Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics
Abstract
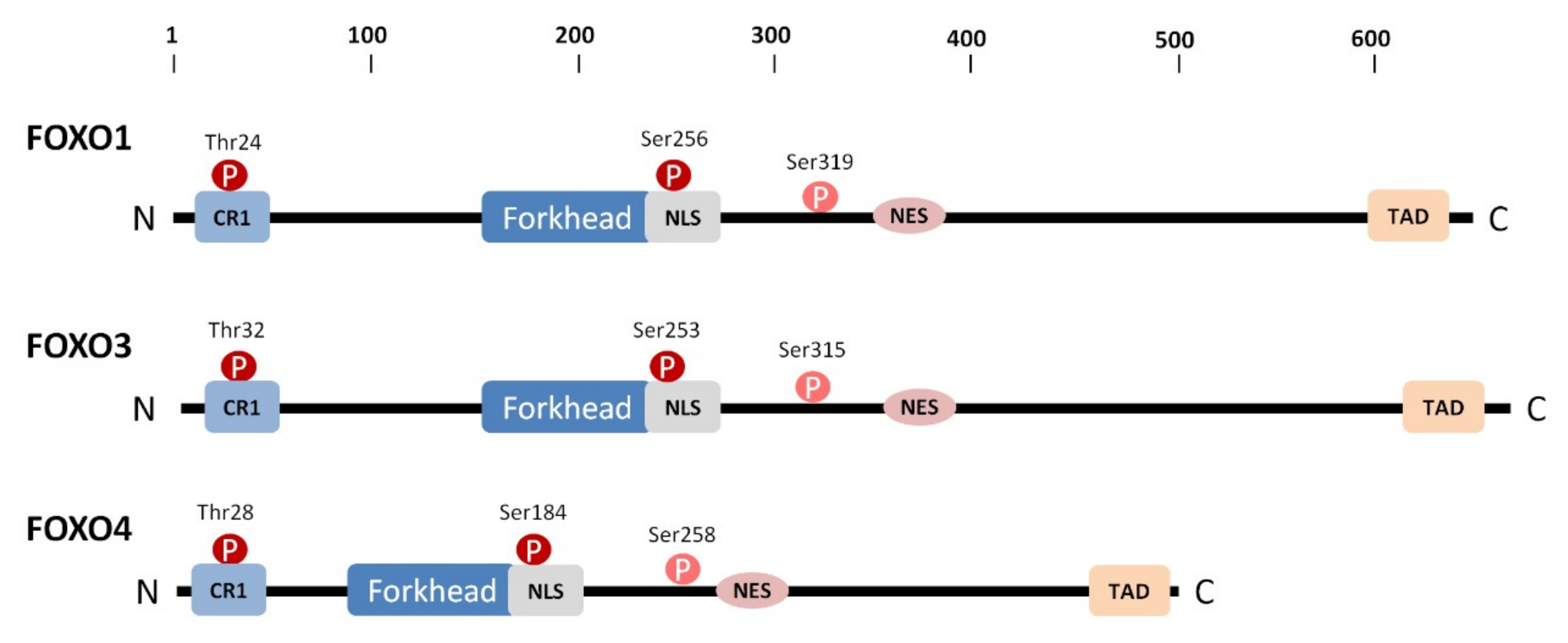
1. Introduction
2. Materials and Methods
2.1. FOXO1-DBD Expression and Purification
2.2. NMR Spectroscopy
2.3. NMR Structure Calculation
3. Results and Discussion
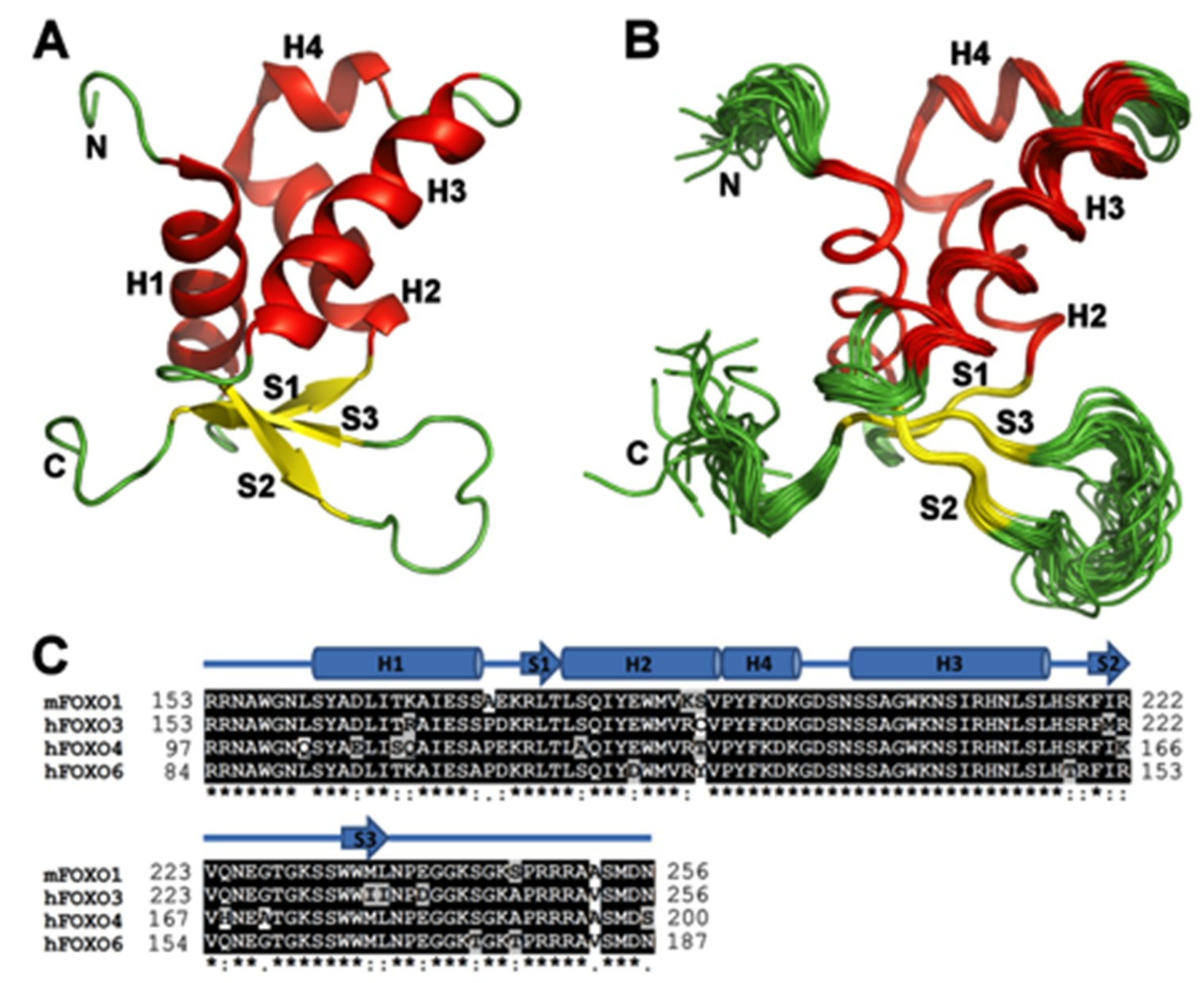
3.1. Solution Structure of the DNA-binding Domain of FOXO1
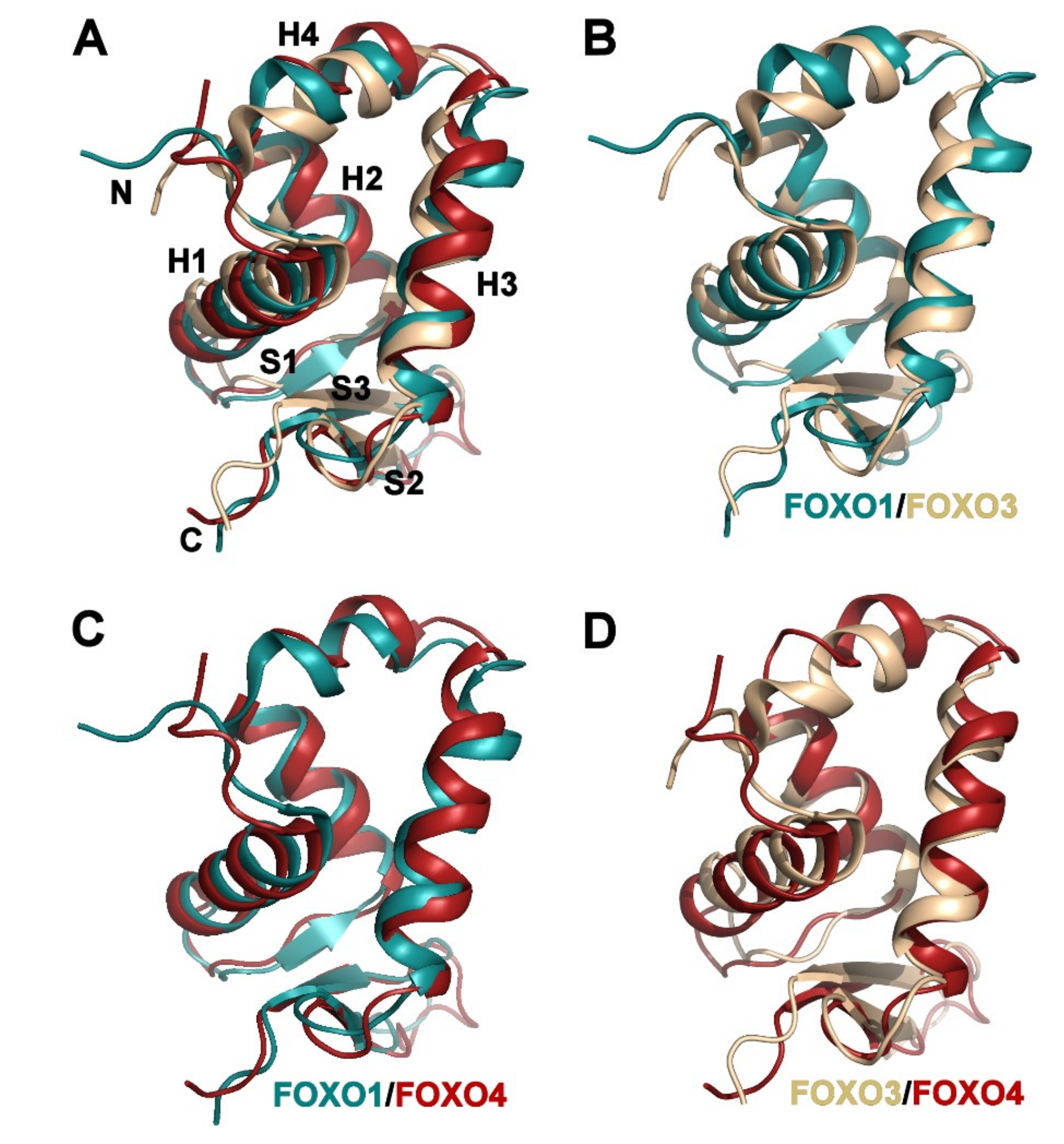
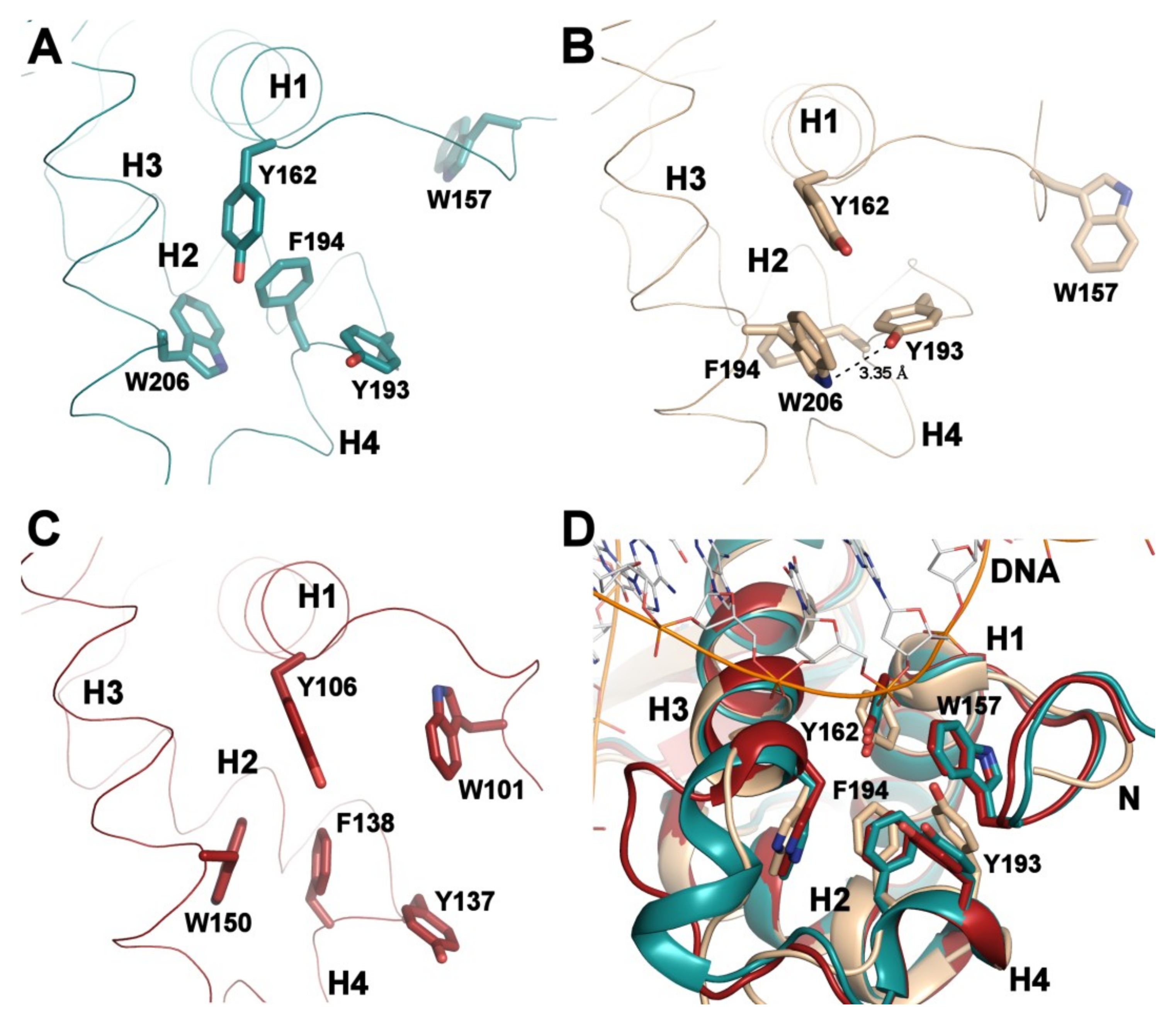
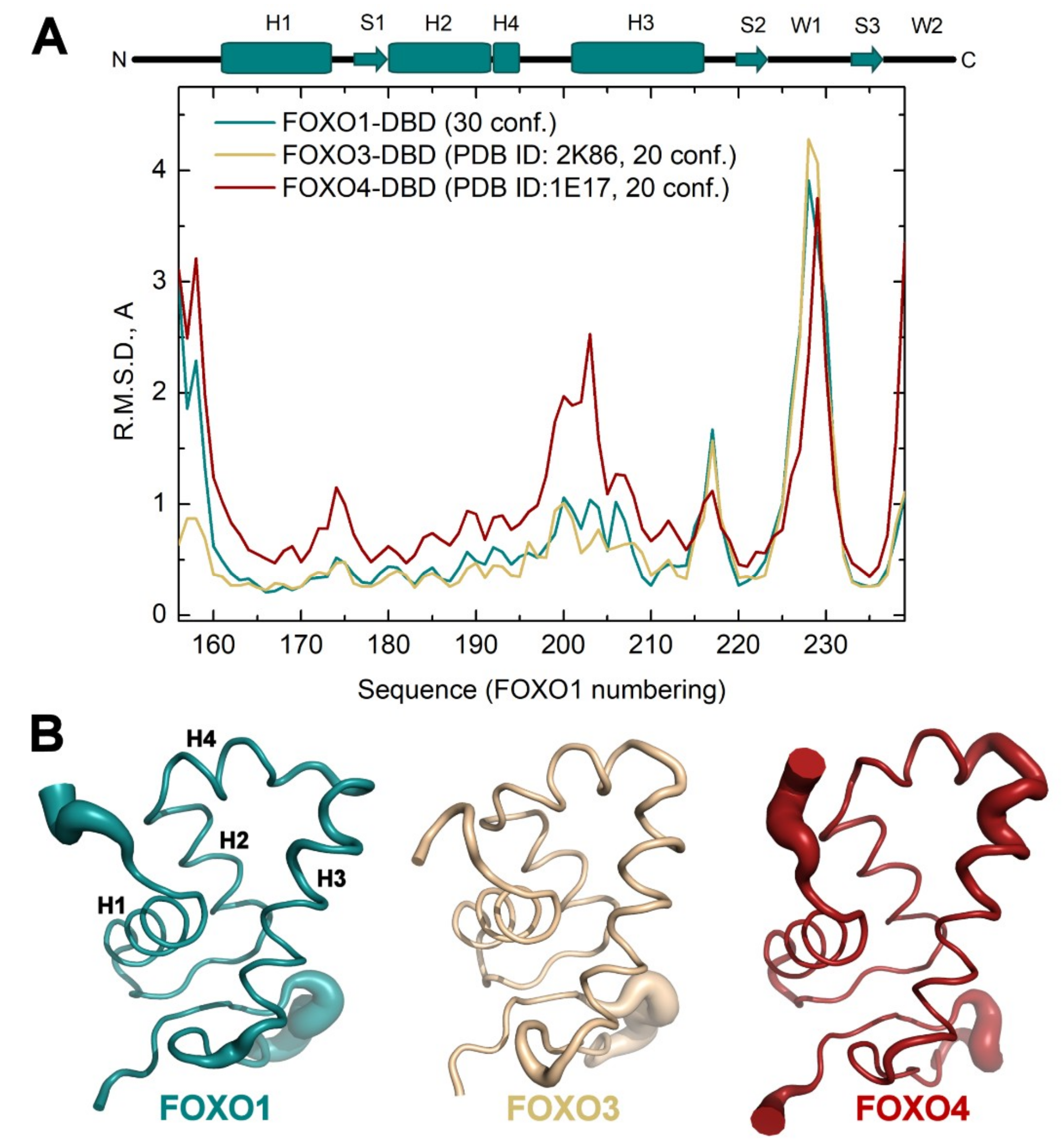
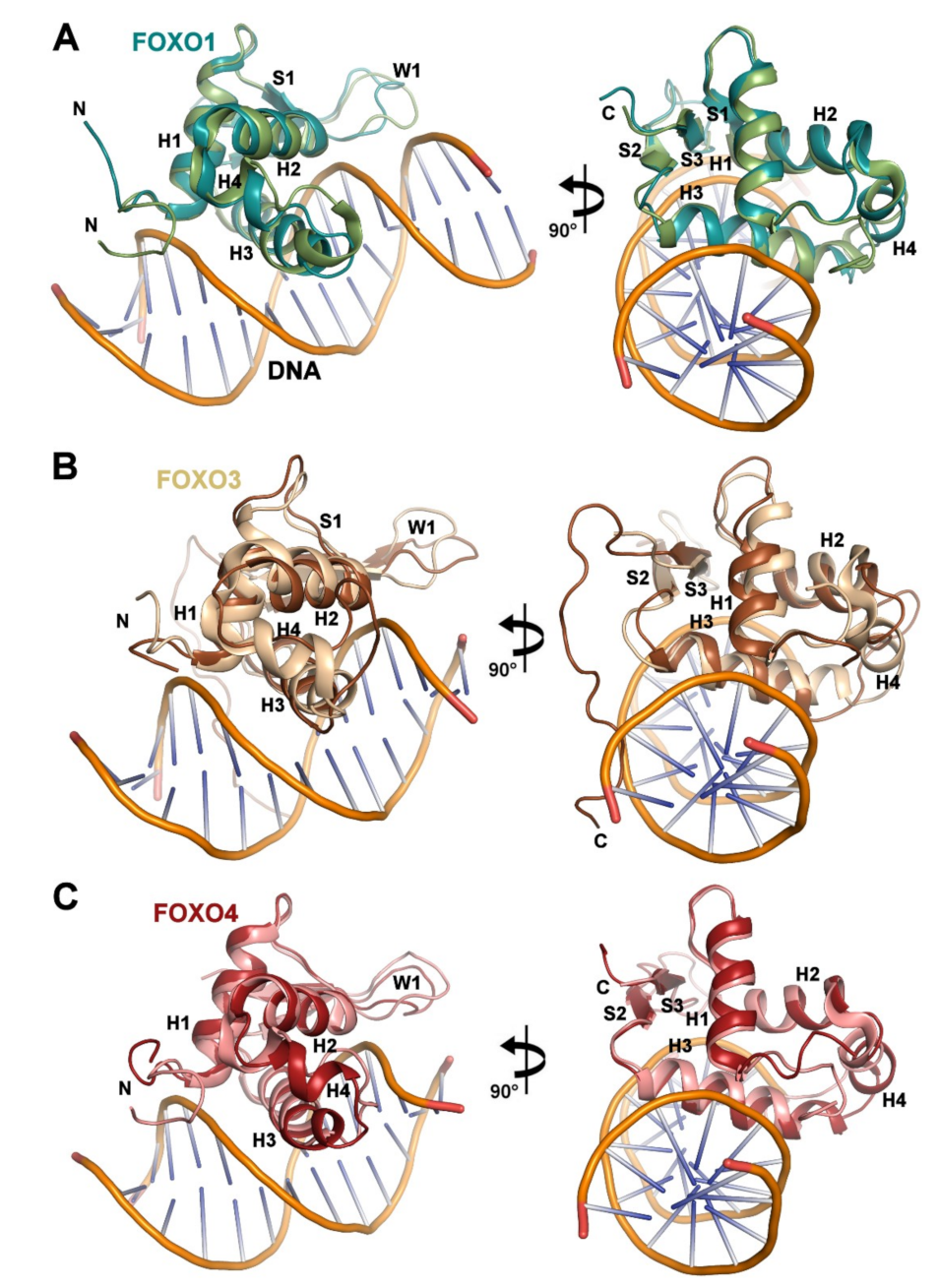
3.2. Comparison of FOXO1-DBD with Forkhead Domains of Other FOXO Proteins
3.3. Structural Differences Between Apo and DNA-bound Forms of FOXO1-DBD
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weigel, D.; Jackle, H. The Fork Head Domain—A Novel DNA-Binding Motif of Eukaryotic Transcription Factors. Cell 1990, 63, 455–456. [Google Scholar] [CrossRef]
- Kaestner, K.H.; Knochel, W.; Martinez, D.E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000, 14, 142–146. [Google Scholar] [PubMed]
- Clark, K.L.; Halay, E.D.; Lai, E.; Burley, S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 1993, 364, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.L.; Huang, C.Y.; Chang, C.H.; Sun, Y.J.; Chuang, W.J.; Hsiao, C.D. Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. J. Biol. Chem. 2006, 281, 17400–17409. [Google Scholar] [CrossRef]
- Stroud, J.C.; Wu, Y.; Bates, D.L.; Han, A.; Nowick, K.; Paabo, S.; Tong, H.; Chen, L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure 2006, 14, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.L.; Sun, Y.J.; Huang, C.Y.; Yang, J.Y.; Hung, M.C.; Hsiao, C.D. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007, 35, 6984–6994. [Google Scholar] [CrossRef] [PubMed]
