Agonist Antibody Converts Stem Cells into Migrating Brown Adipocyte-Like Cells in Heart
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mouse Strains and Cell Lines
2.2. Human Heart Tissues
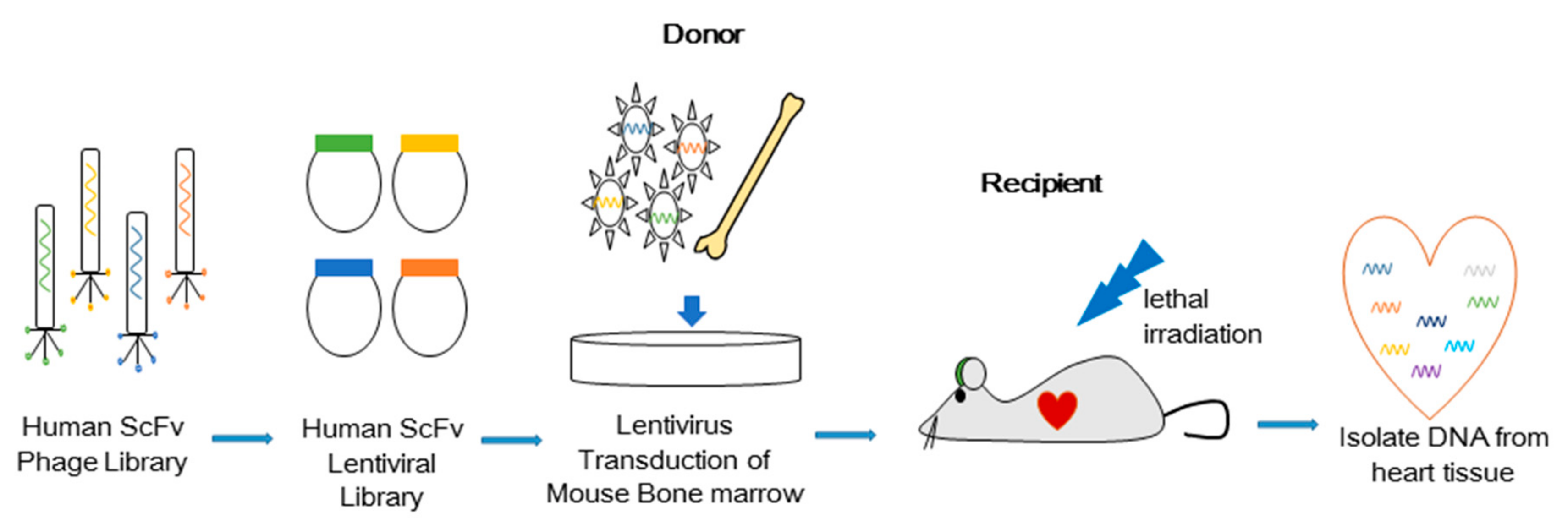
2.3. Combinatorial Antibody Library
2.4. Bone Marrow Transduction and Transplantation
2.5. Purification of Single-Chain Variable Fragment—Fc Proteins
2.6. Immunoprecipitation and Mass Spectrometry
2.7. Western Blot
2.8. Flow Cytometry
2.9. Real Time Quantitative (RT-q) PCR
2.10. Microscopy
2.11. Bioluminescence Imaging
2.12. Electron Microscopy
2.13. RNA Sequencing and Data Analysis
3. Results
3.1. In Vivo Selection of Antibodies that Induce Cell Migration
3.2. Isolated Antibody H3 Induces Migration of Cells to the Heart
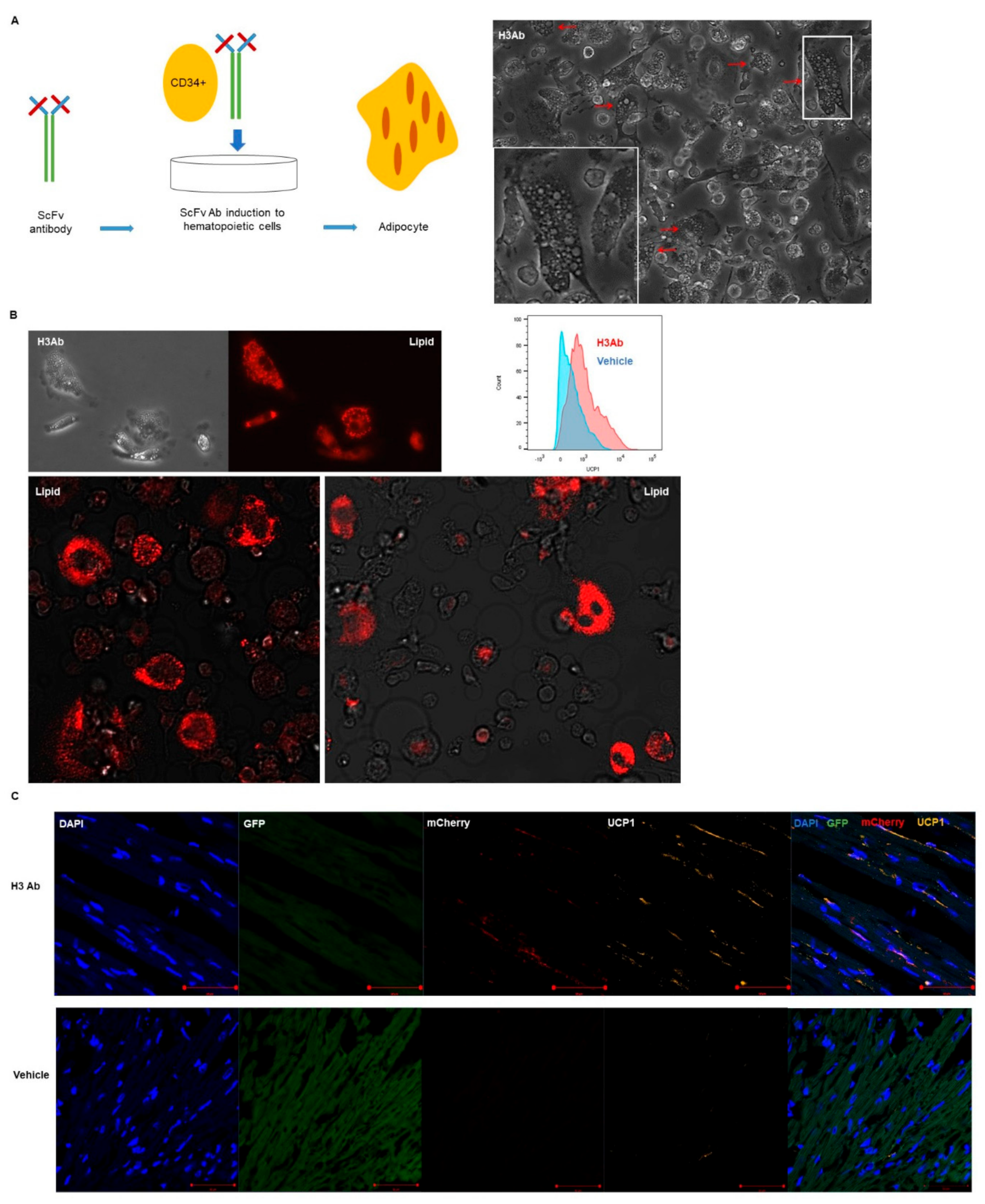
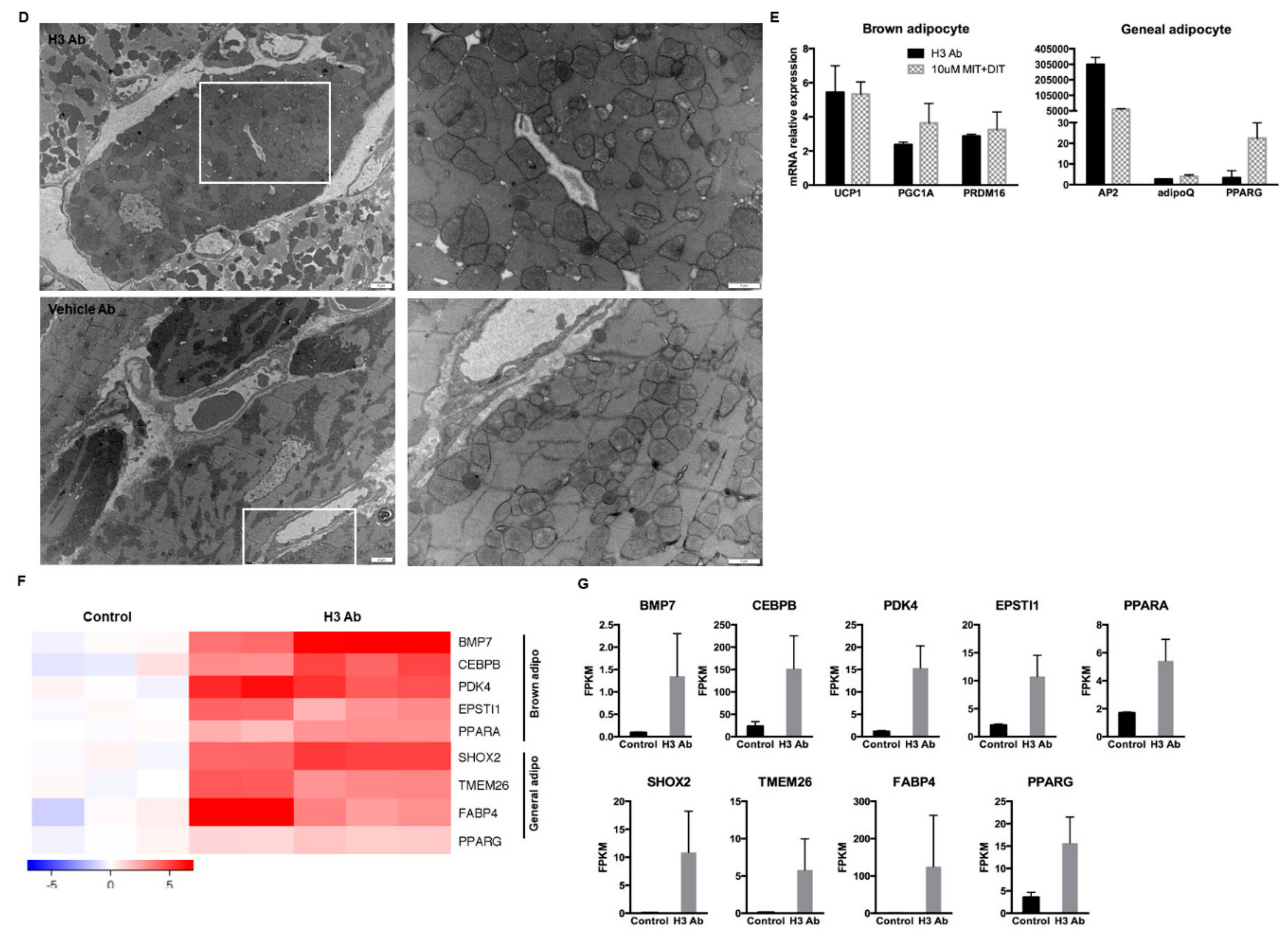
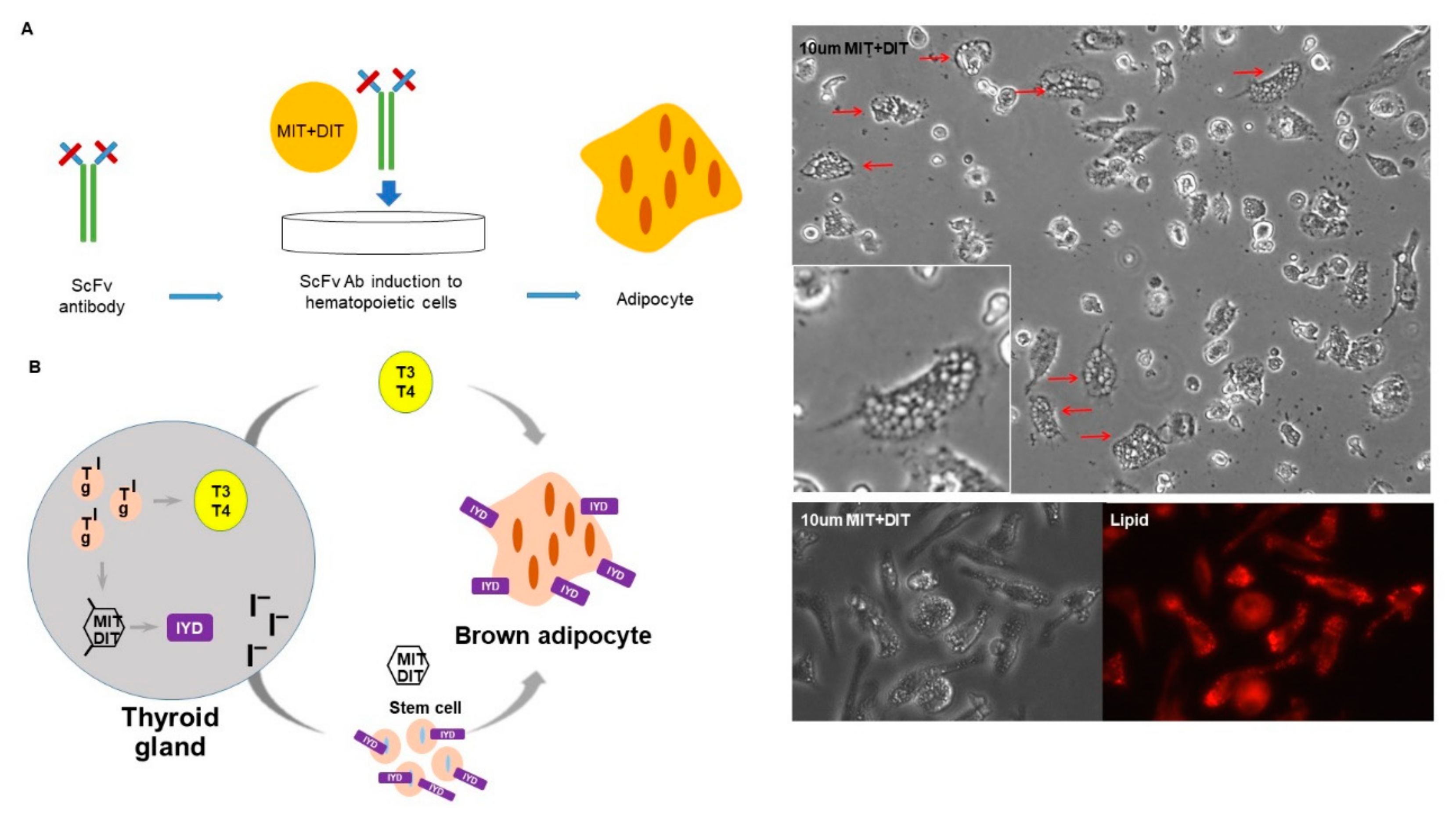
3.3. Purified H3 Antibody Transforms Human Hematopoietic Stem Cells into Brown Adipocyte-Like Cells
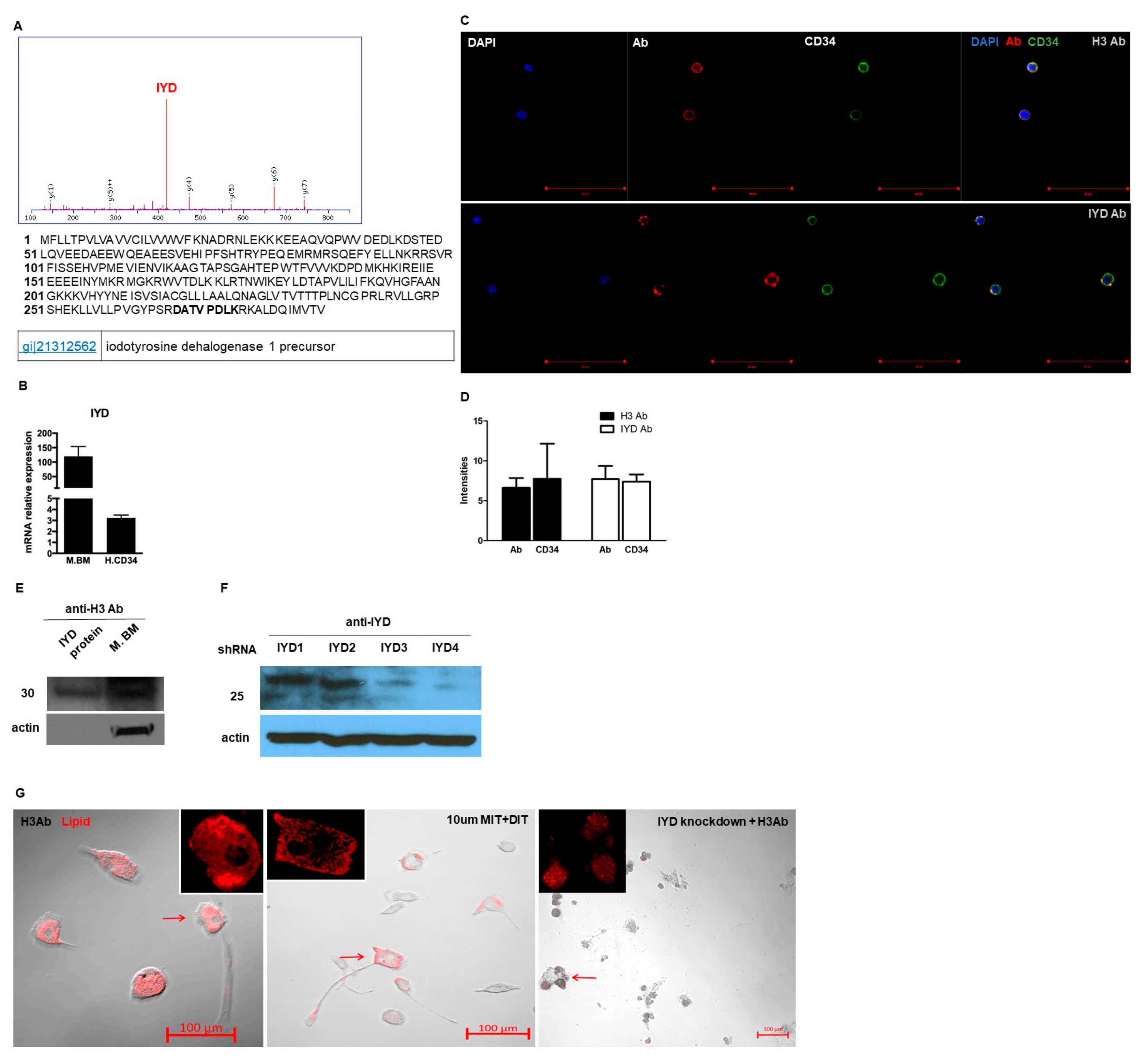
3.4. H3 Ab has a Novel Target
3.5. H3 Ab Detects IYD in Human Heart
3.6. IYD Substrates MIT and DIT Differentiate HSCs into Brown Adipocyte-Like Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Han, K.H.; Arlian, B.M.; Macauley, M.S.; Paulson, J.C.; Lerner, R.A. Migration-based selections of antibodies that convert bone marrow into