Phycocyanin Exerts Anti-Proliferative Effects through Down-Regulating TIRAP/NF-κB Activity in Human Non-Small Cell Lung Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell line and Culture Condition
2.2. siRNA Transfection Assay
2.3. Cell Survival Rate Assay
2.4. Cell Proliferation Assay
2.5. Cell Colony Formation Assay
2.6. Cell Cycle Assay
2.7. Cell Apoptosis Assay
2.8. RNA-Seq Analysis
2.9. Quantitative RT-PCR (qRT-PCR) Analysis
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
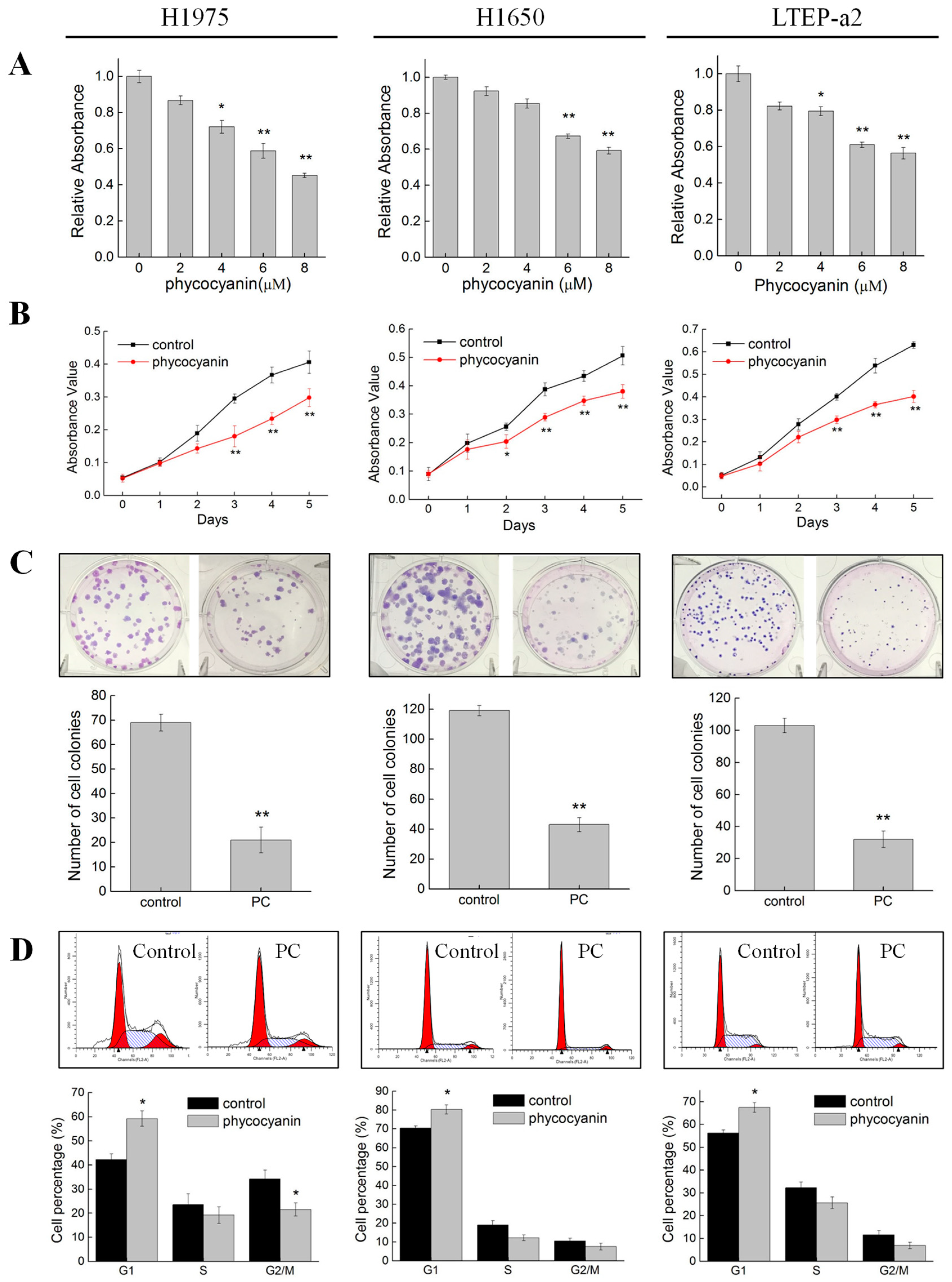
3.1. Phycocyanin Suppressed the Growth and Viability of Non-Small Cell Lung Cancer Cells
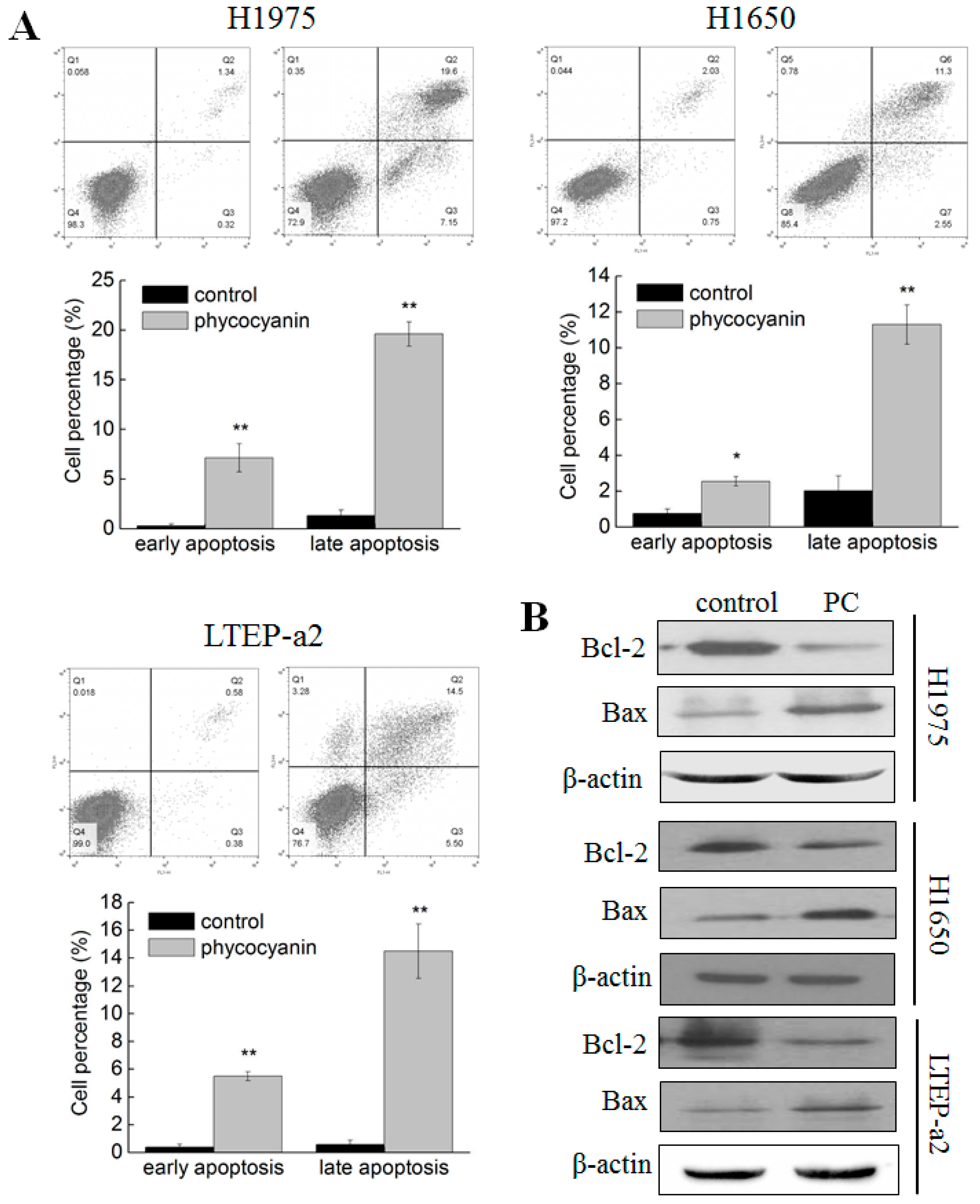
3.2. Phycocyanin Induced Apoptosis of Non-Small Cell Lung Cancer Cells
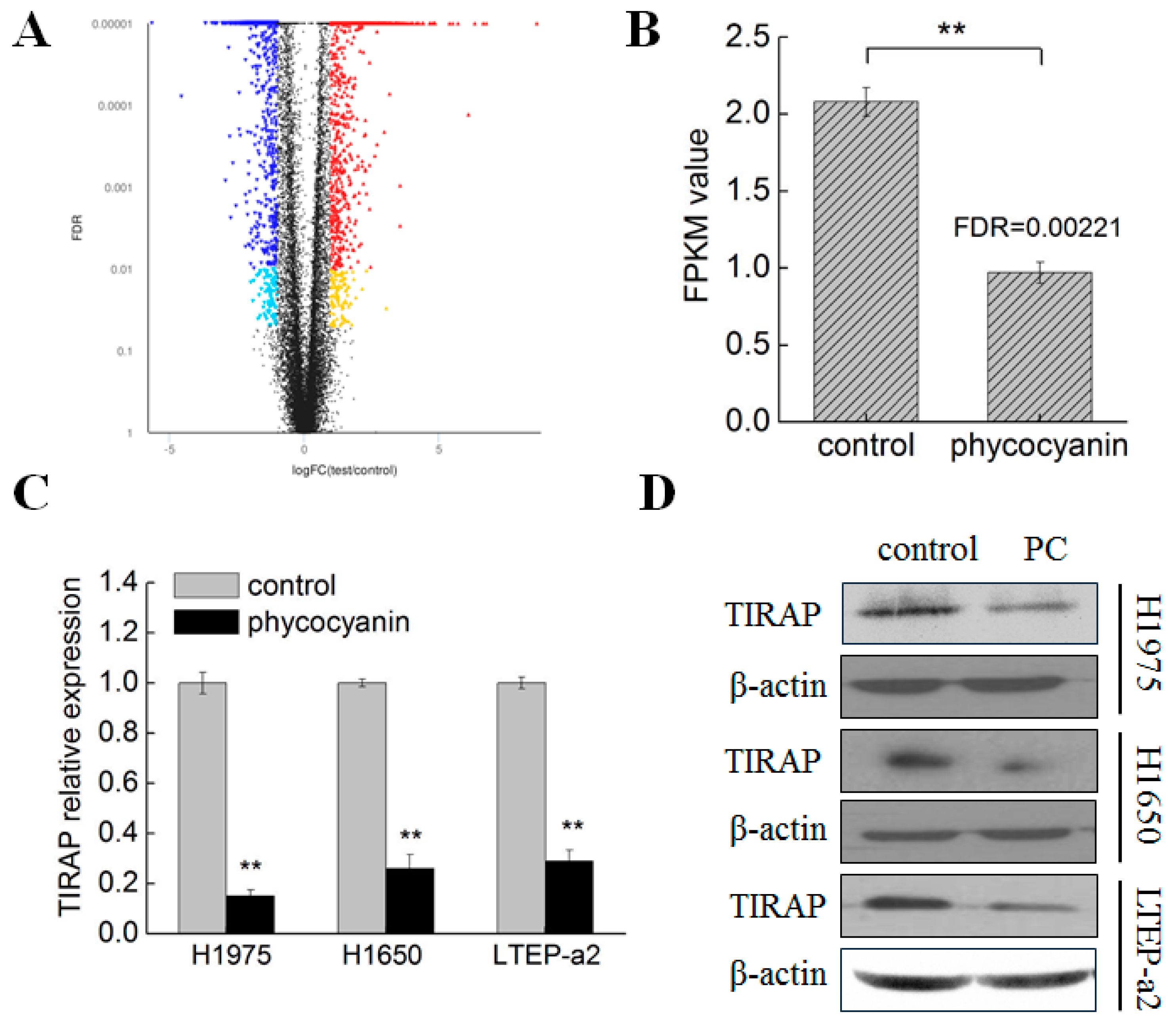
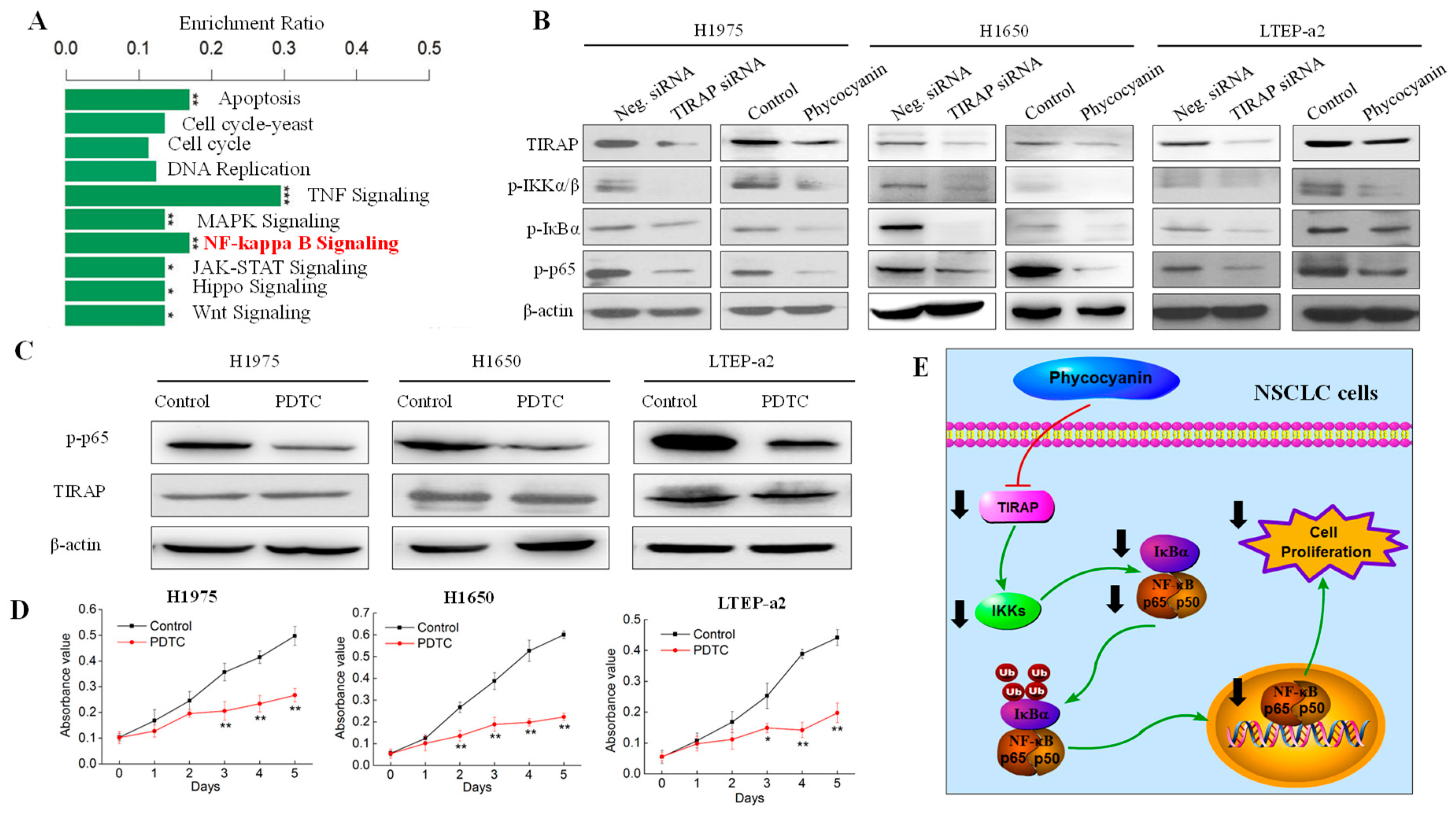
3.3. Transcriptome Analysis Suggested TIRAP Was Down-Regulated by Phycocyanin in Non-Small Cell Lung Cancer Cells
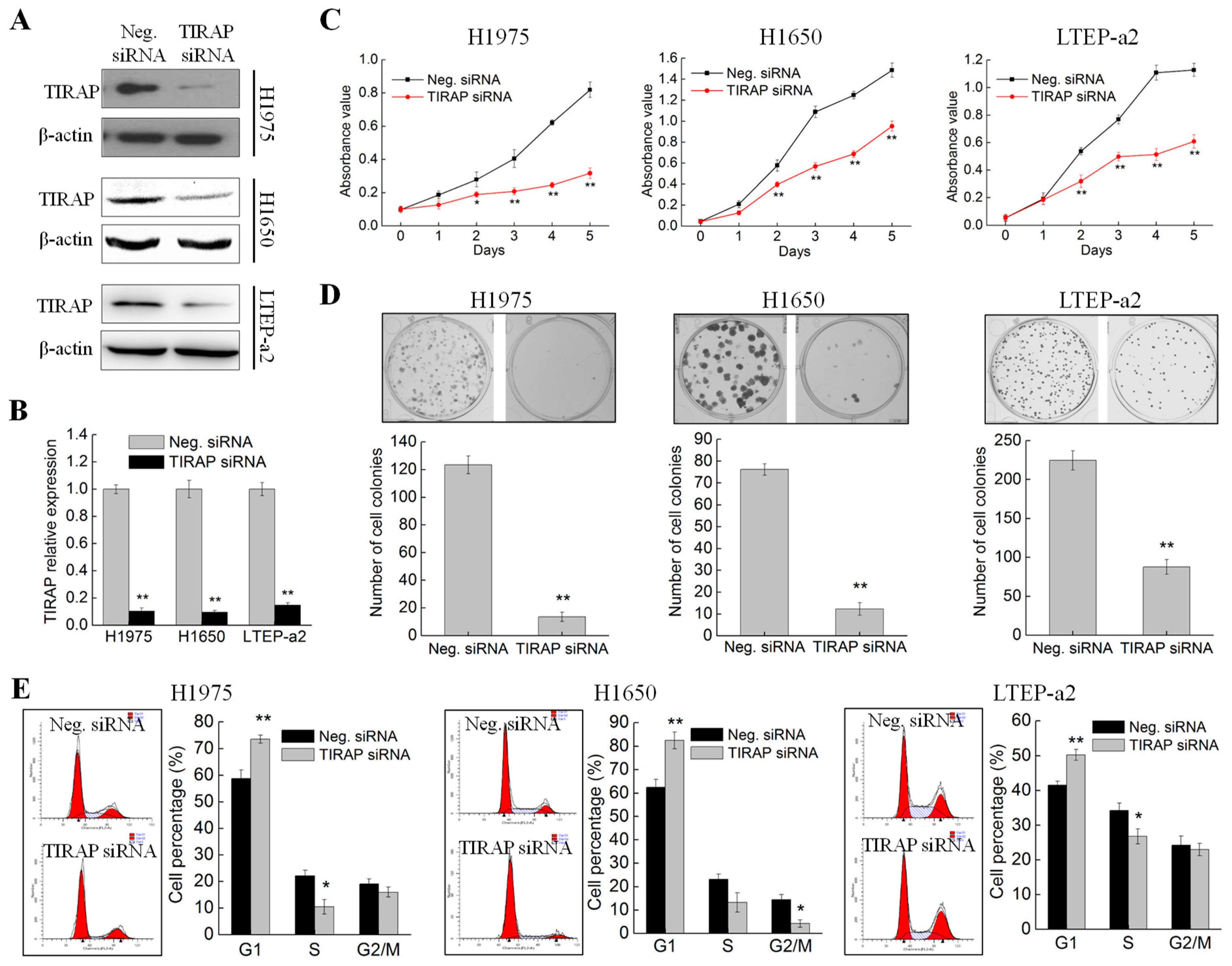
3.4. Knockdown of TIRAP Expression Suppressed Proliferation of Non-Small Cell Lung Cancer Cells
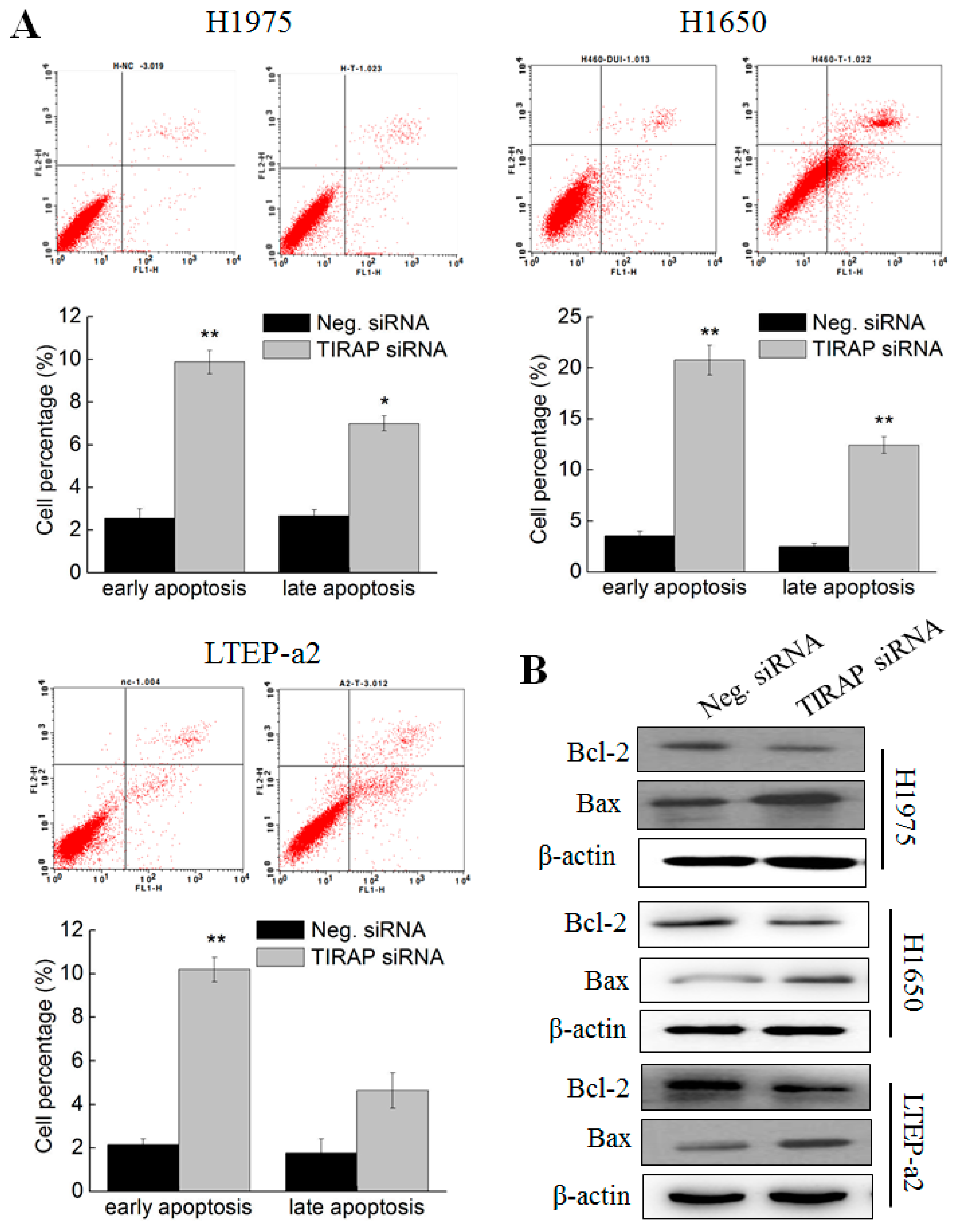
3.5. Knockdown of TIRAP Expression Induced Apoptosis of Non-Small Cell Lung Cancer Cells
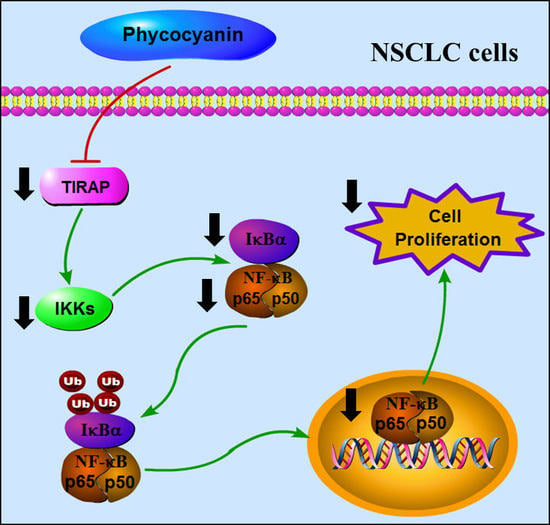
3.6. Phycocyanin Exerted Anti-Proliferative Effects through Down-Regulating TIRAP/NF-κB Activity in Non-Small Cell Lung Cancer Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Carbone, D.P.; Gandara, D.R.; Antonia, S.J.; Zielinski, C.; Paz-Ares, L. Non-Small-Cell Lung Cancer: Role of the Immune System and Potential for Immunotherapy. J. Thorac. Oncol. 