Histone Modifications and the Maintenance of Telomere Integrity
Abstract
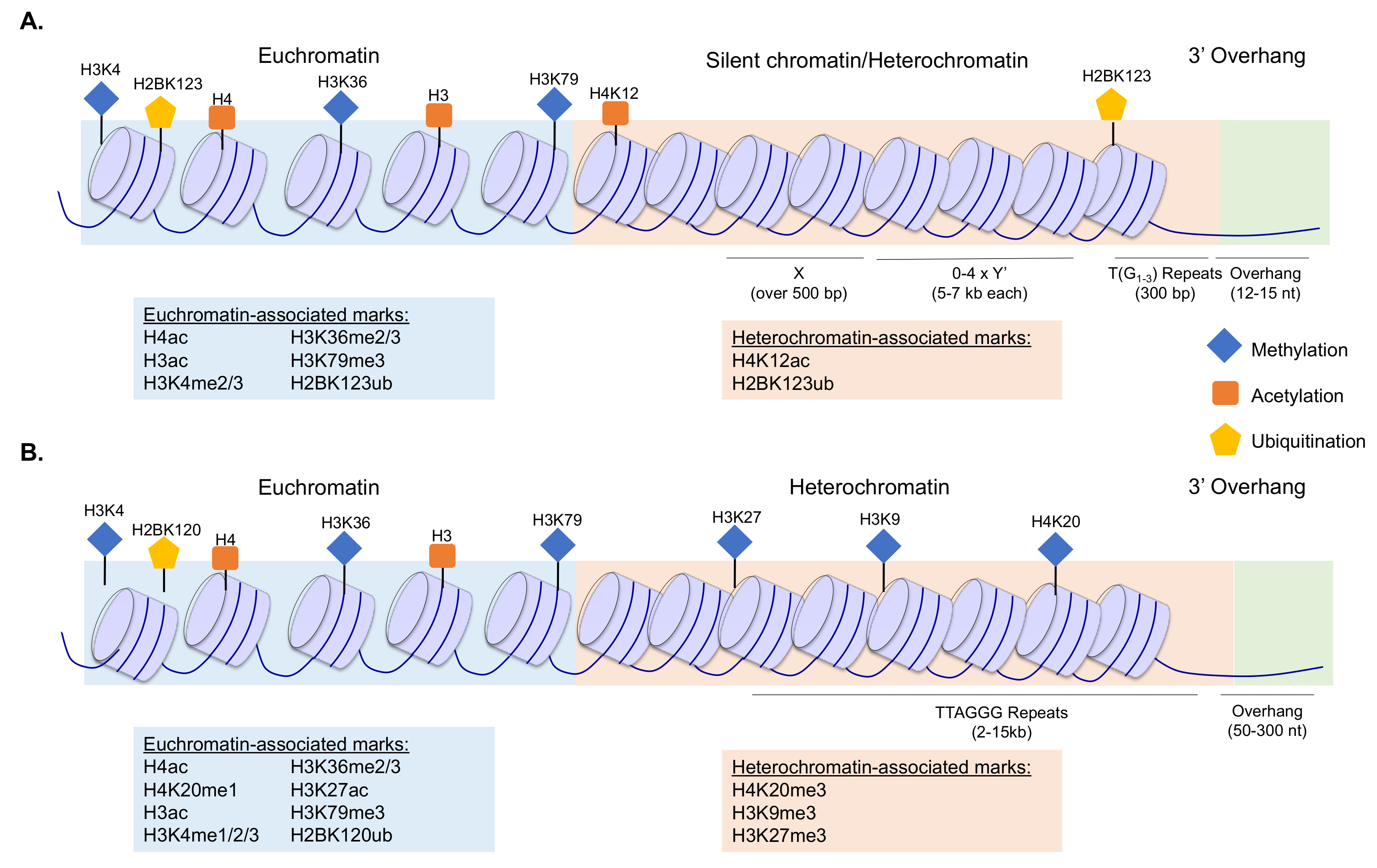
1. Introduction
2. Histone Modifications and Chromatin Dynamics
2.1. Chromatin Dynamics at Telomeres
2.1.1. Histone Acetylation at Telomeres
2.1.2. Histone Methylation at Telomeres
2.1.3. Histone Ubiquitination at Telomeres
3. Chromatin Regulation at Telomeres and Links to Disease
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoch, N.C.; Lai, X.; Heierhorst, J. Genomic stability disorders: From budding yeast to humans. Front. Biosci. Sch. Ed. 2013, 5, 396–411. [Google Scholar] [CrossRef]
- Oeseburg, H.; de Boer, R.A.; van Gilst, W.H.; van der Harst, P. Telomere biology in healthy aging and disease. Pflug. Arch. 2010, 459, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Greider, C.W.; Henderson, E.; Lee, M.S.; Shampay, J.; Shippen-Lentz, D. Recognition and elongation of telomeres by telomerase. Genome 1989, 31, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Kupiec, M. Biology of telomeres: lessons from budding yeast. FEMS Microbiol. Rev. 2014, 38, 144–171. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. How telomeres solve the end-protection problem. Science 2009, 326, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Berger, S.L. Epigenetics of aging and aging-related disease. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 Suppl 1, S17–S20. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Rusche, L.N.; Kirchmaier, A.L.; Rine, J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003, 72, 481–516. [Google Scholar] [CrossRef] [PubMed]
- Grunstein, M.; Gasser, S.M. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb. Perspect. Biol. 2013, 5, a017491. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, Y.; Nishimura, Y.; Kurumizaka, H.; Shimizu, M. Nucleosome organization and chromatin dynamics in telomeres. Biomol. Concepts 2015, 6, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Schneider, J.; Shilatifard, A. Cross-talking histones: Implications for the regulation of gene expression and DNA repair. Biochem. Cell Biol. 2005, 83, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Musselman, C.A.; Lalonde, M.E.; Côté, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012, 19, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Taverna, S.D.; Li, H.; Ruthenburg, A.J.; Allis, C.D.; Patel, D.J. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007, 14, 1025–1040. [Google Scholar] [CrossRef] [PubMed]
- Tardat, M.; Déjardin, J. Telomere chromatin establishment and its maintenance during mammalian development. Chromosoma 2018, 127, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Bitterman, K.J.; Medvedik, O.; Sinclair, D.A. Longevity regulation in Saccharomyces cerevisiae: Linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 2003, 67, 376–399. [Google Scholar] [CrossRef] [PubMed]
- Lombard, D.B.; Schwer, B.; Alt, F.W.; Mostoslavsky, R. SIRT6 in DNA repair, metabolism and ageing. J. Intern. Med. 2008, 263, 128–141. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell. Biol. 2004, 5, 323–329. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Wellinger, R.J.; Zakian, V.A. Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: Beginning to end. Genetics 2012, 191, 1073–1105. [Google Scholar] [CrossRef] [PubMed]
