Exosomes in Bone Sarcomas: Key Players in Metastasis
Abstract
:1. Bone Sarcomas
2. Exosomes
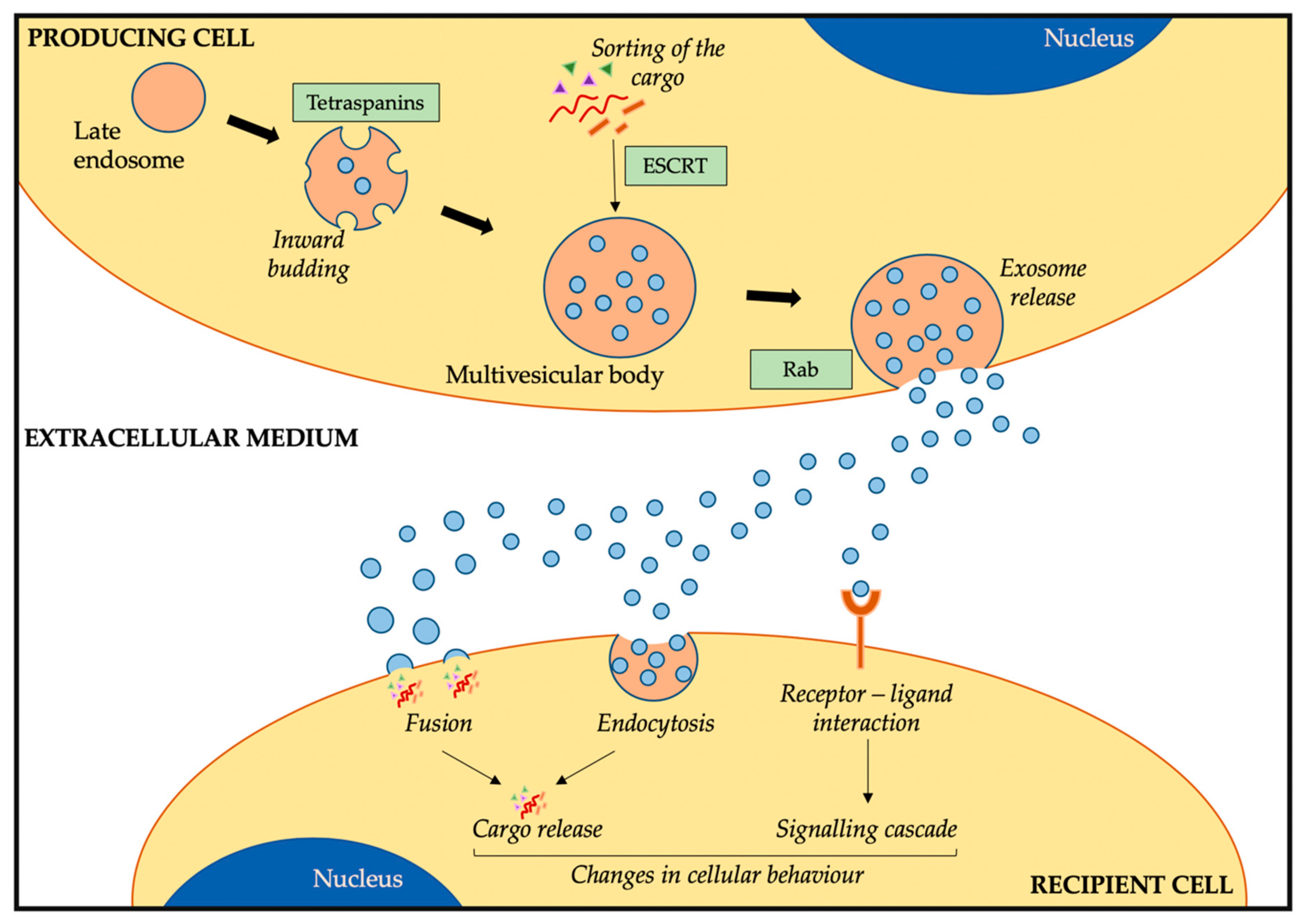
2.1. Exosome Biogenesis, Release, and Uptake
2.2. Exosome Cargo and Composition
2.3. Function of Exosomes
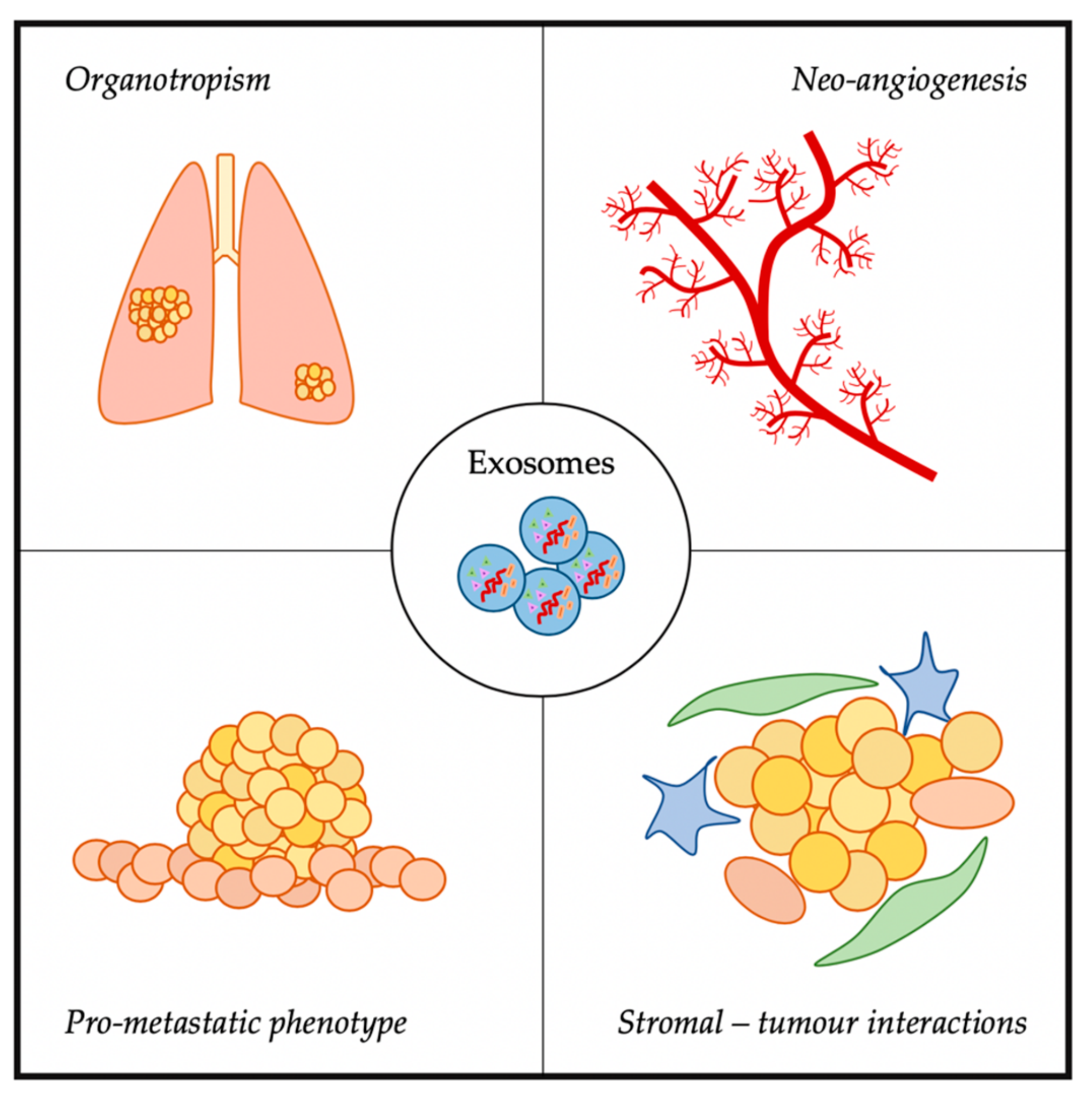
3. Exosomes and Metastasis
3.1. Organostropism
3.2. Neo-Angiogenesis
3.3. Induction of Pro-Metastatic Phenotypes
3.4. Stromal–Tumour Interactions
4. The Role of Exosomes in Bone Sarcoma Metastasis
4.1. Exosomes in OS
4.2. Exosomes in ES
4.3. Exosomes in CS
4.4. Other Roles of Exosomes in Bone Sarcomas
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BC | Breast cancer |
| BM | Bone marrow |
| CAF | Cancer associated fibroblast |
| CRC | Colorectal cancer |
| CS | Chondrosarcoma |
| ECM | Extracellular matrix |
| ES | Ewing sarcoma |
| ESCRT | Endosomal sorting complex required for transport |
| GC | Gastric cancer |
| LAC | Lung adenocarcinoma |
| MET | Mesenchymal-to-epithelial transition |
| MSC | Mesenchymal stem cell |
| OS | Osteosarcoma |
| OSR | Overall survival rate |
| PDA | Pancreatic ductal adenocarcinoma |
| SCC | Squamous cell carcinoma |
| TME | Tumour microenvironment |
| uPA | Urokinase plasminogen activator |
References
- Casali, P.G.; Bielack, S.; Abecassis, N.; Aro, H.T.; Bauer, S.; Biagini, R.; Bonvalot, S.; Boukovinas, I.; Bovee, J.V.M.G.; Brennan, B.; et al. Bone sarcomas: ESMO-PaedCan-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv79–iv95. [Google Scholar] [CrossRef] [PubMed]
- Rozeman, L.B.; Cleton-Jansen, A.M.; Hogendoorn, P.C.W. Pathology of primary malignant bone and cartilage tumours. Int. Orthop. 2006, 30, 437–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiller, C.A.; Trama, A.; Serraino, D.; Rossi, S.; Navarro, C.; Chirlaque, M.D.; Zielonk, N.; Van Eycken, E.; Sundseth, H.; Hedelin, G.; et al. Descriptive epidemiology of sarcomas in Europe: Report from the RARECARE project. Eur. J. Cancer 2013, 49, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Eyre, R.; Feltbower, R.G.; Mubwandarikwa, E.; Eden, T.O.; McNally, R.J. Epidemiology of Bone Tumours in Children and Young Adults. Pediatr. Blood Cancer 2009, 53, 941–952. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar]
- Miller, I.V.; Grunewald, T.G.P. Tumour-derived exosomes: Tiny envelopes for big stories. Biol. Cell 2015, 107, 287–305. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Kharaziha, P.; Ceder, S.; Li, Q.; Panaretakis, T. Tumor cell-derived exosomes: A message in a bottle. Biochim. Biophys. Acta Rev. Cancer 2012, 1826, 103–111. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347. [Google Scholar] [CrossRef] [PubMed]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Dev. Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Laulagnier, K.; Motta, C.; Hamdi, S.; Roy, S.; Fauvelle, F.; Pageaux, J.-F.; Kobayashi, T.; Salles, J.-P.; Perret, B.; Bonnerot, C.; et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004, 380, 161–171. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19. [Google Scholar] [CrossRef] [Green Version]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [Green Version]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Tian, T.; Zhu, Y.L.; Zhou, Y.Y.; Liang, G.F.; Wang, Y.Y.; Hu, F.H.; Xiao, Z.D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Mörgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munich, S.; Sobo-Vujanovic, A.; Buchser, W.J.; Beer-Stolz, D.; Vujanovic, N.L. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology 2012, 1, 1074–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Buchkovich, N.J.; Zhao, Y.; Emr, S.D. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 2012, 151, 356–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Thery, C.; Boussac, M.; Veron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, O.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of doublestranded genomic dna spanning all chromosomes with mutated KRAS and P53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [Green Version]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol. Cell. Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef] [Green Version]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [Green Version]
- Dias, M.V.S.; Costa, C.S.; DaSilva, L.L.P. The Ambiguous Roles of Extracellular Vesicles in HIV Replication and Pathogenesis. Front. Microbiol. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in cancer development and clinical applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, M.; Haider, A.; Rashid, S.; Al-Nabet, A.D.M.H. Paget’s “Seed and Soil” Theory of Cancer Metastasis. Adv. Anat. Pathol. 2018, 26, 69–74. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019, 79, 4557–4567. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 2016, 30, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Chen, M.; Jiang, R.; Guo, Y.; Wu, M.; Zhang, X. Exosome-related tumor microenvironment. J. Cancer 2018, 9, 3084–3092. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Ji, X.; Liu, J.; Fan, D.; Zhou, Q.; Chen, C.; Wang, W.; Wang, G.; Wang, H.; Yuan, W.; et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 2019, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hood, J.L.; San Roman, S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef] [Green Version]
- Costa-silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; Xiang, J.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver Bruno. Nat. Cell Biol. 2018, 17, 816–826. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, T.; Liu, R.; Bai, M.; Zhou, L.; Wang, X.; Li, S.; Wang, X.; Yang, H.; Li, J.; et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017, 8, 15016. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.F.; Ma, J.; Huang, L.; Yi, H.Y.; Zhang, Y.M.; Wu, X.G.; Yan, R.M.; Liang, L.; Zhong, M.; Yu, Y.H.; et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene 2019, 38, 1256–1268. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-P.; Tang, Y.-Y.; Fan, C.-M.; Guo, C.; Zhou, Y.-H.; Li, Z.; Li, X.-L.; Li, Y.; Li, G.-Y.; Xiong, W.; et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget 2018, 9, 12487–12502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Yen, Y.; Wang, Y.; Marcusson, E.G.; Chu, P.; Wu, J.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2015, 25, 501–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [Green Version]
- Lawson, J.; Dickman, C.; Towle, R.; Jabalee, J.; Javer, A.; Garnis, C. Extracellular vesicle secretion of miR-142-3p from lung adenocarcinoma cells induces tumor promoting changes in the stroma through cell-cell communication. Mol. Carcinog. 2019, 58, 376–387. [Google Scholar] [CrossRef]
- Ghayad, S.E.; Rammal, G.; Ghamloush, F.; Basma, H.; Nasr, R.; Diab-Assaf, M.; Chelala, C.; Saab, R. Exosomes derived from embryonal and alveolar rhabdomyosarcoma carry differential miRNA cargo and promote invasion of recipient fibroblasts. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.J.; Lin, X.J.; Tang, X.Y.; Zheng, T.T.; Lin, Y.Y.; Hua, K.Q. Exosomal metastasis-associated lung adenocarcinoma transcript 1 promotes angiogenesis and predicts poor prognosis in epithelial ovarian cancer. Int. J. Biol. Sci. 2018, 14, 1960–1973. [Google Scholar] [CrossRef] [Green Version]