trafficking microglia-like cells that reduce brain amyloid beta. Proc. Natl. Acad. Sci. USA 2018, 115, E372–E381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.H.; Gonzalez-Quintial, R.; Peng, Y.; Baccala, R.; Theofilopoulos, A.N.; Lerner, R.A. An agonist antibody that blocks autoimmunity by inducing anti-inflammatory macrophages. FASEB J. 2016, 30, 738–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerner, R.A.; Grover, R.K.; Zhang, H.; Xie, J.; Han, K.H.; Peng, Y.; Yea, K. Antibodies from combinatorial libraries use functional receptor pleiotropism to regulate cell fates. Q. Rev. Biophys. 2015, 48, 389–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Zeng, W.; Ye, H.; Han, K.H.; Dharmarajan, V.; Novick, S.; Wilson, I.A.; Griffin, P.R.; Friedman, J.M.; Lerner, R.A. A General Method for Insertion of Functional Proteins within Proteins via Combinatorial Selection of Permissive Junctions. Chem. Biol. 2015, 22, 1134–1143. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Xie, J.; Lerner, R.A. A proximity based general method for identification of ligand and receptor interactions in living cells. Biochem. Biophys. Res. Commun. 2014, 454, 251–255. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, H.; Yea, K.; Lerner, R.A. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 8099–8104. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yea, K.; Xie, J.; Ruiz, D.; Wilson, I.A.; Lerner, R.A. Selecting agonists from single cells infected with combinatorial antibody libraries. Chem. Biol. 2013, 20, 734–741. [Google Scholar] [CrossRef] [Green Version]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Daniel, K.W.; Robidoux, J.; Puigserver, P.; Medvedev, A.V.; Bai, X.; Floering, L.M.; Spiegelman, B.M.; Collins, S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell Biol. 2004, 24, 3057–3067. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Lu, Y.; Czernik, P.J.; Rosen, C.J.; Enerback, S.; Lecka-Czernik, B. Inducible brown adipose tissue, or beige fat, is anabolic for the skeleton. Endocrinology 2013, 154, 2687–2701. [Google Scholar] [CrossRef] [Green Version]
- Gnidehou, S.; Caillou, B.; Talbot, M.; Ohayon, R.; Kaniewski, J.; Noel-Hudson, M.S.; Morand, S.; Agnangji, D.; Sezan, A.; Courtin, F.; et al. Iodotyrosine dehalogenase 1 (DEHAL1) is a transmembrane protein involved in the recycling of iodide close to the thyroglobulin iodination site. FASEB J. 2004, 18, 1574–1576. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, X.; Jiang, Y.; Bao, S.; Shan, Z.; Teng, W. Expression of Iodotyrosine Deiodinase in Thyroid and Other Organs in Iodine-Deficient and Iodine-Excess Rats. Biol. Trace Elem. Res. 2015, 167, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Burniat, A.; Pirson, I.; Vilain, C.; Kulik, W.; Afink, G.; Moreno-Reyes, R.; Corvilain, B.; Abramowicz, M. Iodotyrosine deiodinase defect identified via genome-wide approach. J. Clin. Endocrinol. Metab. 