- Tennen, R.I.; Chua, K.F. Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem. Sci. 2011, 36, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Galati, A.; Micheli, E.; Cacchione, S. Chromatin structure in telomere dynamics. Front. Oncol. 2013, 3, 46. [Google Scholar] [CrossRef] [PubMed]
- Cubiles, M.D.; Barroso, S.; Vaquero-Sedas, M.I.; Enguix, A.; Aguilera, A.; Vega-Palas, M.A. Epigenetic features of human telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef] [PubMed]
- Chow, T.T.; Shi, X.; Wei, J.H.; Guan, J.; Stadler, G.; Huang, B.; Blackburn, E.H. Local enrichment of HP1alpha at telomeres alters their structure and regulation of telomere protection. Nat. Commun. 2018, 9, 3583. [Google Scholar] [CrossRef] [PubMed]
- Gottschling, D.E.; Aparicio, O.M.; Billington, B.L.; Zakian, V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 1990, 63, 751–762. [Google Scholar] [CrossRef]
- Luke, B.; Panza, A.; Redon, S.; Iglesias, N.; Li, Z.; Lingner, J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 2008, 32, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.Z.; Gerstein, A.C.; Wigen, L.; Baller, J.A.; Berman, J. Silencing is noisy: Population and cell level noise in telomere-adjacent genes is dependent on telomere position and sir2. PLoS Genet. 2014, 10, e1004436. [Google Scholar] [CrossRef] [PubMed]
- Power, P.; Jeffery, D.; Rehman, M.A.; Chatterji, A.; Yankulov, K. Sub-telomeric core X and Y′ elements in S. cerevisiae suppress extreme variations in gene silencing. PLoS ONE 2011, 6, e17523. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Zou, Y.; Shay, J.W.; Wright, W.E. Telomere position effect in human cells. Science 2001, 292, 2075–2077. [Google Scholar] [CrossRef] [PubMed]
- Koering, C.E.; Pollice, A.; Zibella, M.P.; Bauwens, S.; Puisieux, A.; Brunori, M.; Brun, C.; Martins, L.; Sabatier, L.; Pulitzer, J.F.; et al. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002, 3, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Pedram, M.; Sprung, C.N.; Gao, Q.; Lo, A.W.; Reynolds, G.E.; Murnane, J.P. Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol. Cell. Biol. 2006, 26, 1865–1878. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T. The long reach of telomeres. Genes Dev. 2014, 28, 2445–2446. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Magdinier, F.; Stadler, G.; Wagner, K.R.; Shay, J.W.; Wright, W.E. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014, 28, 2464–2476. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Sharma, S.; Sengupta, S.; Saha, D.; Kumar, P.; Hussain, T.; Srivastava, V.; Roy, S.D.; Shay, J.W.; Chowdhury, S. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 2018, 14, e1007782. [Google Scholar] [CrossRef] [PubMed]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Gaillard, M.C.; Stadler, G.; Magdinier, F.; Wright, W.E.; Shay, J.W. SORBS2 transcription is activated by telomere position effect-over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 2015, 25, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ludlow, A.T.; Min, J.; Robin, J.D.; Stadler, G.; Mender, I.; Lai, T.P.; Zhang, N.; Wright, W.E.; Shay, J.W. Regulation of the human telomerase gene TERT by telomere position effect-over long distances (TPE-OLD): Implications for aging and cancer. PLoS Biol. 2016, 14, e2000016. [Google Scholar] [CrossRef] [PubMed]
