Down-Regulation of Phosphoribosyl Pyrophosphate Synthetase 1 Inhibits Neuroblastoma Cell Proliferation
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Data Analysis
2.2. Cell Culture and Treatment
2.3. Vector Construction, Transfection, and Infection
2.4. Cell Proliferation Analysis
2.5. Bromodeoxyuridine (BrdU) Staining
2.6. Cell Cycle Analysis
2.7. Western Blot Analysis
2.8. Quantitative RT-PCR
2.9. Soft Agar Clonogenic Assay
2.10. Tumor Xenograft Assays
2.11. Histology and Immunohistochemistry
2.12. Statistical Analyses
3. Results
3.1. PRPS1 is Associated with Poor Neuroblastoma Patient Progress and is Commonly Expressed in Neuroblastoma Cells
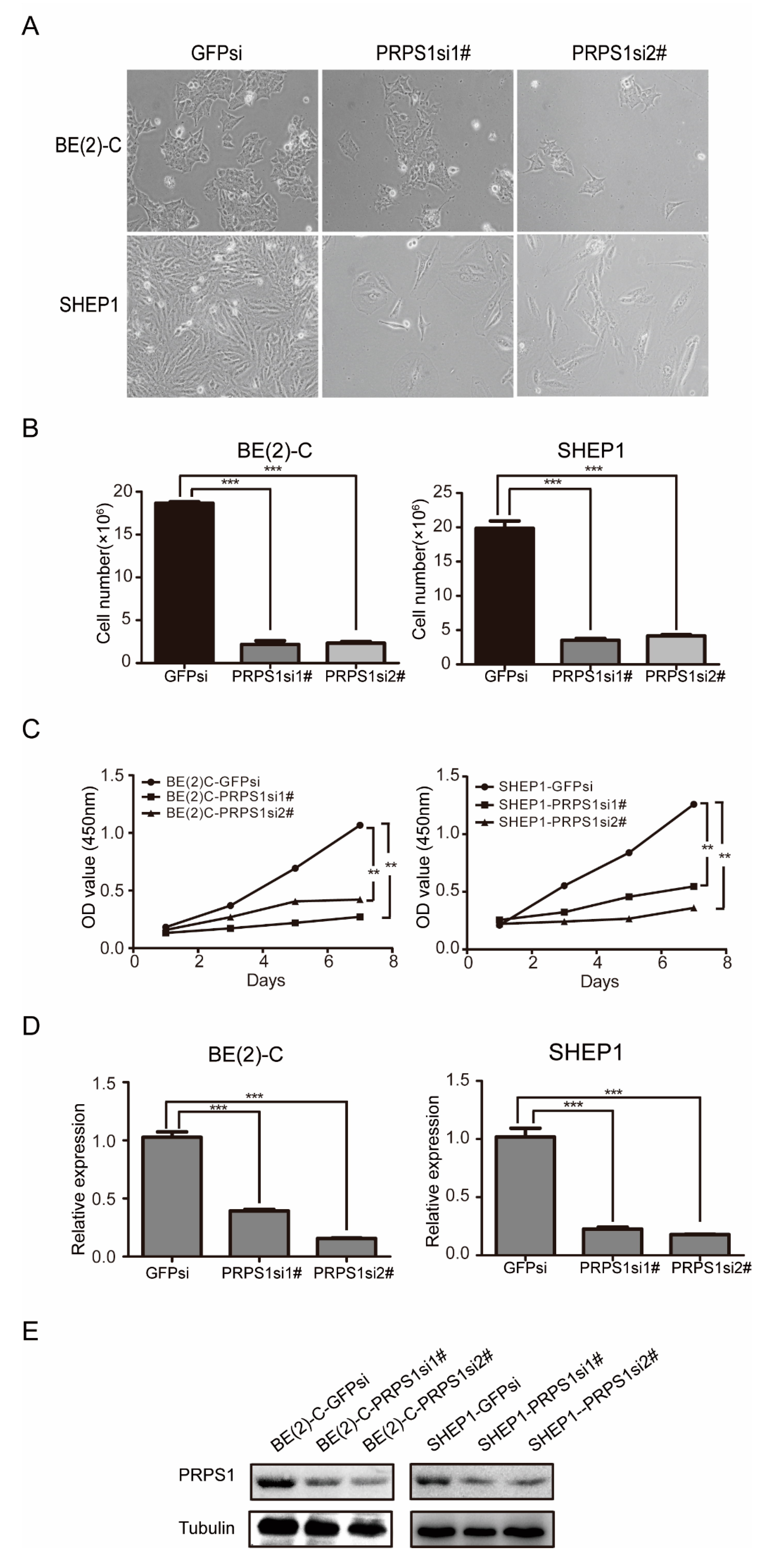
3.2. Down-Regulated PRPS1 Inhibits Proliferation of Neuroblastoma Cells
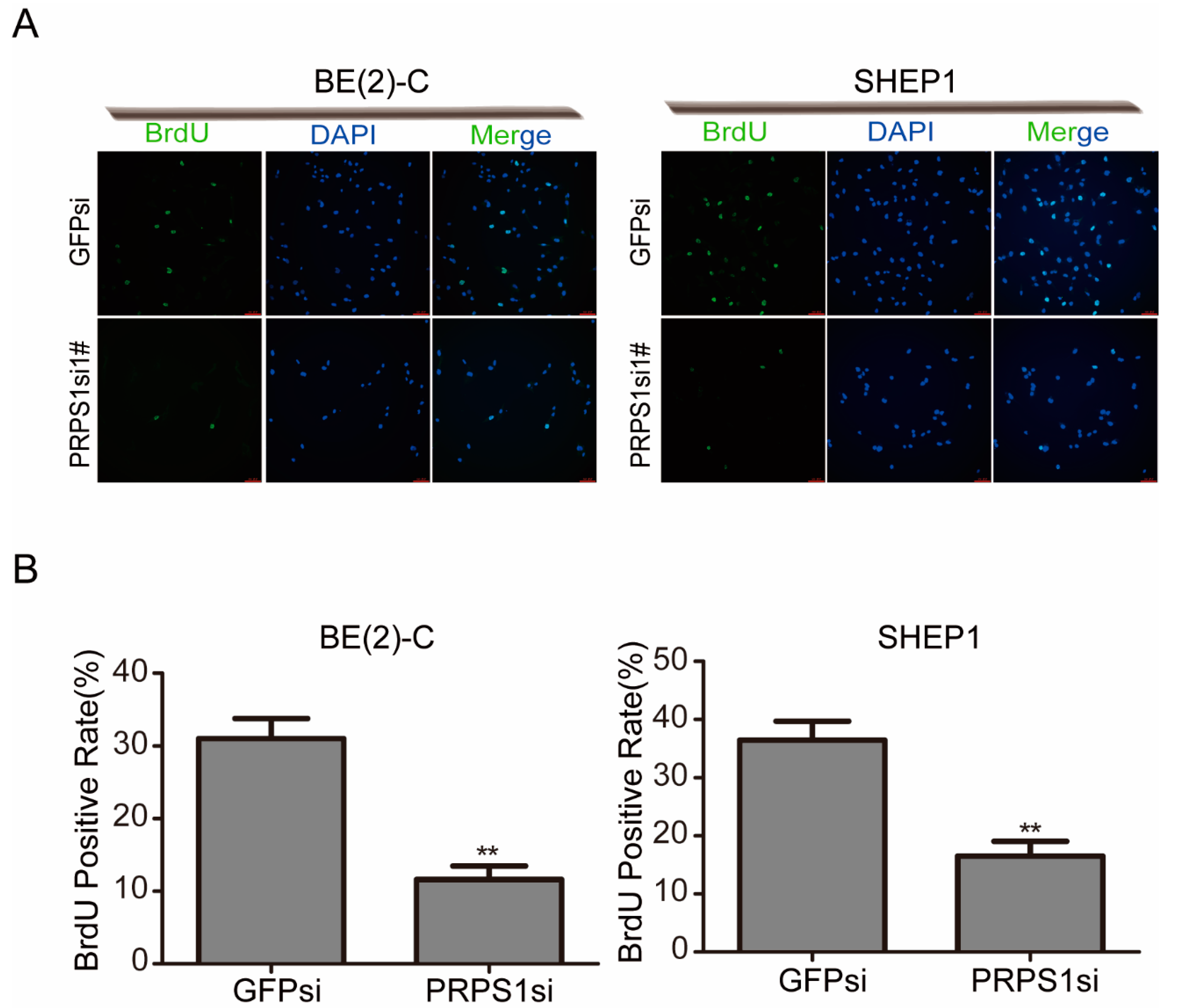
3.3. Down-Regulated PRPS1 Disturbs DNA Synthesis in Neuroblastoma Cells
3.4. PRPS1 Inhibition Decreases Tumorigenicity of Neuroblastoma Cells In Vitro and In Vivo
3.5. PRPS1 Expression Is Consistent with MYCN Amplification in Neuroblastoma Patient
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gerardi, C.; Banzi, R.; Bertele, V.; Garattini, S. Clinical research on rare diseases of children: Neuroblastoma. Cancer Chemother. Pharmacol. 2017, 79, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Tolbert, V.P.; Matthay, K.K. Neuroblastoma: Clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018, 372, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Kaczowka, P.; Wieczorek, A.; Czogala, M.; Ksiazek, T.; Szewczyk, K.; Balwierz, W. The role of N-Myc gene amplification in neuroblastoma childhood tumour-single-centre experience. Contemp. Oncol. 2018, 22, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Parmar, R.; Wadia, F.; Yassa, R.; Zenios, M. Neuroblastoma: A rare cause of a limping child. How to avoid a delayed diagnosis? J. Pediatr. Orthop. 2013. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M. Spontaneous regression of neuroblastoma. Cell Tissue Res. 2018, 372, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Gershan, J.A.; Barr, K.M.; Weber, J.J.; Jing, W.; Johnson, B.D. Immune modulating effects of cyclophosphamide and treatment with tumor lysate/CpG synergize to eliminate murine neuroblastoma. J. Immunother. Cancer 2015. [Google Scholar] [CrossRef]
