JNK1 Induces Notch1 Expression to Regulate Genes Governing Photoreceptor Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Cell Culture
2.3. Antibodies and Reagents
2.4. Real-Time PCR
- β-actin: sense (AAAGACCTGTACGCCAACAC),
- antisense (GTCATACTCCTGCTTGCTGAT);
- c-Jun: sense (GCAAGCCCTGAAGGAAGAG),
- antisense (GTCAT ACTCCTGCTTGCTGAT);
- Neurod1: sense (GACGGGGTCCCAAAAAGAAAA),
- antisense (GCCAAGCGCAGTGTCTCTATT);
- Crx: sense (GTTCAAGAATCGTAGGGCGAA),
- antisense (TGAGATGCCCAAAGGATCTGT);
- Rxrg: sense (CATGAGCCCTTCAGTAGCCTT),
- antisense (CGGAGAGCCAAGAGCATTGAG);
- Notch1: sense (CCGTGTAAGAATGCTGGAACG),
- antisense (AGCGACAGATGTATGAAGACTCA);
- Opn1sw: sense (CAGCCTTCATGGGATTTGTCT),
- antisense (CAAAGAGGAAGTATCCGTGACAG);
- Opn1mw: sense (ATGGCCCAAAGGCTTACAGG),
- antisense (AAGGGACCTTTGGTGCTGTT);
- Notch2: sense (GAGAAAAACCGCTGTCAGAATGG),
- antisense (GGTGGAGTATTGGCAGTCCTC);
- Notch3: sense (TGCCAGAGTTCAGTGGTGG),
- antisense (CACAGGCAAATCGGCCATC);
- Wnt2b: sense (CCGACGTGTCCCCATCTTC),
- antisense (GCCCCTATGTACCACCAGGA);
- Wnt5a: sense (CAACTGGCAGGACTTTCTCAA),
- antisense (CATCTCCGATGCCGGAACT);
- Rho: sense (CCCTTCTCCAACGTCACAGG),
- antisense (TGAGGAAGTTGATGGGGAAGC);
- Nrl: sense (TCCCAGTCCCTTGGCTATGG),
- antisense (CACCGAGCTGTATGGTGTG);
- Onecut2: sense (GGCTTCCGTCCATGAACAAC),
- antisense (CGAAATTGGGGCTGAGCATTTT);
- Chx10: sense (CTGAGCAAGCCCAAATCCGA),
- antisense (CGCAGCTAACAAATGCCCAG);
- Stx1a: sense (AGAGATCCGGGGCTTTATTGA),
- antisense (AATGCTCTTTAGCTTGGAGCG);
- Vim: sense (CGGCTGCGAGAGAAATTGC),
- antisense (CCACTTTCCGTTCAAGGTCAAG);
- Sox9: sense (CGGAACAGACTCACATCTCTCC),
- antisense (GCTTGCACGTCGGTTTTGG);
- Rbpms: sense (CCCGTAGGCTTTGTCAGTTTT),
- antisense (GTAAAGTGCAGGTACTGTGAGC);
- Prkca: sense (GTTTACCCGGCCAACGACT),
- antisense (GGGCGATGAATTTGTGGTCTT);
2.5. Western Blot
2.6. Subcellular Fractionation
2.7. Histology and Immunohistochemistry
2.8. Immunofluorescence
2.9. Chromatin Immunoprecipitation Assay
