagnoni, C.; Vernì, F.; Bonaccorsi, S.; Pasquero, S.; Berto, G.E.; Sgrò, F.; Chiotto, A.A.; Annaratone, L.; et al. ASPM and CITK regulate spindle orientation by affecting the dynamics of astral microtubules. EMBO Rep. 2017, 18, 1870. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.T.; Gai, M.; Berto, G.E.; Di Cunto, F. Of rings and spines: The multiple facets of Citron proteins in neural development. Small GTPases 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P. Citron kinase—Renaissance of a neglected mitotic kinase. J. Cell Sci. 2017, 130, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.O.; Garcez, P.P. Dissecting the Toxic Effects of Zika Virus Proteins on Neural Progenitor Cells. Neuron 2019, 101, 989–991. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.; Matsumoto, Y.; Morishima, K.; Izumi, H.; Matsumoto, H.; Ito, E.; Tsutsui, K.; Kobayashi, J.; Tauchi, H.; Kajiwara, Y.; et al. MonoallelicBUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am. J. Med. Genet. Part A 2006, 140A, 358–367. [Google Scholar] [CrossRef] [PubMed]
- de Wolf, B.; Kops, G.J.P.L. Kinetochore malfunction in human pathologies. In Advances in Experimental Medicine and Biology; Springer New York LLC: New York, NY, USA, 2017; Volume 1002, pp. 69–91. [Google Scholar]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Fukagawa, T. Kinetochore assembly and disassembly during mitotic entry and exit. Curr. Opin. Cell Biol. 2018, 52, 73–81. [Google Scholar] [CrossRef]
- Vukušić, K.; Buđa, R.; Tolić, I.M. Force-generating mechanisms of anaphase in human cells. J. Cell Sci. 2019, 132, jcs231985. [Google Scholar] [CrossRef]
- Cimini, D.; Degrassi, F. Aneuploidy: A matter of bad connections. Trends Cell Biol. 2005, 15, 442–451. [Google Scholar] [CrossRef]
- Gregan, J.; Polakova, S.; Zhang, L.; Tolić-Nørrelykke, I.M.; Cimini, D. Merotelic kinetochore attachment: Causes and effects. Trends Cell Biol. 2011, 21, 374–381. [Google Scholar] [CrossRef]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Etemad, B.; Kops, G.J.P.L. Attachment issues: Kinetochore transformations and spindle checkpoint silencing. Curr. Opin. Cell Biol. 2016, 39, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Vanden Beldt, K.J.; Meng, X.; Khodjakov, A.; McEwen, B.F. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 2007, 9, 516–522. [Google Scholar] [CrossRef]
- Lampson, M.A.; Cheeseman, I.M. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011, 21, 133–140. [Google Scholar] [CrossRef]
- Hauf, S.; Cole, R.W.; LaTerra, S.; Zimmer, C.; Schnapp, G.; Walter, R.; Heckel, A.; van Meel, J.; Rieder, C.L.; Peters, J.-M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003, 161, 281–294. [Google Scholar] [CrossRef]
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef]
- Pesenti, M.E.; Weir, J.R.; Musacchio, A. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 2016, 37, 152–163. [Google Scholar] [CrossRef]
- Saurin, A.T. Kinase and phosphatase cross-talk at the kinetochore. Front. Cell Dev. Biol. 2018, 6, 62. [Google Scholar] [CrossRef]