- Dauphin, G.; de Araujo, P.C.; Forget, P.; Leroy, P.; Rausin, L.; Demarche, M. Atypical clinical presentation of a neuroblastoma in an infant. Rev. Med. Liege 2013, 68, 56–60. [Google Scholar]
- Nakagawara, A.; Li, Y.; Izumi, H.; Muramori, K.; Inada, H.; Nishi, M. Neuroblastoma. Jpn. J. Clin. Oncol. 2018, 48, 214–241. [Google Scholar] [CrossRef]
- Ackermann, S.; Cartolano, M.; Hero, B.; Welte, A.; Kahlert, Y.; Roderwieser, A.; Bartenhagen, C.; Walter, E.; Gecht, J.; Kerschke, L.; et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018, 362, 1165–1170. [Google Scholar] [CrossRef]
- Newman, E.A.; Abdessalam, S.; Aldrink, J.H.; Austin, M.; Heaton, T.E.; Bruny, J.; Ehrlich, P.; Dasgupta, R.; Baertschiger, R.M.; Lautz, T.B.; et al. Update on neuroblastoma. J. Pediatr. Surg. 2019, 54, 383–389. [Google Scholar] [CrossRef]
- Hove-Jensen, B.; Andersen, K.R.; Kilstrup, M.; Martinussen, J.; Switzer, R.L.; Willemoes, M. Phosphoribosyl Diphosphate (PRPP): Biosynthesis, Enzymology, Utilization, and Metabolic Significance. Microbiol. Mol. Biol. Rev. 2017. [Google Scholar] [CrossRef]
- Roessler, B.J.; Nosal, J.M.; Smith, P.R.; Heidler, S.A.; Palella, T.D.; Switzer, R.L.; Becker, M.A. Human X-linked phosphoribosylpyrophosphate synthetase superactivity is associated with distinct point mutations in the PRPS1 gene. J. Biol. Chem. 1993, 268, 26476–26481. [Google Scholar]
- Ahmed, M.; Taylor, W.; Smith, P.R.; Becker, M.A. Accelerated transcription of PRPS1 in X-linked overactivity of normal human phosphoribosylpyrophosphate synthetase. J. Biol. Chem. 1999, 274, 7482–7488. [Google Scholar] [CrossRef]
- Becker, M.A. Phosphoribosylpyrophosphate synthetase and the regulation of phosphoribosylpyrophosphate production in human cells. Prog. Nucleic Acid Res. Mol. Biol. 2001, 69, 115–148. [Google Scholar]
- Mittal, R.; Patel, K.; Mittal, J.; Chan, B.; Yan, D.; Grati, M.; Liu, X.Z. Association of PRPS1 Mutations with Disease Phenotypes. Dis. Markers 2015. [Google Scholar] [CrossRef]
- Liu, X.Z.; Xie, D.; Yuan, H.J.; de Brouwer, A.P.; Christodoulou, J.; Yan, D. Hearing loss and PRPS1 mutations: Wide spectrum of phenotypes and potential therapy. Int. J. Audiol. 2013, 52, 23–28. [Google Scholar] [CrossRef][Green Version]