2.10. Plasmid Constructs
2.11. Transfection and Luciferase Assay
2.12. Cell Isolation and Flow Cytometry
2.13. Electroretinogram (ERG) Recording
2.14. Statistical Analysis
3. Results
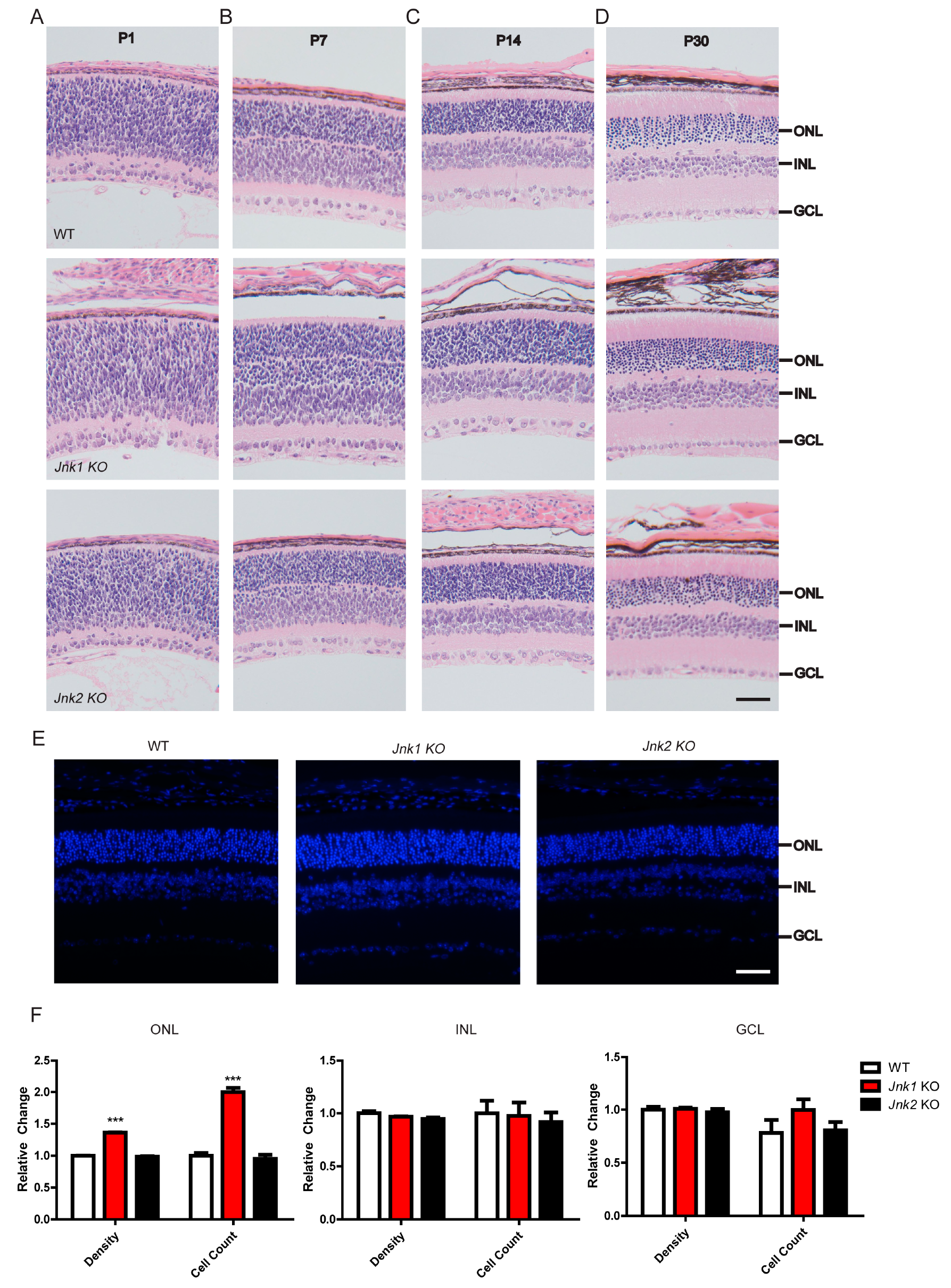
3.1. Knocking Out JNK1 Promoted Retinogenesis
3.2. The Number of Photoreceptors Increased in Jnk1 KO Mice
3.3. Enhanced ERG Responses in the Jnk1 KO Mice
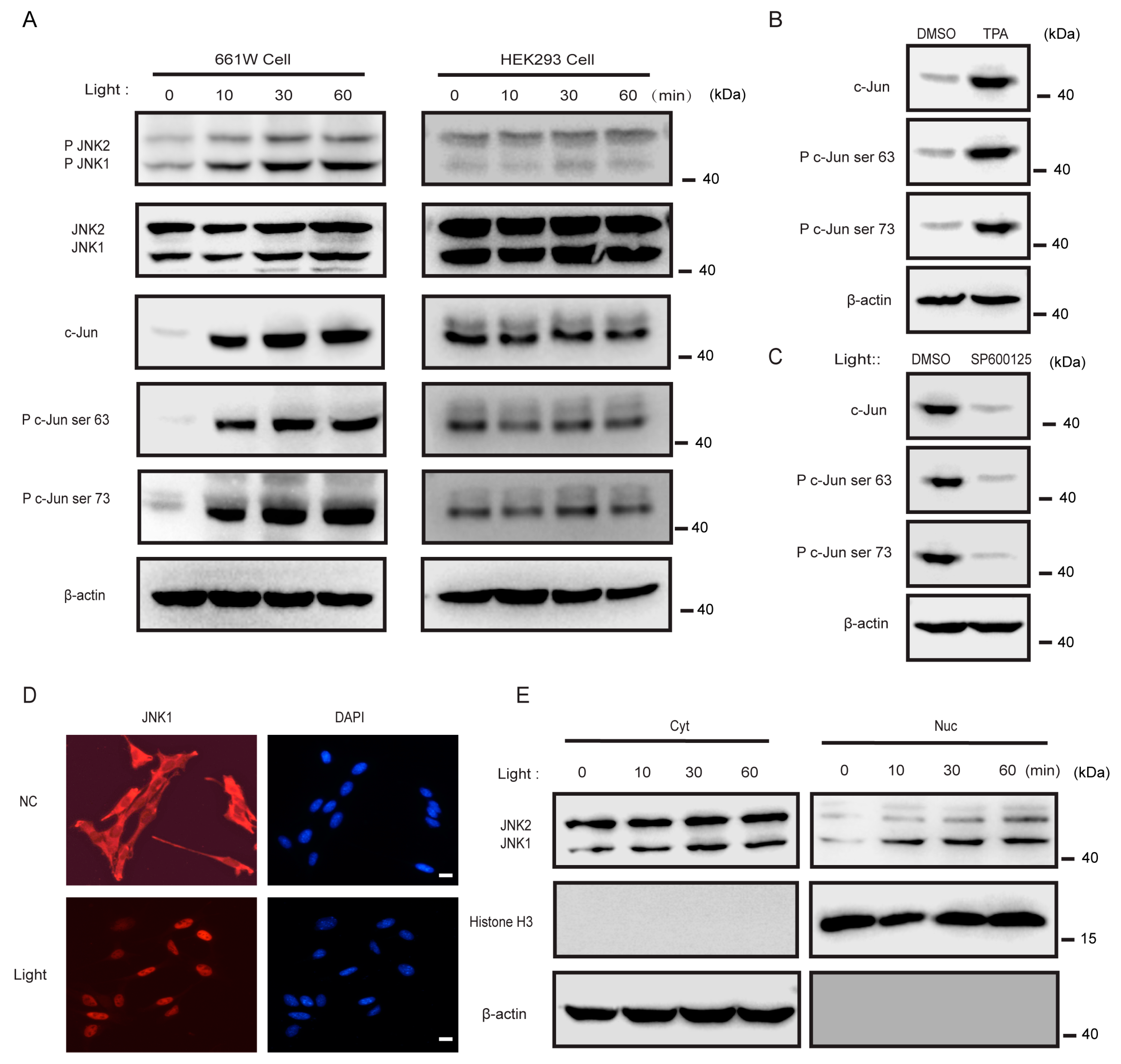
3.4. Light Activated JNK1, but Not JNK2, in 661w Cells
3.5. JNK1 Attenuated the Transcription of Genes Important for Photoreceptors
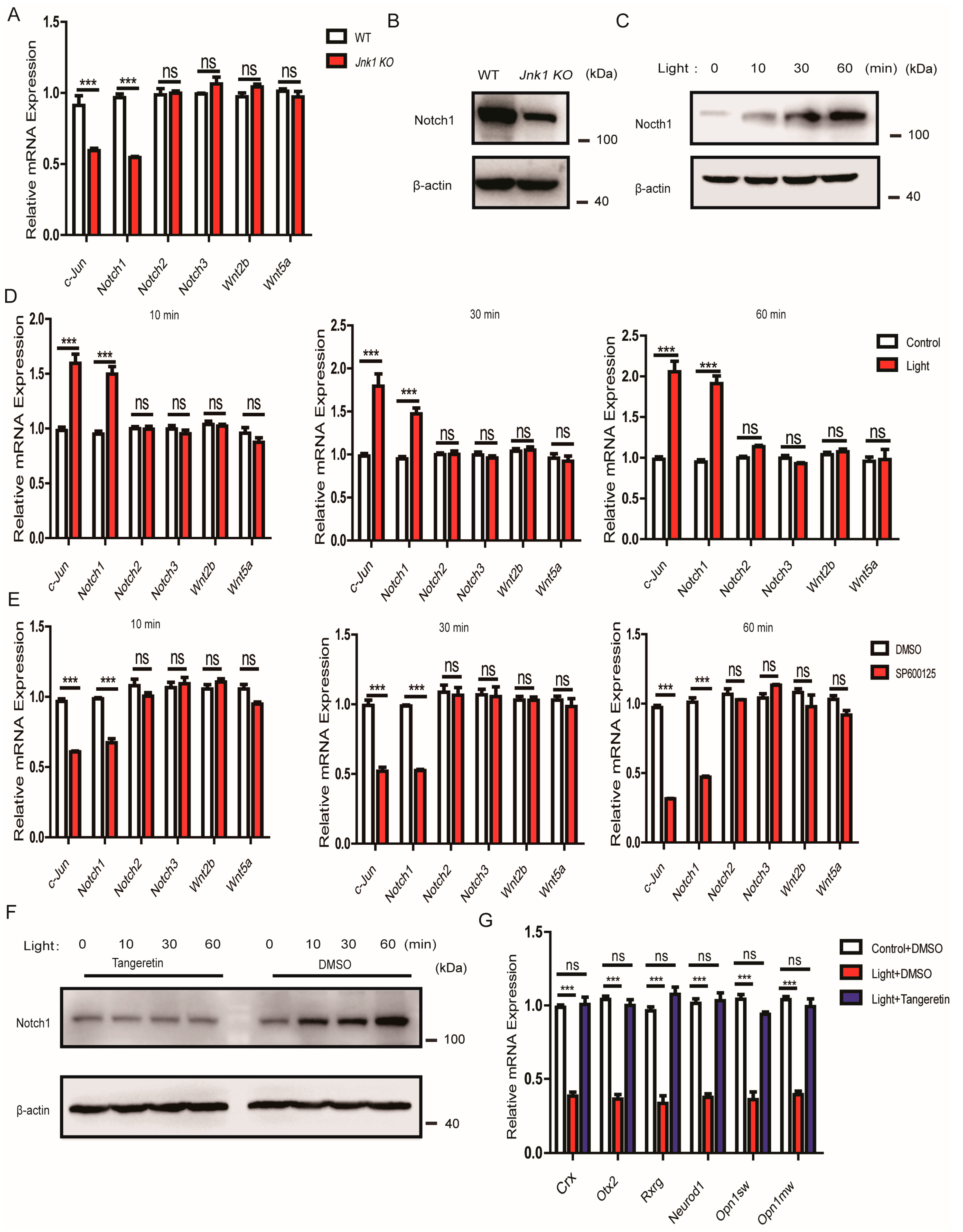
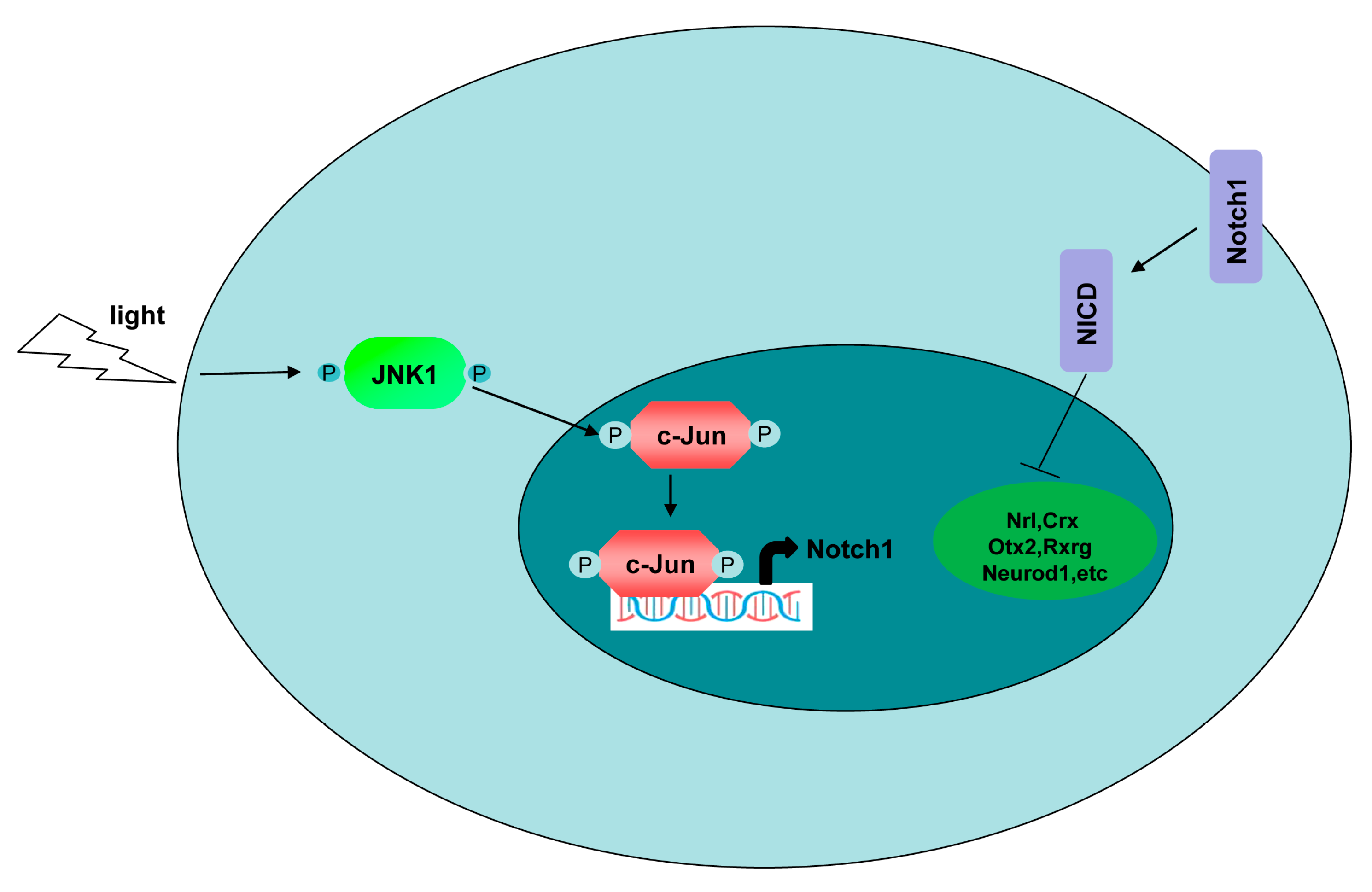
3.6. The Suppressive Effect of JNK1 Was Dependent on Notch1
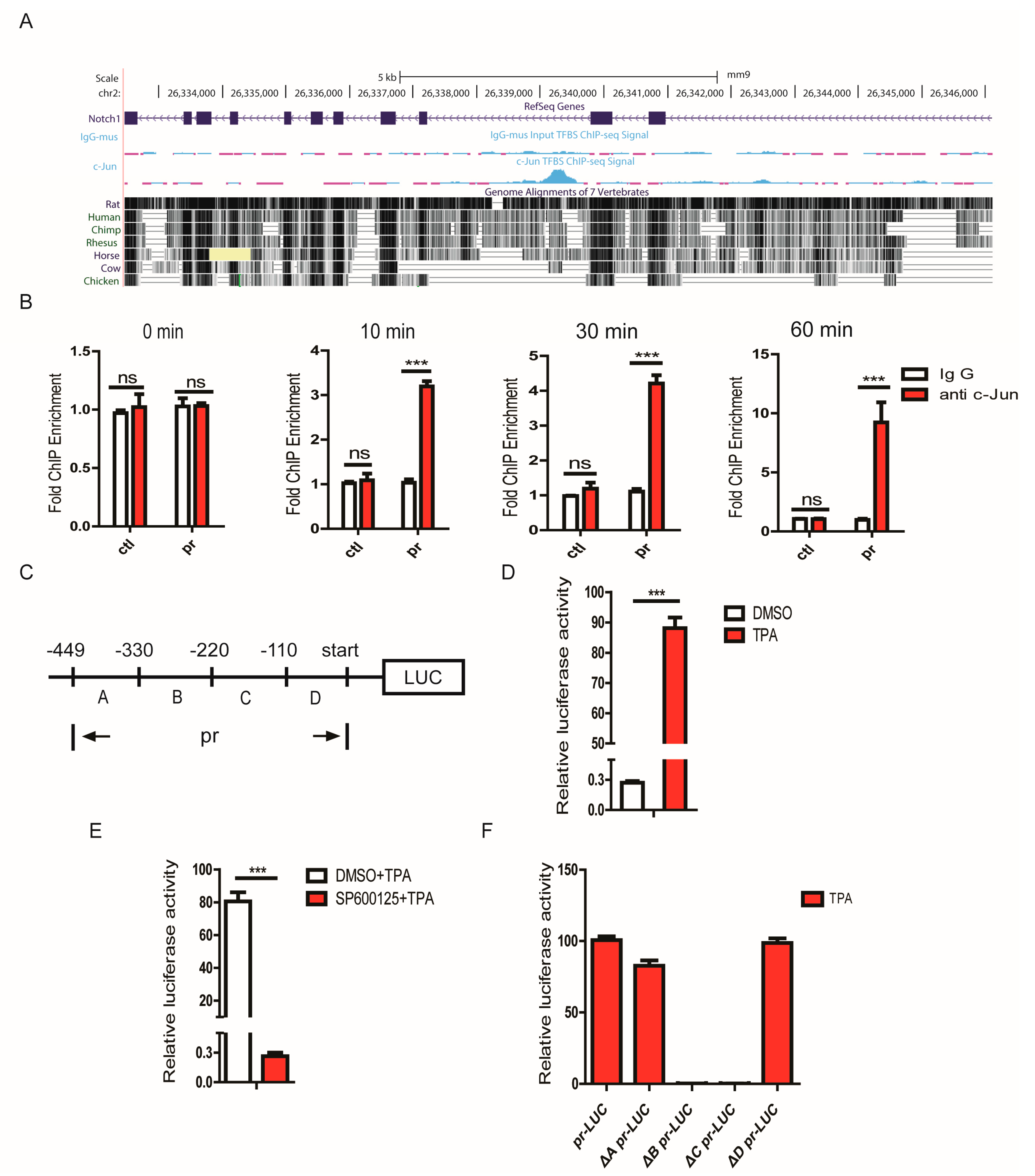
3.7. JNK1 Regulated Notch1 Transcription via Activating c-Jun
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wassle, H.; Boycott, B.B. Functional architecture of the mammalian retina. Physiol. Rev. 1991, 71, 447–480. [Google Scholar] [CrossRef] [PubMed]
- Masland, R.H. The fundamental plan of the retina. Nat. Neurosci. 2001, 4, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.G.; Xue, T.; Yau, K.W. How vision begins: An odyssey. Proc. Natl. Acad. Sci. USA 2008, 105, 9855–9862. [Google Scholar] [CrossRef] [PubMed]
- Bassett, E.A.; Wallace, V.A. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012, 35, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Harada, C.; Parada, L.F. Molecular regulation of visual system development: More than meets the eye. Genes Dev. 2007, 21, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Carter-Dawson, L.D.; LaVail, M.M. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J. Comp. Neurol. 1979, 188, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, Q.L.; Nie, Z.; Sun, H.; Lennon, G.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Zack, D.J. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 1997, 19, 1017–1030. [Google Scholar] [CrossRef]