- Lang, H.L.; Hu, G.W.; Zhang, B.; Kuang, W.; Chen, Y.; Wu, L.; Xu, G.H. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol. Rep. 2017, 38, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.O.; Youn, H.; Lee, C.H.; Kang, K.W.; Chung, J.K. Visualization of exosome-mediated miR-210 transfer from hypoxic tumor cells. Oncotarget 2017, 8, 9899–9910. [Google Scholar] [CrossRef] [Green Version]
- Hsu, Y.L.; Hung, J.Y.; Chang, W.A.; Lin, Y.S.; Pan, Y.C.; Tsai, P.H.; Wu, C.Y.; Kuo, P.L. Hypoxic lung cancer-secreted exosomal MIR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef]
- Huang, Z.; Yang, M.; Li, Y.; Yang, F.; Feng, Y. Exosomes derived from hypoxic colorectal cancer cells transfer wnt4 to normoxic cells to elicit a prometastatic phenotype. Int. J. Biol. Sci. 2018, 14, 2094–2102. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Salomon, C.; Bustos, R.; Díaz, J.; Martínez, S.; Silva, V.; Reyes, C.; Díaz-Valdivia, N.; Varas-Godoy, M.; Lobos-González, L.; et al. Caveolin-1-containing extracellular vesicles transport adhesion proteins and promote malignancy in breast cancer cell lines. Nanomedicine 2018, 13, 2597–2609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, M.T.; Hamar, P.; Guo, C.; Basar, E.; Perdigão-Henriques, R.; Balaj, L.; Lieberman, J. MiR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, D.; Lui, G.Y.L.; Lai, S.L.; Wilmott, J.S.; Tikoo, S.; Jackett, L.A.; Quek, C.; Brown, D.L.; Sharp, D.M.; Kwan, R.Y.Q.; et al. RAB27A promotes melanoma cell invasion and metastasis via regulation of pro-invasive exosomes. Int. J. Cancer 2019, 144, 3070–3085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Q.; Zhang, Q.; Lou, Y.; Yang, J.; Nie, G.; Chen, Q.; Chen, Y.; Zhang, J.; Wang, J.; Wei, T.; et al. Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer. Oncogene 2018, 37, 6105–6118. [Google Scholar] [CrossRef] [Green Version]
- Alharbi, M.; Lai, A.; Guanzon, D.; Palma, C.; Zuñiga, F.; Perrin, L.; He, Y.; Hooper, J.D.; Salomon, C. Ovarian cancer-derived exosomes promote tumour metastasis in vivo: An effect modulated by the invasiveness capacity of their originating cells. Clin. Sci. 2019, 133, 1401–1419. [Google Scholar] [CrossRef]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Chen, T.; Zheng, X.; Yang, S.; Xu, K.; Chen, X.; Xu, F.; Wang, L.; Shen, Y.; Wang, T.; et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis 2018, 39, 1368–1379. [Google Scholar] [CrossRef]
- Languino, L.R.; Singh, A.; Prisco, M.; Inman, G.J.; Luginbuhl, A.; Curry, J.M.; South, A.P. Exosome-mediated transfer from the tumor microenvironment increases TGFβ signaling in squamous cell carcinoma. Am. J. Transl. Res. 2016, 8, 2432–2437. [Google Scholar]
- Liu, Y.; Gu, Y.; Han, Y.; Zhang, Q.; Jiang, Z.; Zhang, X.; Huang, B.; Xu, X.; Zheng, J.; Cao, X. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell 2016, 30, 243–256. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takasugi, M.; Okada, R.; Takahashi, A.; Virya Chen, D.; Watanabe, S.; Hara, E. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Somlo, G.; Palomares, M.; Li, Z.; Tremblay, J.R.; Tsuyada, A.; et al. Breast cancer-secreted miR-122 reprograms glucose metabolism in pre-metastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grünewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; de Álava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Primers 2018, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Kansara, M.; Teng, M.W.; Smyth, M.J.; Thomas, D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing sarcoma: Current management and future approaches through collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef]