- Synofzik, M.; Muller vom Hagen, J.; Haack, T.B.; Wilhelm, C.; Lindig, T.; Beck-Wodl, S.; Nabuurs, S.B.; van Kuilenburg, A.B.; de Brouwer, A.P.; Schols, L. X-linked Charcot-Marie-Tooth disease, Arts syndrome, and prelingual non-syndromic deafness form a disease continuum: Evidence from a family with a novel PRPS1 mutation. Orphanet J. Rare Dis. 2014. [Google Scholar] [CrossRef]
- De Brouwer, A.P.; Williams, K.L.; Duley, J.A.; van Kuilenburg, A.B.; Nabuurs, S.B.; Egmont-Petersen, M.; Lugtenberg, D.; Zoetekouw, L.; Banning, M.J.; Roeffen, M.; et al. Arts syndrome is caused by loss-of-function mutations in PRPS1. Am. J. Hum. Genet. 2007, 81, 507–518. [Google Scholar] [CrossRef]
- De Brouwer, A.P.; van Bokhoven, H.; Nabuurs, S.B.; Arts, W.F.; Christodoulou, J.; Duley, J. PRPS1 mutations: Four distinct syndromes and potential treatment. Am. J. Hum. Genet. 2010, 86, 506–518. [Google Scholar] [CrossRef]
- Al-Maawali, A.; Dupuis, L.; Blaser, S.; Heon, E.; Tarnopolsky, M.; Al-Murshedi, F.; Marshall, C.R.; Paton, T.; Scherer, S.W.; Consortium, F.C.; et al. Prenatal growth restriction, retinal dystrophy, diabetes insipidus and white matter disease: Expanding the spectrum of PRPS1-related disorders. Eur. J. Hum. Genet. 2015, 23, 310–316. [Google Scholar] [CrossRef]
- Liu, X.; Han, D.; Li, J.; Han, B.; Ouyang, X.; Cheng, J.; Li, X.; Jin, Z.; Wang, Y.; Bitner-Glindzicz, M.; et al. Loss-of-function mutations in the PRPS1 gene cause a type of nonsyndromic X-linked sensorineural deafness, DFN2. Am. J. Hum. Genet. 2010, 86, 65–71. [Google Scholar] [CrossRef]
- Li, C.; Yan, Z.; Cao, X.; Zhang, X.; Yang, L. Phosphoribosylpyrophosphate Synthetase 1 Knockdown Suppresses Tumor Formation of Glioma CD133+ Cells Through Upregulating Cell Apoptosis. J. Mol. Neurosci. 2016, 60, 145–156. [Google Scholar] [CrossRef]
- Mullighan, C.G. Mutant PRPS1: A new therapeutic target in relapsed acute lymphoblastic leukemia. Nat. Med. 2015, 21, 553–554. [Google Scholar] [CrossRef]
- Ma, Y.; An, X.; Guan, X.; Kong, Q.; Wang, Y.; Li, P.; Meng, Y.; Cui, Y.; Wen, X.; Guo, Y.; et al. High expression of PRPS1 induces an anti-apoptotic effect in B-ALL cell lines and predicts an adverse prognosis in Chinese children with B-ALL. Oncol. Lett. 2018, 15, 4314–4322. [Google Scholar] [CrossRef]
- Qiu, Z.; Guo, W.; Wang, Q.; Chen, Z.; Huang, S.; Zhao, F.; Yao, M.; Zhao, Y.; He, X. MicroRNA-124 reduces the pentose phosphate pathway and proliferation by targeting PRPS1 and RPIA mRNAs in human colorectal cancer cells. Gastroenterology 2015, 149, 1587–1598. [Google Scholar] [CrossRef]