- Perrod, S.; Gasser, S.M. Long-range silencing and position effects at telomeres and centromeres: Parallels and differences. Cell Mol. Life Sci. 2003, 60, 2303–2318. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, D.; Turniansky, R.; Green, E.M. Choose your own adventure: The role of histone modifications in yeast cell fate. J. Mol. Biol. 2016, 429, 1946–1957. [Google Scholar] [CrossRef] [PubMed]
- Hecht, A.; Strahl-Bolsinger, S.; Grunstein, M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 1996, 383, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.G.; Tanny, J.C.; Kruger, R.G.; Walz, T.; Moazed, D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 2005, 121, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Gotta, M.; Strahl-Bolsinger, S.; Renauld, H.; Laroche, T.; Kennedy, B.K.; Grunstein, M.; Gasser, S.M. Localization of Sir2p: The nucleolus as a compartment for silent information regulators. EMBO J. 1997, 16, 3243–3255. [Google Scholar] [CrossRef] [PubMed]
- Kueng, S.; Oppikofer, M.; Gasser, S.M. SIR proteins and the assembly of silent chromatin in budding yeast. Annu. Rev. Genet. 2013, 47, 275–306. [Google Scholar] [CrossRef] [PubMed]
- Ellahi, A.; Thurtle, D.M.; Rine, J. The chromatin and transcriptional landscape of native Saccharomyces cerevisiae telomeres and subtelomeric domains. Genetics 2015, 200, 505–521. [Google Scholar] [CrossRef] [PubMed]
- Maicher, A.; Kastner, L.; Dees, M.; Luke, B. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic Acids Res. 2012, 40, 6649–6659. [Google Scholar] [CrossRef] [PubMed]
- Maicher, A.; Kastner, L.; Luke, B. Telomeres and disease: Enter TERRA. RNA Biol. 2012, 9, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Wierman, M.B.; Smith, J.S. Yeast sirtuins and the regulation of aging. FEMS Yeast Res. 2014, 14, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, L.; Lu, S. Role of TERRA in the regulation of telomere length. Int. J. Biol. Sci. 2015, 11, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, N.; Redon, S.; Pfeiffer, V.; Dees, M.; Lingner, J.; Luke, B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011, 12, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Tasselli, L.; Xi, Y.; Zheng, W.; Tennen, R.I.; Odrowaz, Z.; Simeoni, F.; Li, W.; Chua, K.F. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016, 23, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; McCord, R.A.; Boxer, L.D.; Barber, M.F.; Hong, T.; Gozani, O.; Chua, K.F. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009, 8, 2664–2666. [Google Scholar] [CrossRef] [PubMed]
- Mostoslavsky, R.; Chua, K.F.; Lombard, D.B.; Pang, W.W.; Fischer, M.R.; Gellon, L.; Liu, P.; Mostoslavsky, G.; Franco, S.; Murphy, M.M.; et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006, 124, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Tennen, R.I.; Bua, D.J.; Wright, W.E.; Chua, K.F. SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2011, 2, 433. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.O.; Wang, S.S.; Zhang, Y.; Fu, X.H.; Dang, W.; Lenzmeier, B.A.; Zhou, J.Q. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genet. 2011, 7, e1001272. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, A.; Bedford, M.T. Histone arginine methylation. FEBS Lett. 2011, 585, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Udugama, M.; Chang, F.T.M.; Chan, F.L.; Tang, M.C.; Pickett, H.A.; McGhie, J.D.R.; Mayne, L.; Collas, P.; Mann, J.R.; Wong, L.H. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015, 43, 10227–10237. [Google Scholar] [CrossRef] [PubMed]