2015, 10, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Wakelee, H.; Kelly, K.; Edelman, M.J. 50 Years of progress in the systemic therapy of non-small cell lung cancer. Am. Soc. Clin. Oncol. Educ. Book 2014, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Gettinger, S.; Lynch, T. A decade of advances in treatment for advanced non-small cell lung cancer. Clin. Chest Med. 2011, 32, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.R.; Kucuk, O.; Khuri, F.R.; Shin, D.M. Perspectives for cancer prevention with natural compounds. J. Clin. Oncol. 2009, 27, 2712–2725. [Google Scholar] [CrossRef]
- Jung, I.L. Soluble extract from Moringa oleifera leaves with a new anticancer activity. PLoS ONE 2014, 9, e95492. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, N.T. Production of phycocyanin--a pigment with applications in biology, biotechnology, foods and medicine. Appl. Microbiol. Biotechnol. 2008, 80, 1–14. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, Y.; Zhang, R.; Cai, T.; Cai, Y. Medical Application of Spirulina platensis Derived C-Phycocyanin. Evid. Based Complement Alternat. Med. 2016, 2016, 7803846. [Google Scholar]
- Gdara, N.B.; Belgacem, A.; Khemiri, I.; Mannai, S.; Bitri, L. Protective effects of phycocyanin on ischemia/reperfusion liver injuries. Biomed. Pharmacother. 2018, 102, 196–202. [Google Scholar] [CrossRef]
- Hao, S.; Li, S.; Wang, J.; Zhao, L.; Zhang, C.; Huang, W.; Wang, C. Phycocyanin Reduces Proliferation of Melanoma Cells through Downregulating GRB2/ERK Signaling. J. Agric. Food Chem. 2018, 66, 10921–10929. [Google Scholar] [CrossRef]
- Zhu, C.; Ling, Q.; Cai, Z.; Wang, Y.; Zhang, Y.; Hoffmann, P.R.; Zheng, W.; Zhou, T.; Huang, Z. Selenium-Containing Phycocyanin from Se-Enriched Spirulina platensis Reduces Inflammation in Dextran Sulfate Sodium-Induced Colitis by Inhibiting NF-kappaB Activation. J. Agric. Food Chem. 2016, 64, 5060–5070. [Google Scholar] [CrossRef] [PubMed]