- Li, X.; Qian, X.; Peng, L.X.; Jiang, Y.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Lee, J.H.; Cote, G.; Wang, H.; et al. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat. Cell Biol. 2016, 18, 561–571. [Google Scholar] [CrossRef]
- Yang, L.; Yan, Z.; Wang, Y.; Ma, W.; Li, C. Down-expression of miR-154 suppresses tumourigenesis in CD133(+) glioblastoma stem cells. Cell Biochem. Funct. 2016, 34, 404–413. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Chamaa, F.; Assi, S.; Chalhoub, R.M.; Abou-Antoun, T.; Abou-Kheir, W. Cancer Stem Cells in Neuroblastoma: Expanding the Therapeutic Frontier. Front. Mol. Neurosci. 2019. [Google Scholar] [CrossRef]
- Ross, R.A.; Spengler, B.A.; Domenech, C.; Porubcin, M.; Rettig, W.J.; Biedler, J.L. Human neuroblastoma I-type cells are malignant neural crest stem cells. Cell Growth Differ. 1995, 6, 449–456. [Google Scholar]
- Zhou, Y.; Dai, R.; Mao, L.; Xia, Y.; Yao, Y.; Yang, X.; Hu, B. Activation of Sonic hedgehog signaling pathway in S-type neuroblastoma cell lines. J. Huazhong Univ. Sci. Technol. Med. Sci. 2010, 30, 271–277. [Google Scholar] [CrossRef]
- Mao, L.; Xia, Y.P.; Zhou, Y.N.; Dai, R.L.; Yang, X.; Duan, S.J.; Qiao, X.; Mei, Y.W.; Hu, B.; Cui, H. A critical role of Sonic Hedgehog signaling in maintaining the tumorigenicity of neuroblastoma cells. Cancer Sci. 2009, 100, 1848–1855. [Google Scholar] [CrossRef]
- Jing, X.; Wang, X.J.; Zhang, T.; Zhu, W.; Fang, Y.; Wu, H.; Liu, X.; Ma, D.; Ji, X.; Jiang, Y.; et al. Cell cycle-dependent phosphorylation of PRPS1 fuels nucleotide synthesis and promotes tumorigenesis. Cancer Res. 2019. [Google Scholar] [CrossRef]
- Cui, H.; Li, T.; Ding, H.F. Linking of N-Myc to death receptor machinery in neuroblastoma cells. J. Biol. Chem. 2005, 280, 9474–9481. [Google Scholar] [CrossRef]
- Cui, H.J.; Hu, B.; Li, T.; Ma, J.; Alam, G.; Gunning, W.T.; Ding, H.F. Bmi-1 is essential for the tumorigenicity of neuroblastoma cells. Am. J. Pathol. 2007, 170, 1370–1378. [Google Scholar] [CrossRef]