- Zhuo, B.; Li, Y.; Gu, F.; Li, Z.; Sun, Q.; Shi, Y.; Shen, Y.; Zhang, F.; Wang, R.; Wang, X. Overexpression of CD155 relates to metastasis and invasion in osteosarcoma. Oncol. Lett. 2018, 15, 7312–7318. [Google Scholar] [CrossRef]
- Xu, H.; Zhu, X.; Bao, H.; Wh Shek, T.; Huang, Z.; Wang, Y.; Wu, X.; Wu, Y.; Chang, Z.; Wu, S.; et al. Genetic and clonal dissection of osteosarcoma progression and lung metastasis. Int. J. Cancer 2018, 143, 1134–1142. [Google Scholar] [CrossRef]
- Mu, X.; Isaac, C.; Greco, N.; Huard, J.; Weiss, K. Notch signaling is associated with ALDH activity and an aggressive metastatic phenotype in murine osteosarcoma cells. Front. Oncol. 2013, 3, 143. [Google Scholar] [CrossRef] [Green Version]
- Potratz, J.; Tillmanns, A.; Berning, P.; Korsching, E.; Schaefer, C.; Lechtape, B.; Schleithoff, C.; Unland, R.; Schäfer, K.L.; Müller-Tidow, C.; et al. Receptor tyrosine kinase gene expression profiles of Ewing sarcomas reveal ROR1 as a potential therapeutic target in metastatic disease. Mol. Oncol. 2016, 10, 677–692. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhang, P.; Zhang, W.; Lei, Z.; He, J.; Meng, J.; Di, T.; Yan, W. Identification of key genes and pathways in Ewing’s sarcoma patients associated with metastasis and poor prognosis. Onco Targets Ther. 2019, 12, 4153–4165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Xu, C.; Ayello, J.; Dela Cruz, F.; Rosenblum, J.M.; Lessnick, S.L.; Cairo, M.S. Protein phosphatase 1 regulatory subunit 1A in ewing sarcoma tumorigenesis and metastasis. Oncogene 2018, 37, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Choo, S.; Wang, P.; Newbury, R.; Roberts, W.; Yang, J. Reactivation of TWIST1 contributes to Ewing sarcoma metastasis. Pediatr. Blood Cancer 2018, 65, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatano, M.; Matsumoto, Y.; Fukushi, J.I.; Matsunobu, T.; Endo, M.; Okada, S.; Iura, K.; Kamura, S.; Fujiwara, T.; Iida, K.; et al. Cadherin-11 regulates the metastasis of Ewing sarcoma cells to bone. Clin. Exp. Metastasis 2015, 32, 579–591. [Google Scholar] [CrossRef]
- Mendoza-Naranjo, A.; El-Naggar, A.; Wai, D.H.; Mistry, P.; Lazic, N.; Ayala, F.R.R.; da Cunha, I.W.; Rodriguez-Viciana, P.; Cheng, H.; Tavares Guerreiro Fregnani, J.H.; et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol. Med. 2013, 5, 1019–1034. [Google Scholar] [CrossRef]
- El-Naggar, A.M.; Veinotte, C.J.; Cheng, H.; Grunewald, T.G.P.; Negri, G.L.; Somasekharan, S.P.; Corkery, D.P.; Tirode, F.; Mathers, J.; Khan, D.; et al. Translational Activation of HIF1α by YB-1 Promotes Sarcoma Metastasis. Cancer Cell 2015, 27, 682–697. [Google Scholar] [CrossRef] [Green Version]
- Krook, M.A.; Nicholls, L.A.; Scannell, C.A.; Chugh, R.; Thomas, D.G.; Lawlor, E.R. Stress-induced CXCR4 promotes migration and invasion of Ewing Sarcoma. Mol. Cancer Res. 2014, 12, 953–964. [Google Scholar] [CrossRef] [Green Version]
- Chow, W.A. Chondrosarcoma: Biology, genetics, and epigenetics. F1000Research 2018, 7, 1826. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.M.; Li, T.M.; Hsu, S.F.; Su, Y.C.; Kao, S.T.; Fong, Y.C.; Tang, C.H. IGF-I enhances α5β1 integrin expression and cell motility in human chondrosarcoma cells. J. Cell. Physiol. 2011, 226, 3270–3277. [Google Scholar] [CrossRef]