- Brent, M.M.; Anand, R.; Marmorstein, R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 2008, 16, 1407–1416. [Google Scholar] [CrossRef]
- Boura, E.; Rezabkova, L.; Brynda, J.; Obsilova, V.; Obsil, T. Structure of the human FOXO4-DBD-DNA complex at 1.9 A resolution reveals new details of FOXO binding to the DNA. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Furuyama, T.; Nakazawa, T.; Nakano, I.; Mori, N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 2000, 349, 629–634. [Google Scholar] [CrossRef]
- Biggs, W.H., 3rd; Cavenee, W.K.; Arden, K.C. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm. Genome. 2001, 12, 416–425. [Google Scholar] [CrossRef]
- O’Brien, R.M.; Granner, D.K. Regulation of gene expression by insulin. Physiol. Rev. 1996, 76, 1109–1161. [Google Scholar] [CrossRef]
- Streeper, R.S.; Svitek, C.A.; Chapman, S.; Greenbaum, L.E.; Taub, R.; O’Brien, R.M. A multicomponent insulin response sequence mediates a strong repression of mouse glucose-6-phosphatase gene transcription by insulin. J. Biol. Chem. 1997, 272, 11698–11701. [Google Scholar] [CrossRef]
- Overdier, D.G.; Porcella, A.; Costa, R.H. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell Biol. 1994, 14, 2755–2766. [Google Scholar] [CrossRef]
- Kaufmann, E.; Muller, D.; Knochel, W. DNA recognition site analysis of Xenopus winged helix proteins. J. Mol. Biol. 1995, 248, 239–254. [Google Scholar] [CrossRef]
- Hornsveld, M.; Dansen, T.B.; Derksen, P.W.; Burgering, B.M.T. Re-evaluating the role of FOXOs in cancer. Semin Cancer Biol. 2018, 50, 90–100. [Google Scholar] [CrossRef]
- Ma, J.; Matkar, S.; He, X.; Hua, X. FOXO family in regulating cancer and metabolism. Semin Cancer Biol. 2018, 50, 32–41. [Google Scholar] [CrossRef]