- Cian, R.E.; Lopez-Posadas, R.; Drago, S.R.; de Medina, F.S.; Martinez-Augustin, O. Immunomodulatory properties of the protein fraction from Phorphyra columbina. J. Agric. Food Chem. 2012, 60, 8146–8154. [Google Scholar] [CrossRef] [PubMed]
- Romay, C.; Gonzalez, R.; Ledon, N.; Remirez, D.; Rimbau, V. C-phycocyanin: A biliprotein with antioxidant, anti-inflammatory and neuroprotective effects. Curr. Protein Pept. Sci. 2003, 4, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Fu, X.; Huang, W.; Li, C.; Wang, X.; Huang, B. Photodynamic effect and mechanism study of selenium-enriched phycocyanin from Spirulina platensis against liver tumours. J. Photochem. Photobiol. B 2018, 180, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, Y.; Liu, G.; Liu, H.; Zhu, F.; Ji, H.; Li, B. C-Phycocyanin exerts anti-cancer effects via the MAPK signaling pathway in MDA-MB-231 cells. Cancer Cell Int. 2018, 18, 12. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Lu, R.; Zhang, Y.; Zhu, M.; Zhu, W.; Yang, R.; Zhang, E.; Ying, J.; Xu, T.; Yi, H.; et al. Spirulina phycocyanin induces differential protein expression and apoptosis in SKOV-3 cells. Int. J. Biol. Macromol. 2015, 81, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Saini, M.K.; Sanyal, S.N. Targeting angiogenic pathway for chemoprevention of experimental colon cancer using C-phycocyanin as cyclooxygenase-2 inhibitor. Biochem. Cell Biol. 2014, 92, 206–218. [Google Scholar] [CrossRef]
- Baudelet, P.H.; Gagez, A.L.; Berard, J.B.; Juin, C.; Bridiau, N.; Kaas, R.; Thiery, V.; Cadoret, J.P.; Picot, L. Antiproliferative activity of Cyanophora paradoxa pigments in melanoma, breast and lung cancer cells. Mar. Drugs 2013, 11, 4390–4406. [Google Scholar] [CrossRef]