- Lai, T.H.; Fong, Y.C.; Fu, W.M.; Yang, R.S.; Tang, C.H. Stromal cell-derived factor-1 increase αvβ3 integrin expression and invasion in human chondrosarcoma cells. J. Cell. Physiol. 2009, 218, 334–342. [Google Scholar] [CrossRef]
- Valery, P.C.; Laversanne, M.; Bray, F. Bone cancer incidence by morphological subtype: A global assessment. Cancer Causes Control 2015, 26, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival improvement. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abarrategi, A.; Tornin, J.; Lucia, M.C.; Hamilton, A.; Enrique, M.C.; Rodrigo, J.P.; González, M.V.; Baldini, N.; Javier, G.C.; Rodriguez, R. Osteosarcoma: Cells-of-Origin, Cancer stem cells, and targeted therapies. Stem Cells Int. 2016, 2016, 3631764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayani, J.; Zielenska, M.; Pandita, A.; Al-Romaih, K.; Karaskova, J.; Harrison, K.; Bridge, J.A.; Sorensen, P.; Thorner, P.; Squire, J.A. Spectral karyotyping identifies recurrent complex rearrangements of chromosomes 8, 17, and 20 in osteosarcomas. Genes Chromosoms Cancer 2003, 36, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef]
- Daw, N.C.; Chou, A.J.; Jaffe, N.; Rao, B.N.; Billups, C.A.; Rodriguez-Galindo, C.; Meyers, P.A.; Huh, W.W. Recurrent osteosarcoma with a single pulmonary metastasis: A multi-institutional review. Br. J. Cancer 2015, 112, 278–282. [Google Scholar] [CrossRef] [Green Version]
- Isakoff, M.S.; Bielack, S.S.; Meltzer, P.; Gorlick, R. Osteosarcoma: Current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015, 33, 3029–3035. [Google Scholar] [CrossRef] [Green Version]
- Jerez, S.; Araya, H.; Thaler, R.; Charlesworth, M.C.; López-Solís, R.; Kalergis, A.M.; Céspedes, P.F.; Dudakovic, A.; Stein, G.S.; van Wijnen, A.J.; et al. Proteomic Analysis of Exosomes and Exosome-Free Conditioned Media From Human Osteosarcoma Cell Lines Reveals Secretion of Proteins Related to Tumor Progression. J. Cell. Biochem. 2017, 118, 351–360. [Google Scholar] [CrossRef]
- Jerez, S.; Araya, H.; Hevia, D.; Irarrázaval, C.E.; Thaler, R.; van Wijnen, A.J.; Galindo, M. Extracellular vesicles from osteosarcoma cell lines contain miRNAs associated with cell adhesion and apoptosis. Gene 2019, 710, 246–257. [Google Scholar] [CrossRef]
- Gong, L.; Bao, Q.; Hu, C.; Wang, J.; Zhou, Q.; Wei, L.; Tong, L.; Zhang, W.; Shen, Y. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem. Biophys. Res. Commun. 2018, 500, 170–176. [Google Scholar] [CrossRef]
- Raimondi, L.; De Luca, A.; Gallo, A.; Costa, V.; Russelli, G.; Cuscino, N.; Manno, M.; Raccosta, S.; Carina, V.; Bellavia, D.; et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Baglio, S.R.; Lagerweij, T.; Pérez-Lanzón, M.; Ho, X.D.; Léveillé, N.; Melo, S.A.; Cleton-Jansen, A.M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking tumor-educated MSC paracrine activity halts osteosarcoma progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torreggiani, E.; Roncuzzi, L.; Perut, F.; Zini, N.; Baldini, N. Multimodal transfer of MDR by exosomes in human osteosarcoma. Int. J. Oncol. 2016, 49, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo-Munoz, L.; Cai, N.; Cumming, A.; Macklin, R.; De Long, L.M.; Topkas, E.; Mukhopadhyay, P.; Hill, M.; Saunders, N.A. Progression of osteosarcoma from a non-metastatic to a metastatic phenotype is causally associated with activation of an autocrine and paracrine uPA axis. PLoS ONE 2015, 10, e0133592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.W.; Wu, X.F.; Gu, X.J.; Jiang, X.H. Exosomal miR-1228 from cancer-associated fibroblasts promotes cell migration and invasion of osteosarcoma by directly targeting SCAI. Oncol. Res. 2019, 27, 979–986. [Google Scholar] [CrossRef]