- Tia, N.; Singh, A.K.; Pandey, P.; Azad, C.S.; Chaudhary, P.; Gambhir, I.S. Role of Forkhead Box O (FOXO) transcription factor in aging and diseases. Gene 2018, 648, 97–105. [Google Scholar] [CrossRef]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Kops, G.J.; de Ruiter, N.D.; De Vries-Smits, A.M.; Powell, D.R.; Bos, J.L.; Burgering, B.M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 1999, 398, 630–634. [Google Scholar] [CrossRef]
- Brunet, A.; Kanai, F.; Stehn, J.; Xu, J.; Sarbassova, D.; Frangioni, J.V.; Dalal, S.N.; DeCaprio, J.A.; Greenberg, M.E.; Yaffe, M.B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002, 156, 817–828. [Google Scholar] [CrossRef]
- Obsilova, V.; Vecer, J.; Herman, P.; Pabianova, A.; Sulc, M.; Teisinger, J.; Boura, E.; Obsil, T. 14-3-3 Protein interacts with nuclear localization sequence of forkhead transcription factor FoxO4. Biochemistry 2005, 44, 11608–11617. [Google Scholar] [CrossRef]
- Silhan, J.; Vacha, P.; Strnadova, P.; Vecer, J.; Herman, P.; Sulc, M.; Teisinger, J.; Obsilova, V.; Obsil, T. 14-3-3 protein masks the DNA binding interface of forkhead transcription factor FOXO4. J. Biol. Chem. 2009, 284, 19349–19360. [Google Scholar] [CrossRef]
- So, C.W.; Cleary, M.L. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol. Cell Biol. 2002, 22, 6542–6552. [Google Scholar] [CrossRef]
- Greer, E.L.; Brunet, A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 2005, 24, 7410–7425. [Google Scholar] [CrossRef]
- Vogt, P.K.; Jiang, H.; Aoki, M. Triple layer control: Phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle 2005, 4, 908–913. [Google Scholar] [CrossRef]
- van der Horst, A.; Burgering, B.M.T. Stressing the role of FoxO proteins in lifespan and disease. Nature Reviews Molecular Cell Biol. 2007, 8, 440–450. [Google Scholar] [CrossRef]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef]