- Freund, C.L.; Gregory-Evans, C.Y.; Furukawa, T.; Papaioannou, M.; Looser, J.; Ploder, L.; Bellingham, J.; Ng, D.; Herbrick, J.A.; Duncan, A.; et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 1997, 91, 543–553. [Google Scholar] [CrossRef]
- Furukawa, T.; Morrow, E.M.; Cepko, C.L. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 1997, 91, 531–541. [Google Scholar] [CrossRef]
- Nishida, A.; Furukawa, A.; Koike, C.; Tano, Y.; Aizawa, S.; Matsuo, I.; Furukawa, T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 2003, 6, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.R.; Hendrickson, A.; McGuire, C.R.; Reh, T.A. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2897–2904. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Etter, P.; Hayes, S.; Jones, I.; Nelson, B.; Hartman, B.; Forrest, D.; Reh, T.A. NeuroD1 regulates expression of thyroid hormone receptor 2 and cone opsins in the developing mouse retina. J. Neurosci. 2008, 28, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Morrow, E.M.; Furukawa, T.; Lee, J.E.; Cepko, C.L. NeuroD regulates multiple functions in the developing neural retina in rodent. Development (Cambridge, England) 1999, 126, 23–36. [Google Scholar]
- Ng, L.; Hurley, J.B.; Dierks, B.; Srinivas, M.; Salto, C.; Vennstrom, B.; Reh, T.A.; Forrest, D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 2001, 27, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.R.; Srinivas, M.; Forrest, D.; Morreale de Escobar, G.; Reh, T.A. Making the gradient: Thyroid hormone regulates cone opsin expression in the developing mouse retina. Proc. Natl. Acad. Sci. USA 2006, 103, 6218–6223. [Google Scholar] [CrossRef] [PubMed]