- Jamieson, C.R.; Govaerts, C.; Abramowicz, M.J. Primary autosomal recessive microcephaly: Homozygosity mapping of MCPH4 to chromosome 15. Am. J. Hum. Genet. 1999, 65, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Saadi, A.; Verny, F.; Siquier-Pernet, K.; Bole-Feysot, C.; Nitschke, P.; Munnich, A.; Abada-Dendib, M.; Chaouch, M.; Abramowicz, M.; Colleaux, L. Refining the phenotype associated with CASC5 mutation. Neurogenetics 2016, 17, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Szczepanski, S.; Hussain, M.S.; Sur, I.; Altmüller, J.; Thiele, H.; Abdullah, U.; Waseem, S.S.; Moawia, A.; Nürnberg, G.; Noegel, A.A.; et al. A novel homozygous splicing mutation of CASC5 causes primary microcephaly in a large Pakistani family. Hum. Genet. 2016, 135, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Zarate, Y.A.; Kaylor, J.A.; Bosanko, K.; Lau, S.; Vargas, J.; Gao, H. First clinical report of an infant with microcephaly and CASC5 mutations. Am. J. Med. Genet. Part A 2016, 170, 2215–2218. [Google Scholar] [CrossRef] [PubMed]
- Caldas, G.V.; Deluca, J.G. KNL1: Bringing order to the kinetochore. Chromosoma 2014, 123, 169–181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shi, L.; Qalieh, A.; Lam, M.M.; Keil, J.M.; Kwan, K.Y. Robust elimination of genome-damaged cells safeguards against brain somatic aneuploidy following Knl1 deletion. Nat. Commun. 2019, 10, 2588. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; Van Der Burg, M.; Szuhai, K.; Kops, G.J.P.L.; Medema, R.H. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011, 333, 1895–1898. [Google Scholar] [CrossRef]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Santaguida, S.; Richardson, A.; Iyer, D.R.; M’Saad, O.; Zasadil, L.; Knouse, K.A.; Wong, Y.L.; Rhind, N.; Desai, A.; Amon, A. Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Dev. Cell 2017, 41, 638–651. [Google Scholar] [CrossRef]
- Omer Javed, A.; Li, Y.; Muffat, J.; Su, K.C.; Cohen, M.A.; Lungjangwa, T.; Aubourg, P.; Cheeseman, I.M.; Jaenisch, R. Microcephaly Modeling of Kinetochore Mutation Reveals a Brain-Specific Phenotype. Cell Rep. 2018, 25, 368–382. [Google Scholar] [CrossRef]
- Suijkerbuijk, S.J.E.; Van Osch, M.H.J.; Bos, F.L.; Hanks, S.; Rahman, N.; Kops, G.J.P.L. Molecular causes for BUBR1 dysfunction in the human cancer predisposition syndrome mosaic variegated aneuploidy. Cancer Res. 2010, 70, 4891–4900. [Google Scholar] [CrossRef] [PubMed]
- Overlack, K.; Primorac, I.; Vleugel, M.; Krenn, V.; Maffini, S.; Hoffmann, I.; Kops, G.J.P.L.; Musacchio, A. A molecular basis for the differential roles of Bubl and BubR1 in the spindle assembly checkpoint. eLife 2015, 4, e05269. [Google Scholar]
- Ciossani, G.; Overlack, K.; Petrovic, A.; Huis In ’T Veld, P.J.; Koerner, C.; Wohlgemuth, S.; Maffini, S.; Musacchio, A. The kinetochore proteins CENP-E and CENP-F directly and specifically interact with distinct BUB mitotic checkpoint Ser/Thr kinases. J. Biol. Chem. 2018, 293, 10084–10101. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lin, L.; Liu, X.; Ye, S.; Yao, P.Y.; Wang, W.; Yang, F.; Gao, X.; Li, J.; Zhang, Y.; et al. BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules. Cell Res. 2019, 29, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Suijkerbuijk, S.J.E.; Vleugel, M.; Teixeira, A.; Kops, G.J.P.L. Integration of Kinase and Phosphatase Activities by BUBR1 Ensures Formation of Stable Kinetochore-Microtubule Attachments. Dev. Cell 2012, 23, 745–755. [Google Scholar] [CrossRef]
- Simmons, A.J.; Park, R.; Sterling, N.A.; Jang, M.H.; Van Deursen, J.M.A.; Yen, T.J.; Cho, S.H.; Kim, S. Nearly complete deletion of BubR1 causes microcephaly through shortened mitosis and massive cell death. Hum. Mol. Genet. 2019, 28, 1822–1836. [Google Scholar] [CrossRef]