- van der Horst, A.; de Vries-Smits, A.M.; Brenkman, A.B.; van Triest, M.H.; van den Broek, N.; Colland, F.; Maurice, M.M.; Burgering, B.M. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006, 8, 1064–1073. [Google Scholar] [CrossRef]
- Calnan, D.R.; Brunet, A. The FoxO code. Oncogene 2008, 27, 2276–2288. [Google Scholar] [CrossRef]
- Webb, A.E.; Kundaje, A.; Brunet, A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell 2016, 15, 673–685. [Google Scholar] [CrossRef]
- Hui, R.C.; Francis, R.E.; Guest, S.K.; Costa, J.R.; Gomes, A.R.; Myatt, S.S.; Brosens, J.J.; Lam, E.W. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol. Cancer Ther. 2008, 7, 670–678. [Google Scholar] [CrossRef]
- Han, C.Y.; Cho, K.B.; Choi, H.S.; Han, H.K.; Kang, K.W. Role of FoxO1 activation in MDR1 expression in adriamycin-resistant breast cancer cells. Carcinogenesis 2008, 29, 1837–1844. [Google Scholar] [CrossRef]
- Salcher, S.; Hagenbuchner, J.; Geiger, K.; Seiter, M.A.; Rainer, J.; Kofler, R.; Hermann, M.; Kiechl-Kohlendorfer, U.; Ausserlechner, M.J.; Obexer, P. C10ORF10/DEPP, a transcriptional target of FOXO3, regulates ROS-sensitivity in human neuroblastoma. Mol. Cancer. 2014, 13, 224. [Google Scholar] [CrossRef]
- Karadedou, C.T.; Gomes, A.R.; Chen, J.; Petkovic, M.; Ho, K.K.; Zwolinska, A.K.; Feltes, A.; Wong, S.Y.; Chan, K.Y.; Cheung, Y.N.; et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene 2012, 31, 1845–1858. [Google Scholar] [CrossRef]
- Yao, S.; Fan, L.Y.; Lam, E.W. The FOXO3-FOXM1 axis: A key cancer drug target and a modulator of cancer drug resistance. Semin Cancer Biol. 2018, 50, 77–89. [Google Scholar] [CrossRef]
- Pajvani, U.B.; Accili, D. The new biology of diabetes. Diabetologia 2015, 58, 2459–2468. [Google Scholar] [CrossRef]