- Li, B.; Gao, M.H.; Chu, X.M.; Teng, L.; Lv, C.Y.; Yang, P.; Yin, Q.F. The synergistic antitumor effects of all-trans retinoic acid and C-phycocyanin on the lung cancer A549 cells in vitro and in vivo. Eur. J. Pharmacol. 2015, 749, 107–114. [Google Scholar] [CrossRef]
- Li, B.; Gao, M.H.; Lv, C.Y.; Yang, P.; Yin, Q.F. Study of the synergistic effects of all-transretinoic acid and C-phycocyanin on the growth and apoptosis of A549 cells. Eur. J. Cancer Prev. 2016, 25, 97–101. [Google Scholar] [CrossRef]
- Bingula, R.; Dupuis, C. Study of the Effects of Betaine and/or C-Phycocyanin on the Growth of Lung Cancer A549 Cells In Vitro and In Vivo. J. Oncol. 2016, 2016, 8162952. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Luo, C.; Abukiwan, A.; Wang, G.; He, J.; Huang, L.; Weber, C.E.; Lv, N.; Xiao, X.; Eichmuller, S.B. miR-137 inhibits proliferation of melanoma cells by targeting PAK2. Exp. Dermatol. 2015, 24, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Li, M.; Wang, R.; Xiao, Q.; Wang, J.; Li, M.; He, D.; Xiao, X. Knockdown of CABYR-a/b increases chemosensitivity of human non-small cell lung cancer cells through inactivation of Akt. Mol. Cancer Res. 2014, 12, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Krylov, D.M.; Wolf, Y.I.; Rogozin, I.B.; Koonin, E.V. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003, 13, 2229–2235. [Google Scholar] [CrossRef] [PubMed]
- Madamwar, D.; Patel, D.K.; Desai, S.N.; Upadhyay, K.K.; Devkar, R.V. Apoptotic potential of C-phycoerythrin from Phormidium sp. A27DM and Halomicronema sp. A32DM on human lung carcinoma cells. EXCLI J. 2015, 14, 527–539. [Google Scholar]