2012, 97, E1276–E1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, J.A.; Ribich, S.; Christoffolete, M.A.; Simovic, G.; Correa-Medina, M.; Patti, M.E.; Bianco, A.C. Absence of thyroid hormone activation during development underlies a permanent defect in adaptive thermogenesis. Endocrinology 2010, 151, 4573–4582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S. The adipose organ at a glance. Dis. Model Mech. 2012, 5, 588–594. [Google Scholar] [CrossRef] [Green Version]
- McAninch, E.A.; Bianco, A.C. Thyroid hormone signaling in energy homeostasis and energy metabolism. Ann. N. Y. Acad. Sci. 2014, 1311, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Barreto, P.; Okura, V.K.; Neshich, I.A.; Maia Ide, G.; Arruda, P. Overexpression of UCP1 in tobacco induces mitochondrial biogenesis and amplifies a broad stress response. BMC Plant Biol. 2014, 14, 144. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, S.; Leslie, P.; Hopwood, D.; Illingworth, P.; Jung, R.T.; Nicholls, D.G.; Peden, N.; Rafael, J.; Rial, E. The characterization and energetic potential of brown adipose tissue in man. Clin. Sci. 1985, 69, 343–348. [Google Scholar] [CrossRef] [Green Version]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, M.; Okamatsu-Ogura, Y.; Matsushita, M.; Watanabe, K.; Yoneshiro, T.; Nio-Kobayashi, J.; Iwanaga, T.; Miyagawa, M.; Kameya, T.; Nakada, K.; et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: Effects of cold exposure and adiposity. Diabetes 2009, 58, 1526–1531. [Google Scholar] [CrossRef] [Green Version]
- van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.J.; Enerback, S.; et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Zingaretti, M.C.; Crosta, F.; Vitali, A.; Guerrieri, M.; Frontini, A.; Cannon, B.; Nedergaard, J.; Cinti, S. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 2009, 23, 3113–3120. [Google Scholar] [CrossRef]
- Fitzgibbons, T.P.; Czech, M.P. Epicardial and perivascular adipose tissues and their influence on cardiovascular disease: Basic mechanisms and clinical associations. J. Am. Heart Assoc. 2014, 3, e000582. [Google Scholar] [CrossRef] [Green Version]
- Bartelt, A.; Heeren, J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 2014, 10, 24–36. [Google Scholar] [CrossRef]
- Wang, G.X.; Zhao, X.Y.; Meng, Z.X.; Kern, M.; Dietrich, A.; Chen, Z.; Cozacov, Z.; Zhou, D.; Okunade, A.L.; Su, X.; et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat. Med. 2014, 20, 1436–1443. [Google Scholar] [CrossRef]