- Langlet, F.; Haeusler, R.A.; Linden, D.; Ericson, E.; Norris, T.; Johansson, A.; Cook, J.R.; Aizawa, K.; Wang, L.; Buettner, C.; et al. Selective Inhibition of FOXO1 Activator/Repressor Balance Modulates Hepatic Glucose Handling. Cell 2017, 171, 824–835 e818. [Google Scholar] [CrossRef]
- Lee, W.; Tonelli, M.; Markley, J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 2015, 31, 1325–1327. [Google Scholar] [CrossRef]
- Renshaw, P.S.; Veverka, V.; Kelly, G.; Frenkiel, T.A.; Williamson, R.A.; Gordon, S.V.; Hewinson, R.G.; Carr, M.D. Sequence-specific assignment and secondary structure determination of the 195-residue complex formed by the Mycobacterium tuberculosis proteins CFP-10 and ESAT-6. J. Biomol. NMR 2004, 30, 225–226. [Google Scholar] [CrossRef]
- Veverka, V.; Lennie, G.; Crabbe, T.; Bird, I.; Taylor, R.J.; Carr, M.D. NMR assignment of the mTOR domain responsible for rapamycin binding. J. Biomol. NMR 2006, 36 (Suppl. 1), 3. [Google Scholar] [CrossRef]
- Herrmann, T.; Guntert, P.; Wuthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002, 319, 209–227. [Google Scholar] [CrossRef]
- Guntert, P.; Wuthrich, K. Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J. Biomol. NMR 1991, 1, 447–456. [Google Scholar] [CrossRef]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef]
- Harjes, E.; Harjes, S.; Wohlgemuth, S.; Muller, K.H.; Krieger, E.; Herrmann, C.; Bayer, P. GTP-Ras disrupts the intramolecular complex of C1 and RA domains of Nore1. Structure 2006, 14, 881–888. [Google Scholar] [CrossRef]
- Weigelt, J.; Climent, I.; Dahlman-Wright, K.; Wikstrom, M. Solution structure of the DNA binding domain of the human forkhead transcription factor AFX (FOXO4). Biochemistry 2001, 40, 5861–5869. [Google Scholar] [CrossRef]