- Narayanan, K.B.; Park, H.H. Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis 2015, 20, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Salaun, B.; Romero, P.; Lebecque, S. Toll-like receptors’ two-edged sword: When immunity meets apoptosis. Eur. J. Immunol. 2007, 37, 3311–3318. [Google Scholar] [CrossRef]
- Fekonja, O.; Avbelj, M.; Jerala, R. Suppression of TLR signaling by targeting TIR domain-containing proteins. Curr. Protein Pept. Sci. 2012, 13, 776–788. [Google Scholar] [CrossRef]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Gao, P.; Gao, Y.J.; Liang, H.L. Effect of NF-kappa B inhibitor PDTC on VEGF and endostatin expression of mice with Lewis lung cancer. Asian Pac. J. Trop. Med. 2015, 8, 220–224. [Google Scholar] [CrossRef]
- Czerwonka, A.; Kalawaj, K.; Slawinska-Brych, A.; Lemieszek, M.K.; Bartnik, M.; Wojtanowski, K.K.; Zdzisinska, B.; Rzeski, W. Anticancer effect of the water extract of a commercial Spirulina (Arthrospira platensis) product on the human lung cancer A549 cell line. Biomed. Pharmacother. 2018, 106, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Deniz, I.; Ozen, M.O.; Yesil-Celiktas, O. Supercritical fluid extraction of phycocyanin and investigation of cytotoxicity on human lung cancer cells. J. Supercrit. Fluid 2016, 108, 13–18. [Google Scholar] [CrossRef]
- Thangam, R.; Suresh, V.; Asenath Princy, W.; Rajkumar, M.; Senthilkumar, N.; Gunasekaran, P.; Rengasamy, R.; Anbazhagan, C.; Kaveri, K.; Kannan, S. C-Phycocyanin from Oscillatoria tenuis exhibited an antioxidant and in vitro antiproliferative activity through induction of apoptosis and G0/G1 cell cycle arrest. Food Chem. 2013, 140, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; He, L.; Song, Z.; Chan, L.; He, J.; Huang, W.; Zhou, B.; Chen, T. Phycocyanin-based nanocarrier as a new nanoplatform for efficient overcoming of cancer drug resistance. J. Mater. Chem. B. 2017, 5, 3300–3314. [Google Scholar] [CrossRef]