- Farooq, Z.; Banday, S.; Pandita, T.K.; Altaf, M. The many faces of histone H3K79 methylation. Mutat. Res. Rev. Mutat. Res. 2016, 768, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, K.; McNaught, K.J.; Ormsby, T.; Leggett, N.A.; Honda, S.; Selker, E.U. Telomere repeats induce domains of H3K27 methylation in Neurospora. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Su, H.; Bhat, A.; Lei, H.; Bajko, J.; Hevi, S.; Baltus, G.A.; Kadam, S.; Zhai, H.; Valdez, R.; et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 2008, 4, e1000190. [Google Scholar] [CrossRef] [PubMed]
- Nishibuchi, G.; Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 2017, 25, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Vaquero-Sedas, M.I.; Vega-Palas, M. Targeting cancer through the epigenetic features of telomeric regions. Trends Cell Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Booth, L.N.; Brunet, A. The aging epigenome. Mol. Cell 2016, 62, 728–744. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; López-Silanes, I.; Megías, D.; Fraga, F.M.; Castells-García, Á.; Blasco, M.A. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018, 9, 1548. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.S.; Kahana, A.; Wolf, A.J.; Meisinger, L.L.; Peterson, S.E.; Goggin, C.; Mahowald, M.; Gottschling, D.E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 1998, 150, 613–632. [Google Scholar] [PubMed]
- Ng, H.H.; Ciccone, D.N.; Morshead, K.B.; Oettinger, M.A.; Struhl, K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 2003, 100, 1820–1825. [Google Scholar] [CrossRef] [PubMed]
- Altaf, M.; Utley, R.T.; Lacoste, N.; Tan, S.; Briggs, S.D.; Côté, J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 2007, 28, 1002–1014. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.; Tellier, M.; Murphy, S. DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules 2018, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.H.; Schulze, J.M.; Jackson, J.; Hentrich, T.; Seidel, C.; Jaspersen, S.L.; Kobor, M.S.; Shilatifard, A. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol. Cell 2011, 42, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.P.; Luo, W.; Tsaponina, O.; Chabes, A.; Stillman, B. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol. Cell 2011, 42, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Caslini, C.; Connelly, J.A.; Serna, A.; Broccoli, D.; Hess, J.L. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol. Cell. Biol. 2009, 29, 4519–4526. [Google Scholar] [CrossRef] [PubMed]
- Kanoh, J.; Francesconi, S.; Collura, A.; Schramke, V.; Ishikawa, F.; Baldacci, G.; Géli, V. The fission yeast spSet1p is a histone H3-K4 methyltransferase that functions in telomere maintenance and DNA repair in an ATM kinase Rad3-dependent pathway. J. Mol. Biol. 2003, 326, 1081–1094. [Google Scholar] [CrossRef]
- Mikheyeva, I.V.; Grady, P.J.; Tamburini, F.B.; Lorenz, D.R.; Cam, H.P. Multifaceted genome control by Set1 Dependent and Independent of H3K4 methylation and the Set1C/COMPASS complex. PLoS Genet. 2014, 10, e1004740. [Google Scholar] [CrossRef] [PubMed]
- Castelnuovo, M.; Zaugg, J.B.; Guffanti, E.; Maffioletti, A.; Camblong, J.; Xu, Z.; Clauder-Münster, S.; Steinmetz, L.M.; Luscombe, N.M.; Stutz, F. Role of histone modifications and early termination in pervasive transcription and antisense-mediated gene silencing in yeast. Nucleic Acids Res. 2014, 42, 4348–4362. [Google Scholar] [CrossRef] [PubMed]