- Wang, F.; Marshall, C.B.; Yamamoto, K.; Li, G.Y.; Plevin, M.J.; You, H.; Mak, T.W.; Ikura, M. Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53. J. Mol. Biol. 2008, 384, 590–603. [Google Scholar] [CrossRef]
- Kelley, L.A.; Gardner, S.P.; Sutcliffe, M.J. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng 1996, 9, 1063–1065. [Google Scholar] [CrossRef]
- Lee, B.; Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971, 55, 379–400. [Google Scholar] [CrossRef]
- Pierrou, S.; Hellqvist, M.; Samuelsson, L.; Enerback, S.; Carlsson, P. Cloning and characterization of seven human forkhead proteins: Binding site specificity and DNA bending. Embo J. 1994, 13, 5002–5012. [Google Scholar] [CrossRef]
- Martin, A.J.; Walsh, I.; Tosatto, S.C. MOBI: A web server to define and visualize structural mobility in NMR protein ensembles. Bioinformatics 2010, 26, 2916–2917. [Google Scholar] [CrossRef]
- Baar, M.P.; Brandt, R.M.C.; Putavet, D.A.; Klein, J.D.D.; Derks, K.W.J.; Bourgeois, B.R.M.; Stryeck, S.; Rijksen, Y.; van Willigenburg, H.; Feijtel, D.A.; et al. Targeted Apoptosis of Senescent Cells Restores Tissue Homeostasis in Response to Chemotoxicity and Aging. Cell 2017, 169, 132–147 e116. [Google Scholar] [CrossRef]
- Vacha, P.; Zuskova, I.; Bumba, L.; Herman, P.; Vecer, J.; Obsilova, V.; Obsil, T. Detailed kinetic analysis of the interaction between the FOXO4-DNA-binding domain and DNA. Biophys. Chem. 2013, 184C, 68–78. [Google Scholar] [CrossRef]
- Golson, M.L.; Kaestner, K.H. Fox transcription factors: From development to disease. Development 2016, 143, 4558–4570. [Google Scholar] [CrossRef]
- Benayoun, B.A.; Caburet, S.; Veitia, R.A. Forkhead transcription factors: Key players in health and disease. Trends Genet. 2011, 27, 224–232. [Google Scholar] [CrossRef]