- Senthilkumar, N.; Kurinjimalar, C.; Thangam, R.; Suresh, V.; Kavitha, G.; Gunasekaran, P.; Rengasamy, R. Further studies and biological activities of macromolecular protein R-Phycoerythrin from Portieria hornemannii. Int. J. Biol. Macromol. 2013, 62, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Madamwar, D.; Kaushal, A.; Patel, D.K.; Desai, S.N.; Upadhyay, K.; Devkar, R.V. Cyanobacterial phycoerythrin purified from marine Lyngbya sp induces apoptosis in lung carcinoma cells. Bioresour. Technol. 2015, 10, 770–778. [Google Scholar] [CrossRef]
- Sen, R.; Baltimore, D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986, 46, 705–716. [Google Scholar] [CrossRef]
- Li, C.; Yu, Y.; Li, W.; Liu, B.; Jiao, X.; Song, X.; Lv, C.; Qin, S. Phycocyanin attenuates pulmonary fibrosis via the TLR2-MyD88-NF-kappaB signaling pathway. Sci. Rep. 2017, 7, 5843. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Yan, Y.; Huang, W.; Gai, F.; Wang, J.; Liu, L.; Wang, C. C-phycocyanin reduces inflammation by inhibiting NF-kappa B activity through downregulating PDCD5 in lipopolysaccharide-induced RAW 264.7 macrophages. J. Funct. Foods 2018, 42, 21–29. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef]
- Liao, G.; Gao, B.; Gao, Y.; Yang, X.; Cheng, X.; Ou, Y. Phycocyanin Inhibits Tumorigenic Potential of Pancreatic Cancer Cells: Role of Apoptosis and Autophagy. Sci. Rep. 2016, 6, 34564. [Google Scholar] [CrossRef] [PubMed]