- Mears, A.J.; Kondo, M.; Swain, P.K.; Takada, Y.; Bush, R.A.; Saunders, T.L.; Sieving, P.A.; Swaroop, A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001, 29, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Yang, H.J.; Brooks, M.J.; Zelinger, L.; Karakülah, G.; Gotoh, N.; Boleda, A.; Gieser, L.; Giuste, F.; Whitaker, D.T.; et al. NRL-Regulated Transcriptome Dynamics of Developing Rod Photoreceptors. Cell Rep. 2016, 17, 2460–2473. [Google Scholar] [CrossRef] [PubMed]
- Perron, M.; Harris, W.A. Determination of vertebrate retinal progenitor cell fate by the Notch pathway and basic helix-loop-helix transcription factors. Cell. Mol. Life Sci. 2000, 57, 215–223. [Google Scholar] [CrossRef]
- Luo, H.; Jin, K.; Xie, Z.; Qiu, F.; Li, S.; Zou, M.; Cai, L.; Hozumi, K.; Shima, D.T.; Xiang, M. Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc. Natl. Acad. Sci. USA 2012, 109, E553–E562. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, A.P.; Mason, H.A.; Cepko, C.L. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development (Cambridge, England) 2006, 133, 913–923. [Google Scholar] [CrossRef]
- Mizeracka, K.; DeMaso, C.R.; Cepko, C.L. Notch1 is required in newly postmitotic cells to inhibit the rod photoreceptor fate. Development (Cambridge, England) 2013, 140, 3188–3197. [Google Scholar] [CrossRef] [PubMed]
- Yaron, O.; Farhy, C.; Marquardt, T.; Applebury, M.; Ashery-Padan, R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development (Cambridge, England) 2006, 133, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Hartman, B.H.; Georgi, S.A.; Lan, M.S.; Reh, T.A. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev. Biol. 2007, 304, 479–498. [Google Scholar] [CrossRef] [PubMed]
- Cubero, F.J.; Zhao, G.; Trautwein, C. JNK: A double-edged sword in tumorigenesis. Hepatology (Baltimore, Md.) 2011, 54, 1470–1472. [Google Scholar] [CrossRef] [PubMed]
- Bogoyevitch, M.A.; Kobe, B. Uses for JNK: The many and varied substrates of the c-Jun N-terminal kinases. Microbiol. Mol. Biol. Rev. 2006, 70, 1061–1095. [Google Scholar] [CrossRef] [PubMed]