- Snape, K.; Hanks, S.; Ruark, E.; Barros-Núñez, P.; Elliott, A.; Murray, A.; Lane, A.H.; Shannon, N.; Callier, P.; Chitayat, D.; et al. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat. Genet. 2011, 43, 527–529. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, T.; Zheng, T.; Teng, J.; Chen, J. Cep57 is a Mis12-interacting kinetochore protein involved in kinetochore targeting of Mad1-Mad2. Nat. Commun. 2016, 7, 10151. [Google Scholar] [CrossRef]
- Yost, S.; De Wolf, B.; Hanks, S.; Zachariou, A.; Marcozzi, C.; Clarke, M.; De Voer, R.M.; Etemad, B.; Uijttewaal, E.; Ramsay, E.; et al. Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat. Genet. 2017, 49, 1148–1151. [Google Scholar] [CrossRef]
- Yao, X.; Abrieu, A.; Zheng, Y.; Sullivan, K.F.; Cleveland, D.W. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000, 2, 484–491. [Google Scholar] [CrossRef]
- Weaver, B.A.A.; Bonday, Z.Q.; Putkey, F.R.; Kops, G.J.P.L.; Silk, A.D.; Cleveland, D.W. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 2003, 162, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Kim, C.; Ahmad, S.; Zhang, J.; Mao, Y. CENP-E--dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. J. Cell Biol. 2012, 198, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Taveras, C.; Liu, C.; Mao, Y. A tension-independent mechanism reduces Aurora B-mediated phosphorylation upon microtubule capture by CENP-E at the kinetochore. Cell Cycle 2019, 18, 1349–1363. [Google Scholar] [CrossRef] [PubMed]
- Weaver, B.A.A.; Silk, A.D.; Montagna, C.; Verdier-Pinard, P.; Cleveland, D.W. Aneuploidy Acts Both Oncogenically and as a Tumor Suppressor. Cancer Cell 2007, 11, 25–36. [Google Scholar] [CrossRef]
- Varis, A.; Salmela, A.L.; Kallio, M.J. Cenp-F (mitosin) is more than a mitotic marker. Chromosoma 2006, 115, 288–295. [Google Scholar] [CrossRef]
- Liao, H.; Winkfein, R.J.; Mack, G.; Rattner, J.B.; Yen, T.J. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 1995, 130, 507–518. [Google Scholar] [CrossRef]
- Zhu, X.; Chang, K.H.; He, D.; Mancini, M.A.; Brinkley, W.R.; Lee, W.H. The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J. Biol. Chem. 1995, 270, 19545–19550. [Google Scholar] [CrossRef]
- Gurden, M.D.J.; Holland, A.J.; Van Zon, W.; Tighe, A.; Vergnolle, M.A.; Andres, D.A.; Spielmann, H.P.; Malumbres, M.; Wolthuis, R.M.F.; Cleveland, D.W.; et al. Cdc20 is required for the post-anaphase, KEN-dependent degradation of centromere protein F. J. Cell Sci. 2010, 123, 321–330. [Google Scholar] [CrossRef]
- Feng, J.; Huang, H.; Yen, T.J. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma 2006, 115, 320–329. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Guo, J.; Li, N.; Qian, M.; Wang, S.N.; Zhu, X.L. Mitosin/CENP-F is a conserved kinetochore protein subjected to cytoplasmic dynein-mediated poleward transport. Cell Res. 2003, 13, 275–283. [Google Scholar] [CrossRef]
- Vergnolle, M.A.S.; Taylor, S.S. Cenp-F Links Kinetochores to Ndel1/Nde1/Lis1/Dynein Microtubule Motor Complexes. Curr. Biol. 2007, 17, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Bolhy, S.; Bouhlel, I.; Dultz, E.; Nayak, T.; Zuccolo, M.; Gatti, X.; Vallee, R.; Ellenberg, J.; Doye, V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J. Cell Biol. 2011, 192, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Berto, A.; Doye, V. Regulation of Cenp-F localization to nuclear pores and kinetochores. Cell Cycle 2018, 17, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Zuccolo, M.; Alves, A.; Galy, V.; Bolhy, S.; Formstecher, E.; Racine, V.; Sibarita, J.B.; Fukagawa, T.; Shiekhattar, R.; Yen, T.; et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007, 26, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Filges, I.; Bruder, E.; Brandal, K.; Meier, S.; Undlien, D.E.; Waage, T.R.; Hoesli, I.; Schubach, M.; de Beer, T.; Sheng, Y.; et al. Strømme Syndrome Is a Ciliary Disorder Caused by Mutations in CENPF. Hum. Mutat. 2016, 37, 359–363. [Google Scholar] [CrossRef]