- Dehe, P.M.; Geli, V. The multiple faces of Set1. Biochem. Cell Biol. 2006, 84, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Jezek, M.; Gast, A.; Choi, G.; Kulkarni, R.; Quijote, J.; Graham-Yooll, A.; Park, D.; Green, E.M. The histone methyltransferases Set5 and Set1 have overlapping functions in gene silencing and telomere maintenance. Epigenetics 2017, 12, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Berretta, J.; Pinskaya, M.; Morillon, A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008, 22, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Fingerman, I.M.; Wu, C.L.; Wilson, B.D.; Briggs, S.D. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 28761–28765. [Google Scholar] [CrossRef] [PubMed]
- Nislow, C.; Ray, E.; Pillus, L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 1997, 8, 2421–2436. [Google Scholar] [CrossRef] [PubMed]
- Trelles-Sticken, E.; Bonfils, S.; Sollier, J.; Geli, V.; Scherthan, H.; de La Roche Saint-Andre, C. Set1- and Clb5-deficiencies disclose the differential regulation of centromere and telomere dynamics in Saccharomyces cerevisiae meiosis. J. Cell Sci. 2005, 118, 4985–4994. [Google Scholar] [CrossRef] [PubMed]
- Acquaviva, L.; Drogat, J.; Dehé, P.M.; de La Roche Saint-André, C.; Géli, V. Spp1 at the crossroads of H3K4me3 regulation and meiotic recombination. Epigenetics 2013, 8, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, B.; Drogaris, P.; Lin, H.H.; Armstrong, H.; Hiragami-Hamada, K.; Imhof, A.; Bonneil, E.; Thibault, P.; Verreault, A.; Festenstein, R.J. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011, 7, e1001354. [Google Scholar] [CrossRef] [PubMed]
- Pokholok, D.K.; Harbison, C.T.; Levine, S.; Cole, M.; Hannett, N.M.; Lee, T.I.; Bell, G.W.; Walker, K.; Rolfe, P.A.; Herbolsheimer, E.; et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005, 122, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.H.; Robert, F.; Young, R.A.; Struhl, K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 2003, 11, 709–719. [Google Scholar] [CrossRef]
- Krogan, N.J.; Dover, J.; Khorrami, S.; Greenblatt, J.F.; Schneider, J.; Johnston, M.; Shilatifard, A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002, 277, 10753–10755. [Google Scholar] [CrossRef] [PubMed]
- Bryk, M.; Briggs, S.D.; Strahl, B.D.; Curcio, M.J.; Allis, C.D.; Winston, F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 2002, 12, 165–170. [Google Scholar] [CrossRef]
- Weiner, A.; Chen, H.V.; Liu, C.L.; Rahat, A.; Klien, A.; Soares, L.; Gudipati, M.; Pfeffner, J.; Regev, A.; Buratowski, S.; et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012, 10, e1001369. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, D.; Jezek, M.; Quijote, J.; Lum, J.; Choi, G.; Kulkarni, R.; Park, D.; Green, E.M. Repression of middle sporulation genes in. G3 (Bethesda) 2017, 7, 3971–3982. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Lin, W.; Soustelle, C.; Suhre, K.; Nicolas, A.; Géli, V.; de La Roche Saint-André, C. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J. 2004, 23, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Wood, A.; Lee, J.S.; Schuster, R.; Dueker, J.; Maguire, C.