- Rubie, H.; Hartmann, O.; Michon, J.; Frappaz, D.; Coze, C.; Chastagner, P.; Baranzelli, M.C.; Plantaz, D.; Avet-Loiseau, H.; Benard, J.; et al. N-Myc gene amplification is a major prognostic factor in localized neuroblastoma: Results of the French NBL 90 study. Neuroblastoma Study Group of the Societe Francaise d’Oncologie Pediatrique. J. Clin. Oncol. 1997, 15, 1171–1182. [Google Scholar] [CrossRef]
- Schwab, M. Amplification of N-myc as a prognostic marker for patients with neuroblastoma. Semin. Cancer Biol. 1993, 4, 13–18. [Google Scholar]
- Meitar, D.; Crawford, S.E.; Rademaker, A.W.; Cohn, S.L. Tumor angiogenesis correlates with metastatic disease, N-myc amplification, and poor outcome in human neuroblastoma. J. Clin. Oncol. 1996, 14, 405–414. [Google Scholar] [CrossRef]
- Pession, A.; Trere, D.; Perri, P.; Rondelli, R.; Montanaro, L.; Mantovani, W.; Derenzini, M.; Paolucci, G. N-myc amplification and cell proliferation rate in human neuroblastoma. J. Pathol. 1997, 183, 339–344. [Google Scholar] [CrossRef]
- Galderisi, U.; Di Bernardo, G.; Cipollaro, M.; Peluso, G.; Cascino, A.; Cotrufo, R.; Melone, M.A. Differentiation and apoptosis of neuroblastoma cells: Role of N-myc gene product. J. Cell Biochem. 1999, 73, 97–105. [Google Scholar] [CrossRef]
- Wang, X.; Yang, K.; Xie, Q.; Wu, Q.; Mack, S.C.; Shi, Y.; Kim, L.J.Y.; Prager, B.C.; Flavahan, W.A.; Liu, X.; et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci. 2017, 20, 661–673. [Google Scholar] [CrossRef]
- Sonoda, T.; Taira, M.; Ishijima, S.; Ishizuka, T.; Iizasa, T.; Tatibana, M. Complete nucleotide sequence of human phosphoribosyl pyrophosphate synthetase subunit I (PRS I) cDNA and a comparison with human and rat PRPS gene families. J. Biochem. 1991, 109, 361–364. [Google Scholar]
- Lei, B.; Wan, B.; Peng, J.; Yang, Y.; Lv, D.; Zhou, X.; Shu, F.; Li, F.; Zhong, L.; Wu, H.; et al. PRPS2 Expression Correlates with Sertoli-Cell Only Syndrome and Inhibits the Apoptosis of TM4 Sertoli Cells. J. Urol. 2015, 194, 1491–1497. [Google Scholar] [CrossRef]
- Xue, C.; Yu, D.M.; Gherardi, S.; Koach, J.; Milazzo, G.; Gamble, L.; Liu, B.; Valli, E.; Russell, A.J.; London, W.B.; et al. MYCN promotes neuroblastoma malignancy by establishing a regulatory circuit with transcription factor AP4. Oncotarget 2016, 7, 54937–54951. [Google Scholar] [CrossRef][Green Version]
- Mannava, S.; Grachtchouk, V.; Wheeler, L.J.; Im, M.; Zhuang, D.; Slavina, E.G.; Mathews, C.K.; Shewach, D.S.; Nikiforov, M.A. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 2008, 7, 2392–2400. [Google Scholar] [CrossRef]