- Kyosseva, S.V. Mitogen-activated protein kinase signaling. Int. Rev. Neurobiol. 2004, 59, 201–220. [Google Scholar] [CrossRef] [PubMed]
- Weston, C.R.; Wong, A.; Hall, J.P.; Goad, M.E.; Flavell, R.A.; Davis, R.J. JNK initiates a cytokine cascade that causes Pax2 expression and closure of the optic fissure. Genes Dev. 2003, 17, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Sabapathy, K.; Kallunki, T.; David, J.-P.; Graef, I.; Karin, M.; Wagner, E.F. c-Jun NH2-terminal kinase (JNK)1 and JNK2 have similar and stage-dependent roles in regulating T cell apoptosis and proliferation. J. Exp. Med. 2001, 193, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Sabapathy, K.; Hu, Y.; Kallunki, T.; Schreiber, M.; David, J.P.; Jochum, W.; Wagner, E.F.; Karin, M. JNK2 is required for efficient T-cell activation and apoptosis but not for normal lymphocyte development. Curr. Biol. 1999, 9, 116–125. [Google Scholar] [CrossRef]
- Rao, S.; Chun, C.; Fan, J.; Kofron, J.M.; Yang, M.B.; Hegde, R.S.; Ferrara, N.; Copenhagen, D.R.; Lang, R.A. A direct and melanopsin-dependent fetal light response regulates mouse eye development. Nature 2013, 494, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Philp, N.J.; Chang, W.; Long, K. Light-stimulated protein movement in rod photoreceptor cells of the rat retina. FEBS Lett. 1987, 225, 127–132. [Google Scholar] [CrossRef]
- Tikidji-Hamburyan, A.; Reinhard, K.; Storchi, R.; Dietter, J.; Seitter, H.; Davis, K.E.; Idrees, S.; Mutter, M.; Walmsley, L.; Bedford, R.A.; et al. Rods progressively escape saturation to drive visual responses in daylight conditions. Nat. Commun. 2017, 8, 1813. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.; Ma, J.; Shekhar, S.; Leon-Salas, W.D.; Weake, V.M. Blue light induces a neuroprotective gene expression program in Drosophila photoreceptors. BMC Neurosci. 