- Nishanth, R.P.; Ramakrishna, B.S.; Jyotsna, R.G.; Roy, K.R.; Reddy, G.V.; Reddy, P.K.; Reddanna, P. C-Phycocyanin inhibits MDR1 through reactive oxygen species and cyclooxygenase-2 mediated pathways in human hepatocellular carcinoma cell line. Eur. J. Pharmacol. 2010, 649, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Li, S.; Wang, J.; Zhao, L.; Yan, Y.; Cao, Q.; Wu, T.; Liu, L.; Wang, C. Transcriptome Analysis of Phycocyanin-Mediated Inhibitory Functions on Non-Small Cell Lung Cancer A549 Cell Growth. Mar Drugs 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Potts-Kant, E.N.; Garantziotis, S.; Foster, W.M.; Hollingsworth, J.W. Hyaluronan signaling during ozone-induced lung injury requires TLR4, MyD88, and TIRAP. PLoS ONE 2011, 6, e27137. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Tong, C.; Sun, Z.; Shen, Y.; Yao, C.; Jiang, J.; Yin, J.; Gao, L.; Song, Y.; Bai, C. Genetic variants in the TIRAP gene are associated with increased risk of sepsis-associated acute lung injury. BMC Med. Genet. 2010, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Jeyaseelan, S.; Young, S.K.; Yamamoto, M.; Arndt, P.G.; Akira, S.; Kolls, J.K.; Worthen, G.S. Toll/IL-1R domain-containing adaptor protein (TIRAP) is a critical mediator of antibacterial defense in the lung against Klebsiella pneumoniae but not Pseudomonas aeruginosa. J. Immunol. 2006, 177, 538–547. [Google Scholar] [CrossRef]
- Antosz, H.; Sajewicz, J.; Marzec-Kotarska, B.; Dmoszynska, A.; Baszak, J.; Jargiello-Baszak, M. Aberrant TIRAP and MyD88 expression in B-cell chronic lymphocytic leukemia. Blood Cells Mol. Dis. 2013, 51, 48–55. [Google Scholar] [CrossRef]
- Kennedy, C.L.; Najdovska, M.; Tye, H.; McLeod, L.; Yu, L.; Jarnicki, A.; Bhathal, P.S.; Putoczki, T.; Ernst, M.; Jenkins, B.J. Differential role of MyD88 and Mal/TIRAP in TLR2-mediated gastric tumourigenesis. Oncogene 2014, 33, 2540–2546. [Google Scholar] [CrossRef]