- Shimbo, K.; Miyaki, S.; Ishitobi, H.; Kato, Y.; Kubo, T.; Shimose, S.; Ochi, M. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem. Biophys. Res. Commun. 2014, 445, 381–387. [Google Scholar] [CrossRef]
- Fujiwara, T.; Uotani, K.; Yoshida, A.; Morita, T.; Nezu, Y.; Kobayashi, E.; Yoshida, A.; Uehara, T.; Omori, T.; Sugiu, K.; et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget 2017, 8, 33375–33392. [Google Scholar] [CrossRef]
- Xu, J.-F.; Wang, Y.-P.; Zhang, S.-J.; Chen, Y.; Gu, H.-F.; Dou, X.-F.; Xia, B.; Bi, Q.; Fan, S.-W. Exosomes containing differential expression of microRNA and mRNA in osteosarcoma that can predict response to chemotherapy. Oncotarget 2017, 8, 75968–75978. [Google Scholar] [CrossRef]
- Delattre, O.; Zucman, J.; Plougastel, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; de Jong, P.; Rouleau, G. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef]
- Peter, M.; Gilbert, E.; Delattre, O. A multiplex Real-Time PCR assay for the detection of gene fusions observed in solid tumors. Lab. Investig. 2001, 81, 905–912. [Google Scholar] [CrossRef]
- Pinto, A.; Dickman, P.; Parham, D. Pathobiologic markers of the Ewing sarcoma family of tumors: State of the art and prediction of behaviour. Sarcoma 2011, 2011, 856190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreppel, M.; Aryee, D.N.T.; Schaefern, K.L.; Amann, G.; Kofler, R.; Poremba, C.; Kovar, H. Suppression of KCMF1 by constitutive high CD99 expression is involved in the migratory ability of Ewing’s sarcoma cells. Oncogene 2006, 25, 2795–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocchi, A.; Manara, M. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J. Clin. Investig. 2010, 120, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Balamuth, N.J.; Womer, R.B. Ewing’s sarcoma. Lancet Oncol. 2010, 11, 184–192. [Google Scholar] [CrossRef]
- Rodriguez-Galindo, C.; Spunt, S.L.; Pappo, A.S. Treatment of Ewing sarcoma family of tumors: Current status and outlook for the future. Med. Pediatr. Oncol. 2003, 40, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.V.; Raposo, G.; Welsch, U.; Prazeres da Costa, O.; Thiel, U.; Lebar, M.; Maurer, M.; Bender, H.U.; von Luettichau, I.; Richter, G.H.S.; et al. First identification of Ewing’s sarcoma-derived extracellular vesicles and exploration of their biological and potential diagnostic implications. Biol. Cell 2013, 105, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Tsugita, M.; Yamada, N.; Noguchi, S.; Yamada, K.; Moritake, H.; Shimizu, K.; Akao, Y.; Ohno, T. Ewing sarcoma cells secrete EWS/Fli-1 fusion mRNA via microvesicles. PLoS ONE 2013, 8, e77416. [Google Scholar] [CrossRef] [Green Version]
- Villasante, A.; Marturano-Kruik, A.; Ambati, S.R.; Liu, Z.; Godier-Furnemont, A.; Parsa, H.; Lee, B.W.; Moore, M.A.S.; Vunjak-Novakovic, G. Recapitulating the size and cargo of tumor exosomes in a tissue-engineered model. Theranostics 2016, 6, 1119–1130. [Google Scholar] [CrossRef]