- Jacobs, F.M.; van der Heide, L.P.; Wijchers, P.J.; Burbach, J.P.; Hoekman, M.F.; Smidt, M.P. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 2003, 278, 35959–35967. [Google Scholar] [CrossRef]
- Xing, Y.Q.; Li, A.; Yang, Y.; Li, X.X.; Zhang, L.N.; Guo, H.C. The regulation of FOXO1 and its role in disease progression. Life Sci. 2018, 193, 124–131. [Google Scholar] [CrossRef]
- Lu, H.; Huang, H. FOXO1: A potential target for human diseases. Current drug targets 2011, 12, 1235–1244. [Google Scholar] [CrossRef]
- Nakae, J.; Biggs, W.H., 3rd; Kitamura, T.; Cavenee, W.K.; Wright, C.V.; Arden, K.C.; Accili, D. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 2002, 32, 245–253. [Google Scholar] [CrossRef]
- Matsumoto, M.; Han, S.; Kitamura, T.; Accili, D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J. Clin. Invest. 2006, 116, 2464–2472. [Google Scholar] [CrossRef]
- Matsumoto, M.; Pocai, A.; Rossetti, L.; Depinho, R.A.; Accili, D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007, 6, 208–216. [Google Scholar] [CrossRef]
- Cook, J.R.; Matsumoto, M.; Banks, A.S.; Kitamura, T.; Tsuchiya, K.; Accili, D. A mutant allele encoding DNA binding-deficient FoxO1 differentially regulates hepatic glucose and lipid metabolism. Diabetes 2015, 64, 1951–1965. [Google Scholar] [CrossRef]
| Non-Redundant Distance and Angle Constrains | |
|---|---|
| Total number of NOE constraints | 1712 |
| Short-range NOEs | |
| Intra-residue (i = j) | 445 |
| Sequential (|i − j| = 1) | 402 |
| Medium-range NOEs (1 < | i − j | < 5) | 325 |
| Long-range NOEs (|i − j| ≥ 5) | 530 |
| Torsion angles | 128 |
| Hydrogen bond restrains | - |
| Total number of restricting constraints | 1840 |