- Tahara, N.; Brush, M.; Kawakami, Y. Cell migration during heart regeneration in zebrafish. Dev. Dyn. 2016, 245, 774–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrijsen, K.R.; Sluijter, J.P.; Schuchardt, M.W.; van Balkom, B.W.; Noort, W.A.; Chamuleau, S.A.; Doevendans, P.A. Cardiomyocyte progenitor cell-derived exosomes stimulate migration of endothelial cells. J. Cell. Mol. Med. 2010, 14, 1064–1070. [Google Scholar] [CrossRef] [Green Version]
- Ma, N.; Ladilov, Y.; Kaminski, A.; Piechaczek, C.; Choi, Y.H.; Li, W.; Steinhoff, G.; Stamm, C. Umbilical cord blood cell transplantation for myocardial regeneration. Transpl. Proc. 2006, 38, 771–773. [Google Scholar] [CrossRef] [PubMed]
- Urbich, C.; Aicher, A.; Heeschen, C.; Dernbach, E.; Hofmann, W.K.; Zeiher, A.M.; Dimmeler, S. Soluble factors released by endothelial progenitor cells promote migration of endothelial cells and cardiac resident progenitor cells. J. Mol. Cell. Cardiol. 2005, 39, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.Y.; Ng, W.H.; Ellison-Hughes, G.M.; Tan, J.J. Cardiac Stem Cells for Myocardial Regeneration: They Are Not Alone. Front. Cardiovasc. Med. 2017, 4, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, L.; Gao, H.; Zhang, T.; Jing, L.; Xiao, C.; Xiao, Y.; Luo, N.; Zhu, H.; Meng, W.; Xu, H.; et al. Snail modulates the assembly of fibronectin via alpha5 integrin for myocardial migration in zebrafish