2018, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Pulverer, B.J.; Kyriakis, J.M.; Avruch, J.; Nikolakaki, E.; Woodgett, J.R. Phosphorylation of c-jun mediated by MAP kinases. Nature 1991, 353, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Smeal, T.; Binetruy, B.; Mercola, D.A.; Birrer, M.; Karin, M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature 1991, 354, 494–496. [Google Scholar] [CrossRef] [PubMed]
- Franklin, C.C.; Sanchez, V.; Wagner, F.; Woodgett, J.R.; Kraft, A.S. Phorbol ester-induced amino-terminal phosphorylation of human JUN but not JUNB regulates transcriptional activation. Proc. Natl. Acad. Sci. USA 1992, 89, 7247–7251. [Google Scholar] [CrossRef] [PubMed]
- Bennett, B.L.; Sasaki, D.T.; Murray, B.W.; O’Leary, E.C.; Sakata, S.T.; Xu, W.; Leisten, J.C.; Motiwala, A.; Pierce, S.; Satoh, Y.; et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 2001, 98, 13681–13686. [Google Scholar] [CrossRef] [PubMed]
- Kubo, F.; Takeichi, M.; Nakagawa, S. Wnt2b controls retinal cell differentiation at the ciliary marginal zone. Development (Cambridge, England) 2003, 130, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Kubo, F.; Takeichi, M.; Nakagawa, S. Wnt2b inhibits differentiation of retinal progenitor cells in the absence of Notch activity by downregulating the expression of proneural genes. Development (Cambridge, England) 2005, 132, 2759–2770. [Google Scholar] [CrossRef]
- Nakagawa, S.; Takada, S.; Takada, R.; Takeichi, M. Identification of the laminar-inducing factor: Wnt-signal from the anterior rim induces correct laminar formation of the neural retina in vitro. Dev. Biol. 2003, 260, 414–425. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, L.; Sun, Y.; Wang, T.; Wang, B. Tangeretin enhances radiosensitivity and inhibits the radiation-induced epithelial-mesenchymal transition of gastric cancer cells. Oncol. Rep. 2015, 34, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, R.K.; Euskirchen, G.; Rozowsky, J.; Lamarre-Vincent, N.; Moqtaderi, Z.; Lefrancois, P.; Struhl, K.; Gerstein, M.; Snyder, M. Mapping accessible chromatin regions using Sono-Seq. Proc. Natl. Acad. Sci. USA 2009, 106, 14926–14931. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Cheng, Y.; Breschi, A.; Vierstra, J.; Wu, W.; Ryba, T.; Sandstrom, R.; Ma, Z.; Davis, C.; Pope, B.D.; et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014, 515, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Zaqouras, P.; Artavanis-Tsakonas, S. Involvement of Notch-1 in mammalian retinal neurogenesis: Association of Notch-1 activity with both immature and terminally differentiated cells. Mech. Dev. 1995, 53, 73–85. [Google Scholar] [CrossRef]
- Austin, C.P.; Feldman, D.E.; Ida, J.A., Jr.; Cepko, C.L. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development (Cambridge, England) 1995, 121, 3637–3650. [Google Scholar]
- Bao, Z.Z.; Cepko, C.L. The expression and function of Notch pathway genes in the developing rat eye. J. Neurosci. 1997, 17, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Dorsky, R.I.; Chang, W.S.; Rapaport, D.H.; Harris, W.A. Regulation of neuronal diversity in the Xenopus retina by Delta signalling. Nature 1997, 385, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Mukherjee, S.; Bao, Z.Z.; Morrow, E.M.; Cepko, C.L. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron 2000, 26, 383–394. [Google Scholar] [CrossRef]
- Scheer, N.; Groth, A.; Hans, S.; Campos-Ortega, J.A. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development (Cambridge, England) 2001, 128, 1099–1107. [Google Scholar]
- Kageyama, R.; Ishibashi, M.; Takebayashi, K.; Tomita, K. bHLH transcription factors and mammalian neuronal differentiation. Int. J. Biochem. Cell. Biol. 1997, 29, 1389–1399. [Google Scholar] [CrossRef]