- Cunningham, J.T.; Moreno, M.V.; Lodi, A.; Ronen, S.M.; Ruggero, D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell 2014, 157, 1088–1103. [Google Scholar] [CrossRef]
- Barr, E.K.; Applebaum, M.A. Genetic Predisposition to Neuroblastoma. Children 2018, 5, 119. [Google Scholar] [CrossRef]
- Fornalewicz, K.; Wieczorek, A.; Wegrzyn, G.; Lyzen, R. Silencing of the pentose phosphate pathway genes influences DNA replication in human fibroblasts. Gene 2017, 635, 33–38. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, H.; Zhu, Y.; Liu, S.; Yuan, Z.; Wu, J.; Wen, L. Tannic Acid Induces the Mitochondrial Pathway of Apoptosis and S Phase Arrest in Porcine Intestinal IPEC-J2 Cells. Toxins 2019, 11, 397. [Google Scholar] [CrossRef]
- He, M.; Chao, L.; You, Y.P. PRPS1 silencing reverses cisplatin resistance in human breast cancer cells. Biochem. Cell Biol. 2017, 95, 385–393. [Google Scholar] [CrossRef]
- Chassevent, A.; Benard, J. Oncogen N-myc expression and measurement of DNA ploidy in neuroblastoma: A double staining flow cytometric analysis. Bull Cancer. 1997, 84, 29–34. [Google Scholar]
- Taira, M.; Iizasa, T.; Yamada, K.; Shimada, H.; Tatibana, M. Tissue-differential expression of two distinct genes for phosphoribosyl pyrophosphate synthetase and existence of the testis-specific transcript. Biochim. Biophys. Acta 1989, 1007, 203–208. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Ye, J.; Zhu, S.; Cui, H. Down-Regulation of Phosphoribosyl Pyrophosphate Synthetase 1 Inhibits Neuroblastoma Cell Proliferation. Cells 2019, 8, 955. https://doi.org/10.3390/cells8090955
Li J, Ye J, Zhu S, Cui H. Down-Regulation of Phosphoribosyl Pyrophosphate Synthetase 1 Inhibits Neuroblastoma Cell Proliferation. Cells. 2019; 8(9):955. https://doi.org/10.3390/cells8090955
Chicago/Turabian StyleLi, Jifu, Junhong Ye, Shunqin Zhu, and Hongjuan Cui. 2019. "Down-Regulation of Phosphoribosyl Pyrophosphate Synthetase 1 Inhibits Neuroblastoma Cell Proliferation" Cells 8, no. 9: 955. https://doi.org/10.3390/cells8090955
APA StyleLi, J., Ye, J., Zhu, S., & Cui, H. (2019). Down-Regulation of Phosphoribosyl Pyrophosphate Synthetase 1 Inhibits Neuroblastoma Cell Proliferation. Cells, 8(9), 955. https://doi.org/10.3390/cells8090955