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Shilatifard, A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell 2005, 19, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Mueller, J.E.; Canze, M.; Bryk, M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics 2006, 173, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Walter, D.; Matter, A.; Fahrenkrog, B. Loss of histone H3 methylation at lysine 4 triggers apoptosis in Saccharomyces cerevisiae. PLoS Genet. 2014, 10, e1004095. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rosa, H.; Bannister, A.J.; Dehe, P.M.; Géli, V.; Kouzarides, T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 2004, 279, 47506–47512. [Google Scholar] [CrossRef] [PubMed]
- Venkatasubrahmanyam, S.; Hwang, W.W.; Meneghini, M.D.; Tong, A.H.; Madhani, H.D. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc. Natl. Acad. Sci. USA 2007, 104, 16609–16614. [Google Scholar] [CrossRef] [PubMed]
- Margaritis, T.; Oreal, V.; Brabers, N.; Maestroni, L.; Vitaliano-Prunier, A.; Benschop, J.J.; van Hooff, S.; van Leenen, D.; Dargemont, C.; Géli, V.; et al. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 2012, 8, e1002952. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Chen, Z.; Jiang, R.; Meneghini, M.D. H3K4 Methylation Dependent and Independent Chromatin Regulation by JHD2 and SET1 in Budding Yeast. G3 (Bethesda) 2018, 8, 1829–1839. [Google Scholar] [CrossRef] [PubMed]
- Sayou, C.; Millán-Zambrano, G.; Santos-Rosa, H.; Petfalski, E.; Robson, S.; Houseley, J.; Kouzarides, T.; Tollervey, D. RNA Binding by histone methyltransferases Set1 and Set2. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Luciano, P.; Jeon, J.; El-Kaoutari, A.; Challal, D.; Bonnet, A.; Barucco, M.; Candelli, T.; Jourquin, F.; Lesage, P.; Kim, J.; et al. Binding to RNA regulates Set1 function. Cell Discov. 2017, 3, 17040. [Google Scholar] [CrossRef] [PubMed]
- Trésaugues, L.; Dehé, P.M.; Guérois, R.; Rodriguez-Gil, A.; Varlet, I.; Salah, P.; Pamblanco, M.; Luciano, P.; Quevillon-Cheruel, S.; Sollier, J.; et al. Structural characterization of Set1 RNA recognition motifs and their role in histone H3 lysine 4 methylation. J. Mol. Biol. 2006, 359, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Corda, Y.; Schramke, V.; Longhese, M.P.; Smokvina, T.; Paciotti, V.; Brevet, V.; Gilson, E.; Geli, V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat. Genet. 1999, 21, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Green, E.M.; Mas, G.; Young, N.L.; Garcia, B.A.; Gozani, O. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat. Struct. Mol. Biol. 2012, 19, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Martín, G.M.; King, D.A.; Green, E.M.; Garcia-Nieto, P.E.; Alexander, R.; Collins, S.R.; Krogan, N.J.; Gozani, O.P.; Morrison, A.J. Set5 and Set1 cooperate to repress gene expression at telomeres and retrotransposons. Epigenetics 2014, 9, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Green, E.M.; Morrison, A.J.; Gozani, O. New marks on the block: Set5 methylates H4 lysines 5, 8 and 12. Nucleus 2012, 3, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Zukowski, A.; Johnson, A.M. The interplay of histone H2B ubiquitination with budding and fission yeast heterochromatin. Curr. Genet. 2018, 64, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Cheung, W.L.; Hsu, J.Y.; Diaz, R.L.; Smith, M.M.; Allis, C.D. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 2005, 120, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.W.; Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002, 418, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Holt, M.T.; Ohashi, N.; Zhao, A.; Müller, M.M.; Wang, B.; Muir, T.W. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat. Commun. 2016, 7, 10589. [Google Scholar] [CrossRef] [PubMed]