| Total restricting constraints per restrained residue | 16.3 |
| Residual constraint violations | |
| Distance violations per structure | |
| 0.1–0.2 Å | 7.44 |
| 0.2–0.5 Å | 2.46 |
| > 0.5 Å | 0 |
| Root mean square (r.m.s.) of distance violation per constraint | 0.02 Å |
| Maximum distance violation | 0.48 Å |
| Dihedral angle viol. per structure | |
| 1–10° | 3.12 |
| > 10° | 0 |
| r.m.s. of dihedral violations per constraint | 0.55° |
| Maximum dihedral angle viol. | 5.0° |
| Ramachandran plot summary | |
| Most favored regions | 98.3% |
| Additionally allowed regions | 1.7% |
| Generously allowed regions | 0.0% |
| Disallowed regions | 0.0% |
| r.m.s.d. to the mean structure | all/ordered 1 |
| All backbone atoms | 10.0/0.4 Å |
| All heavy atoms | 10.1/0.8 Å |
| PDB entry | 6QVW |
| BMRB accession code | 34364 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psenakova, K.; Kohoutova, K.; Obsilova, V.; Ausserlechner, M.J.; Veverka, V.; Obsil, T. Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics. Cells 2019, 8, 966. https://doi.org/10.3390/cells8090966
Psenakova K, Kohoutova K, Obsilova V, Ausserlechner MJ, Veverka V, Obsil T. Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics. Cells. 2019; 8(9):966. https://doi.org/10.3390/cells8090966
Chicago/Turabian StylePsenakova, Katarina, Klara Kohoutova, Veronika Obsilova, Michael J. Ausserlechner, Vaclav Veverka, and Tomas Obsil. 2019. "Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics" Cells 8, no. 9: 966. https://doi.org/10.3390/cells8090966
APA StylePsenakova, K., Kohoutova, K., Obsilova, V., Ausserlechner, M. J., Veverka, V., & Obsil, T. (2019). Forkhead Domains of FOXO Transcription Factors Differ in both Overall Conformation and Dynamics. Cells, 8(9), 966. https://doi.org/10.3390/cells8090966