- Ventura, S.; Aryee, D.N.T.; Felicetti, F.; De Feo, A.; Mancarella, C.; Manara, M.C.; Picci, P.; Colombo, M.P.; Kovar, H.; Care, A.; et al. CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch-mediated control of NF-κB signaling. Oncogene 2016, 35, 3944–3954. [Google Scholar] [CrossRef]
- De Feo, A.; Sciandra, M.; Ferracin, M.; Felicetti, F.; Astolfi, A.; Pignochino, Y.; Picci, P.; Carè, A.; Scotlandi, K. Exosomes from CD99-deprived Ewing sarcoma cells reverse tumor malignancy by inhibiting cell migration and promoting neural differentiation. Cell Death Dis. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Kim, M.J.; Cho, K.J.; Ayala, A.G.; Ro, J.Y. Chondrosarcoma: With updates on molecular genetics. Sarcoma 2011, 2011, 405437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cintra, F.F.; Etchebehere, M.; Gonçalves, J.C.B.; Cassone, A.E.; Amstalden, E.M.I. Vascular pattern in enchondroma and chondrosarcoma: Clinical and immunohistologic study. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.T.; Chen, H.T.; Tsou, H.K.; Tan, T.W.; Fong, Y.C.; Chen, P.C.; Yang, W.H.; Wang, S.W.; Chen, J.C.; Tang, C.H. CCL5 promotes VEGF-dependent angiogenesis by downregulating miR-200B through PI3K/Akt signaling pathway in human chondrosarcoma cells. Oncotarget 2014, 5, 10718–10731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.T.; Huang, Y.L.; Tzeng, H.E.; Tsai, C.H.; Wang, S.W.; Tang, C.H. CCL5 promotes vascular endothelial growth factor expression and induces angiogenesis by down-regulating miR-199a in human chondrosarcoma cells. Cancer Lett. 2015, 357, 476–487. [Google Scholar] [CrossRef]
- Sun, X.; Charbonneau, C.; Wei, L.; Chen, Q.; Terek, R.M. MiR-181a targets RGS16 to promote chondrosarcoma growth, angiogenesis, and metastasis. Mol. Cancer Res. 2015, 13, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Li, J.; Song, Y.; Wang, X. MiR-129-5p inhibits proliferation and invasion of chondrosarcoma cells by regulating SOX4/Wnt/β-catenin signaling pathway. Cell. Physiol. Biochem. 2017, 42, 242–253. [Google Scholar] [CrossRef] [Green Version]
- Lu, N.; Lin, T.; Wang, L.; Qi, M.; Liu, Z.; Dong, H.; Zhang, X.; Zhai, C.; Wang, Y.; Liu, L.; et al. Association of SOX4 regulated by tumor suppressor miR-30a with poor prognosis in low-grade chondrosarcoma. Tumor Biol. 2015, 36, 3843–3852. [Google Scholar] [CrossRef]
- Horng, C.T.; Shieh, P.C.; Tan, T.W.; Yang, W.H.; Tang, C.H. Paeonol suppresses chondrosarcoma metastasis through up-regulation of miR-141 by modulating PKCδ and c-Src signaling pathway. Int. J. Mol. Sci. 2014, 15, 11760–11772. [Google Scholar] [CrossRef] [Green Version]
- Tsai, C.H.; Yang, D.Y.; Lin, C.Y.; Chen, T.M.; Tang, C.H.; Huang, Y.L. Sphingosine-1-phosphate suppresses chondrosarcoma metastasis by upregulation of tissue inhibitor of metalloproteinase 3 through suppressing miR-101 expression. Mol. Oncol. 2017, 11, 1380–1398. [Google Scholar] [CrossRef]
- Aghebati-Maleki, A.; Nami, S.; Baghbanzadeh, A.; Karzar, B.H.; Noorolyai, S.; Fotouhi, A.; Aghebati-Maleki, L. Implications of exosomes as diagnostic and therapeutic strategies in cancer. J. Cell. Physiol. 2019, 234, 21694–21706. [Google Scholar] [CrossRef]
- Gudbergsson, J.M.; Jønsson, K.; Simonsen, J.B.; Johnsen, K.B. Systematic review of targeted extracellular vesicles for drug delivery—Considerations on methodological and biological heterogeneity. J. Control. Release 2019, 306, 108–120. [Google Scholar] [CrossRef] [PubMed]
| Origin Cell | Recipient Cell | Cargo | Change | Ref. |
|---|---|---|---|---|
| OS cells and conditioned media | - | Profiling of proteome and secretome | Exosome proteins involved in migration, adhesion, and angiogenesis | [98] |