- Ozkinay, F.; Atik, T.; Isik, E.; Gormez, Z.; Sagiroglu, M.; Sahin, O.A.; Corduk, N.; Onay, H. A further family of Stromme syndrome carrying CENPF mutation. Am. J. Med. Genet. A 2017, 173, 1668–1672. [Google Scholar] [CrossRef]
- Vasu, S.; Shah, S.; Orjalo, A.; Park, M.; Fischer, W.H.; Forbes, D.J. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 2001, 155, 339–353. [Google Scholar] [CrossRef]
- Hu, D.J.K.; Baffet, A.D.; Nayak, T.; Akhmanova, A.; Doye, V.; Vallee, R.B. XDynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell 2013, 154, 1300. [Google Scholar] [CrossRef]
- Splinter, D.; Tanenbaum, M.E.; Lindqvist, A.; Jaarsma, D.; Flotho, A.; Yu, K.L.; Grigoriev, I.; Engelsma, D.; Haasdijk, E.D.; Keijzer, N.; et al. Bicaudal D2, Dynein, and Kinesin-1 Associate with Nuclear Pore Complexes and Regulate Centrosome and Nuclear Positioning during Mitotic Entry. PLoS Biol. 2010, 8, e1000350. [Google Scholar] [CrossRef]
- Fujita, A.; Tsukaguchi, H.; Koshimizu, E.; Nakazato, H.; Itoh, K.; Kuraoka, S.; Komohara, Y.; Shiina, M.; Nakamura, S.; Kitajima, M.; et al. Homozygous splicing mutation in NUP133 causes Galloway-Mowat syndrome. Ann. Neurol. 2018, 84, 814–828. [Google Scholar] [CrossRef]
- Rosti, R.O.; Sotak, B.N.; Bielas, S.L.; Bhat, G.; Silhavy, J.L.; Aslanger, A.D.; Altunoglu, U.; Bilge, I.; Tasdemir, M.; Yzaguirrem, A.D.; et al. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat syndrome. J. Med. Genet. 2017, 54, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Gogendeau, D.; Siudeja, K.; Gambarotto, D.; Pennetier, C.; Bardin, A.J.; Basto, R. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun. 2015, 6, 8894. [Google Scholar] [CrossRef] [PubMed]
- Poulton, J.S.; Cuningham, J.C.; Peifer, M. Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size. J. Cell Biol. 2017, 216, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Quadrato, G.; Arlotta, P. Present and future of modeling human brain development in 3D organoids. Curr. Opin. Cell Biol. 2017, 49, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, C.A.; Muotri, A.R. Brain Organoids and the Study of Neurodevelopment. Trends Mol. Med. 2018, 24, 982–990. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degrassi, F.; Damizia, M.; Lavia, P. The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases. Cells 2020, 9, 49. https://doi.org/10.3390/cells9010049
Degrassi F, Damizia M, Lavia P. The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases. Cells. 2020; 9(1):49. https://doi.org/10.3390/cells9010049
Chicago/Turabian StyleDegrassi, Francesca, Michela Damizia, and Patrizia Lavia. 2020. "The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases" Cells 9, no. 1: 49. https://doi.org/10.3390/cells9010049
APA StyleDegrassi, F., Damizia, M., & Lavia, P. (2020). The Mitotic Apparatus and Kinetochores in Microcephaly and Neurodevelopmental Diseases. Cells, 9(1), 49. https://doi.org/10.3390/cells9010049