embryos. Sci. Rep. 2014, 4, 4470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahill, T.J.; Choudhury, R.P.; Riley, P.R. Heart regeneration and repair after myocardial infarction: Translational opportunities for novel therapeutics. Nat. Rev. Drug Discov. 2017, 16, 699–717. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, K.H.; Arlian, B.M.; Lin, C.-W.; Jin, H.Y.; Kang, G.-H.; Lee, S.; Lee, P.C.-W.; Lerner, R.A. Agonist Antibody Converts Stem Cells into Migrating Brown Adipocyte-Like Cells in Heart. Cells 2020, 9, 256. https://doi.org/10.3390/cells9010256
Han KH, Arlian BM, Lin C-W, Jin HY, Kang G-H, Lee S, Lee PC-W, Lerner RA. Agonist Antibody Converts Stem Cells into Migrating Brown Adipocyte-Like Cells in Heart. Cells. 2020; 9(1):256. https://doi.org/10.3390/cells9010256
Chicago/Turabian StyleHan, Kyung Ho, Britni M. Arlian, Chih-Wei Lin, Hyun Yong Jin, Geun-Hyung Kang, Sahmin Lee, Peter Chang-Whan Lee, and Richard A. Lerner. 2020. "Agonist Antibody Converts Stem Cells into Migrating Brown Adipocyte-Like Cells in Heart" Cells 9, no. 1: 256. https://doi.org/10.3390/cells9010256
APA StyleHan, K. H., Arlian, B. M., Lin, C.-W., Jin, H. Y., Kang, G.-H., Lee, S., Lee, P. C.-W., & Lerner, R. A. (2020). Agonist Antibody Converts Stem Cells into Migrating Brown Adipocyte-Like Cells in Heart. Cells, 9(1), 256. https://doi.org/10.3390/cells9010256