- Wilkinson, G.; Dennis, D.; Schuurmans, C. Proneural genes in neocortical development. Neuroscience 2013, 253, 256–273. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Jones, Y.; Ellisman, M.H.; Goldstein, L.S.; Karin, M. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell 2003, 4, 521–533. [Google Scholar] [CrossRef]
- Amura, C.R.; Marek, L.; Winn, R.A.; Heasley, L.E. Inhibited neurogenesis in JNK1-deficient embryonic stem cells. Mol. Cell. Biol. 2005, 25, 10791–10802. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Jang, S.M.; Kim, C.H.; An, J.H.; Choi, K.H. Transcriptional activity of neural retina leucine zipper (Nrl) is regulated by c-Jun N-terminal kinase and Tip60 during retina development. Mol. Cell. Biol. 2012, 32, 1720–1732. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, C.H.; Huang, S.; Chen, J. Wnt Signaling in vascular eye diseases. Prog. Retin. Eye Res. 2019, 70, 110–133. [Google Scholar] [CrossRef] [PubMed]
- Athanasiou, D.; Aguila, M.; Bellingham, J.; Li, W.; McCulley, C.; Reeves, P.J.; Cheetham, M.E. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog. Retin. Eye Res. 2018, 62, 1–23. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, M.; Hu, H.; Wang, R.; Zhou, Y.; Zhang, L.; Wang, C.; Wang, Q. JNK1 Induces Notch1 Expression to Regulate Genes Governing Photoreceptor Production. Cells 2019, 8, 970. https://doi.org/10.3390/cells8090970
Pan M, Hu H, Wang R, Zhou Y, Zhang L, Wang C, Wang Q. JNK1 Induces Notch1 Expression to Regulate Genes Governing Photoreceptor Production. Cells. 2019; 8(9):970. https://doi.org/10.3390/cells8090970
Chicago/Turabian StylePan, Mingyu, Haiyang Hu, Rui Wang, Yi Zhou, Lele Zhang, Chen Wang, and Quanyi Wang. 2019. "JNK1 Induces Notch1 Expression to Regulate Genes Governing Photoreceptor Production" Cells 8, no. 9: 970. https://doi.org/10.3390/cells8090970
APA StylePan, M., Hu, H., Wang, R., Zhou, Y., Zhang, L., Wang, C., & Wang, Q. (2019). JNK1 Induces Notch1 Expression to Regulate Genes Governing Photoreceptor Production. Cells, 8(9), 970. https://doi.org/10.3390/cells8090970