- Rhie, B.H.; Song, Y.H.; Ryu, H.Y.; Ahn, S.H. Cellular aging is associated with increased ubiquitylation of histone H2B in yeast telomeric heterochromatin. Biochem. Biophys. Res. Commun. 2013, 439, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, J.; Zhang, Q.D.; Lv, D.K.; Wu, N.F.; Zhou, J.Q. Rad6-Bre1-mediated H2B ubiquitination regulates telomere replication by promoting telomere-end resection. Nucleic Acids Res. 2017, 45, 3308–3322. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; He, M.H.; Zhang, L.L.; Liu, J.; Zhang, Q.D.; Zhou, J.Q. Rad6-Bre1 mediated histone H2Bub1 protects uncapped telomeres from exonuclease Exo1 in Saccharomyces cerevisiae. DNA Repair 2018, 72, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Galán, A.; García-Oliver, E.; Nuño-Cabanes, C.; Rubinstein, L.; Kupiec, M.; Rodríguez-Navarro, S. The evolutionarily conserved factor Sus1/ENY2 plays a role in telomere length maintenance. Curr. Genet. 2018, 64, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, F.; Dann, G.P.; Beh, L.Y.; Debelouchina, G.T.; Hofmann, R.; Muir, T.W. Functional crosstalk between histone H2B ubiquitylation and H2A modifications and variants. Nat. Commun. 2018, 9, 1394. [Google Scholar] [CrossRef] [PubMed]
- Whitcomb, S.J.; Fierz, B.; McGinty, R.K.; Holt, M.; Ito, T.; Muir, T.W.; Allis, C.D. Histone monoubiquitylation position determines specificity and direction of enzymatic cross-talk with histone methyltransferases Dot1L and PRC2. J. Biol. Chem. 2012, 287, 23718–23725. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.; Blasco, M.A. Telomere-driven diseases and telomere-targeting therapies. J. Cell. Biol. 2017, 216, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Holohan, B.; Wright, W.E.; Shay, J.W. Cell biology of disease: Telomeropathies: An emerging spectrum disorder. J. Cell Biol. 2014, 205, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Padberg, G.W.; Frants, R.R.; Brouwer, O.F.; Wijmenga, C.; Bakker, E.; Sandkuijl, L.A. Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve Suppl. 1995, 2, S81–S84. [Google Scholar] [CrossRef] [PubMed]
- Stadler, G.; Rahimov, F.; King, O.D.; Chen, J.C.; Robin, J.D.; Wagner, K.R.; Shay, J.W.; Emerson, C.P.; Wright, W.E. Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nat. Struct. Mol. Biol. 2013, 20, 671–678. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jezek, M.; Green, E.M. Histone Modifications and the Maintenance of Telomere Integrity. Cells 2019, 8, 199. https://doi.org/10.3390/cells8020199
Jezek M, Green EM. Histone Modifications and the Maintenance of Telomere Integrity. Cells. 2019; 8(2):199. https://doi.org/10.3390/cells8020199
Chicago/Turabian StyleJezek, Meagan, and Erin M. Green. 2019. "Histone Modifications and the Maintenance of Telomere Integrity" Cells 8, no. 2: 199. https://doi.org/10.3390/cells8020199
APA StyleJezek, M., & Green, E. M. (2019). Histone Modifications and the Maintenance of Telomere Integrity. Cells, 8(2), 199. https://doi.org/10.3390/cells8020199