| Metastatic and non-metastatic OS cell lines | - | Profiling of miRNAs and target genes | miRNA of metastatic OS exosomes target metastasis-associated genes, cell adhesion, and apoptosis | [99] |
| Metastatic and non-metastatic OS cell lines | Osteoblasts | miR-675 (miRNA profiling) | Metastasis-associated exosomes induce migration and invasion in osteoblasts via miR-675/CALN1 axis | [100] |
| Metastatic OS cell lines | Macrophages, osteoclasts, endothelial cells | miR-148a, miR-21-5p (RNA profiling) | Induction of osteoclast-like gene expression (macrophage), increase in bone resorption (osteoclasts) and angiogenesis (endothelial cells) via miRNA transfer | [101] |
| Metastatic and non-metastatic OS cell lines | MSC | TGFβ (induction of IL6) | Internalization of TGFβ induces IL6 production, cell growth, and lung metastasis in vivo | [102] |
| Doxorubicin-resistant OS cell lines | Sensitive OS cell lines | MRP1, Pgp(multidrug resistant proteins) | Increase in doxorubicin resistance in recipient cells; increase in MRP1 and Pgp mRNA levels. | [103] |
| Bone marrow (conditioned media) | Metastatic and non-metastatic OS cells | uPA (secreted, paracrine loop) | Increase in migration on recipient cells, induction of OS metastasis in vivo | [104] |
| CAF | OS cell lines | miR-1228 (miRNA profiling) | Increase in migration and invasion via miR-1228 transfer | [105] |
| MSC | OS cell lines | miR-143 (synthetic introduction) | Reduction of migration via exosome transfer (better than transfection) | [106] |
| OS cell lines | - | Profiling of miRNA as OS biomarkers | Better biomarker than ALP or patient stratification according to chemotherapy response | [107,108], |
| Origin cell | Recipient cell | Cargo | Change | Ref. |
|---|---|---|---|---|
| ES cell lines | - | EWSR1-FLI1, EZH2, and 10 more mRNAs (mRNA profiling of known ES targets) | Suitable as circulating biomarkers for ES, detectable in spike-in healthy blood samples | [116] |
| ES cell lines | ES cell lines | EWSR1-FLI1 mRNA | Labelled EWSR1-FLI1 transferred to other ES cells, not to OS | [117] |
| ES cell lines | Osteoblasts, osteoclasts in 3D scaffold | EZH2 mRNA (target of interest) | Transfer of EZH2 mRNA to MSC (increase expression), osteoblasts (no change), and osteoclasts (reduction expression) | [118] |
| ESCD99neg cell line model | ES cell lines (normal CD99) | Increased miR-34a | Regulation of NFκB via miR-34a through reduction of Notch. Increase in neural differentiation (similar to direct CD99 silencing) | [119] |
| ESCD99neg cell line model | ES cell lines (normal CD99) | miR-199a-3p (miRNA profiling) miR-199a-3p | Induction of different gene expression profiles, neural differentiation, and neurogenesis; reduction of cell growth and migration (similar to miR mimic) | [120] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chicón-Bosch, M.; Tirado, O.M. Exosomes in Bone Sarcomas: Key Players in Metastasis. Cells 2020, 9, 241. https://doi.org/10.3390/cells9010241
Chicón-Bosch M, Tirado OM. Exosomes in Bone Sarcomas: Key Players in Metastasis. Cells. 2020; 9(1):241. https://doi.org/10.3390/cells9010241
Chicago/Turabian StyleChicón-Bosch, Mariona, and Oscar M. Tirado. 2020. "Exosomes in Bone Sarcomas: Key Players in Metastasis" Cells 9, no. 1: 241. https://doi.org/10.3390/cells9010241
APA StyleChicón-Bosch, M., & Tirado, O. M. (2020). Exosomes in Bone Sarcomas: Key Players in Metastasis. Cells, 9(1), 241. https://doi.org/10.3390/cells9010241