- Aviello, G.; Corr, S.C.; Johnston, D.G.; O’Neill, L.A.; Fallon, P.G. MyD88 adaptor-like (Mal) regulates intestinal homeostasis and colitis-associated colorectal cancer in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G769–G778. [Google Scholar] [CrossRef]
- Martinez-Montemayor, M.M.; Otero-Franqui, E.; Martinez, J.; De La Mota-Peynado, A.; Cubano, L.A.; Dharmawardhane, S. Individual and combined soy isoflavones exert differential effects on metastatic cancer progression. Clin. Exp. Metastasis 2010, 27, 465–480. [Google Scholar] [CrossRef][Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, S.; Li, S.; Wang, J.; Yan, Y.; Ai, X.; Zhang, J.; Ren, Y.; Wu, T.; Liu, L.; Wang, C. Phycocyanin Exerts Anti-Proliferative Effects through Down-Regulating TIRAP/NF-κB Activity in Human Non-Small Cell Lung Cancer Cells. Cells 2019, 8, 588. https://doi.org/10.3390/cells8060588
Hao S, Li S, Wang J, Yan Y, Ai X, Zhang J, Ren Y, Wu T, Liu L, Wang C. Phycocyanin Exerts Anti-Proliferative Effects through Down-Regulating TIRAP/NF-κB Activity in Human Non-Small Cell Lung Cancer Cells. Cells. 2019; 8(6):588. https://doi.org/10.3390/cells8060588
Chicago/Turabian StyleHao, Shuai, Shuang Li, Jing Wang, Yan Yan, Xin Ai, Jiawen Zhang, Yuqing Ren, Tingting Wu, Liyun Liu, and Chengtao Wang. 2019. "Phycocyanin Exerts Anti-Proliferative Effects through Down-Regulating TIRAP/NF-κB Activity in Human Non-Small Cell Lung Cancer Cells" Cells 8, no. 6: 588. https://doi.org/10.3390/cells8060588
APA StyleHao, S., Li, S., Wang, J., Yan, Y., Ai, X., Zhang, J., Ren, Y., Wu, T., Liu, L., & Wang, C. (2019). Phycocyanin Exerts Anti-Proliferative Effects through Down-Regulating TIRAP/NF-κB Activity in Human Non-Small Cell Lung Cancer Cells. Cells, 8(6), 588. https://doi.org/10.3390/cells8060588