Tristetraprolin/ZFP36 Regulates the Turnover of Autoimmune-Associated HLA-DQ mRNAs
Abstract
1. Introduction
2. Results
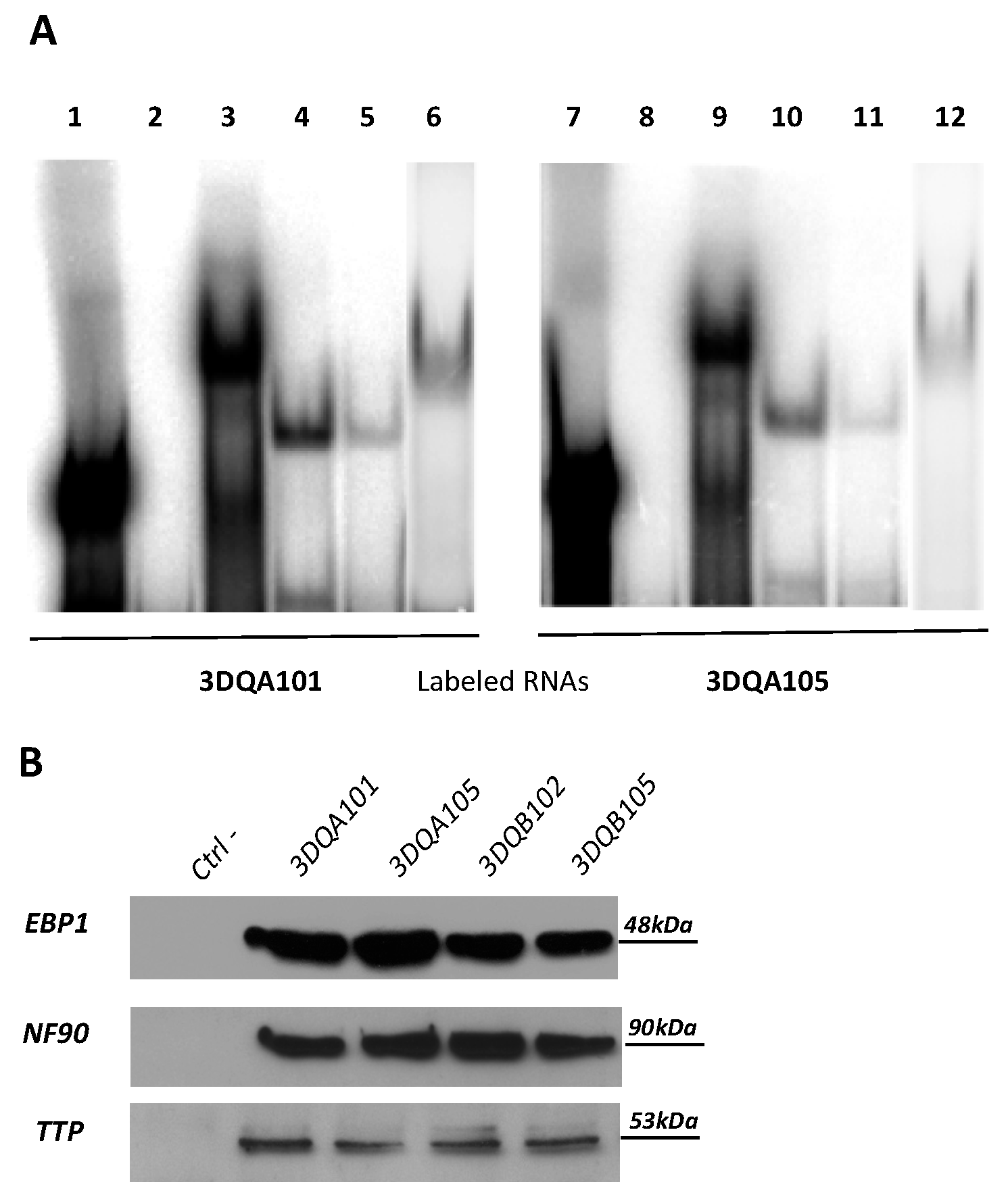
2.1. TTP Binds to CD—Associated and Non-Associated Transcripts
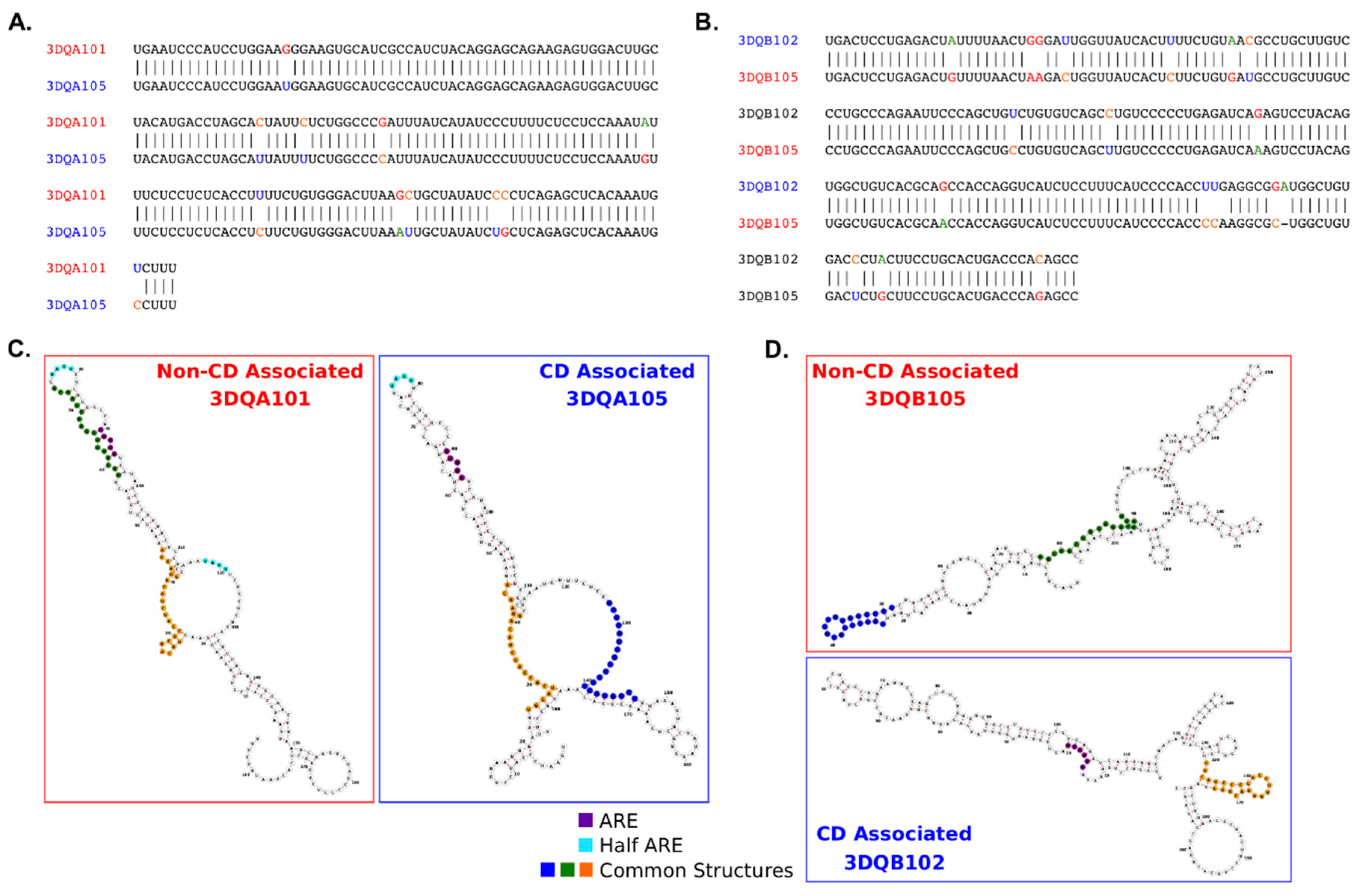
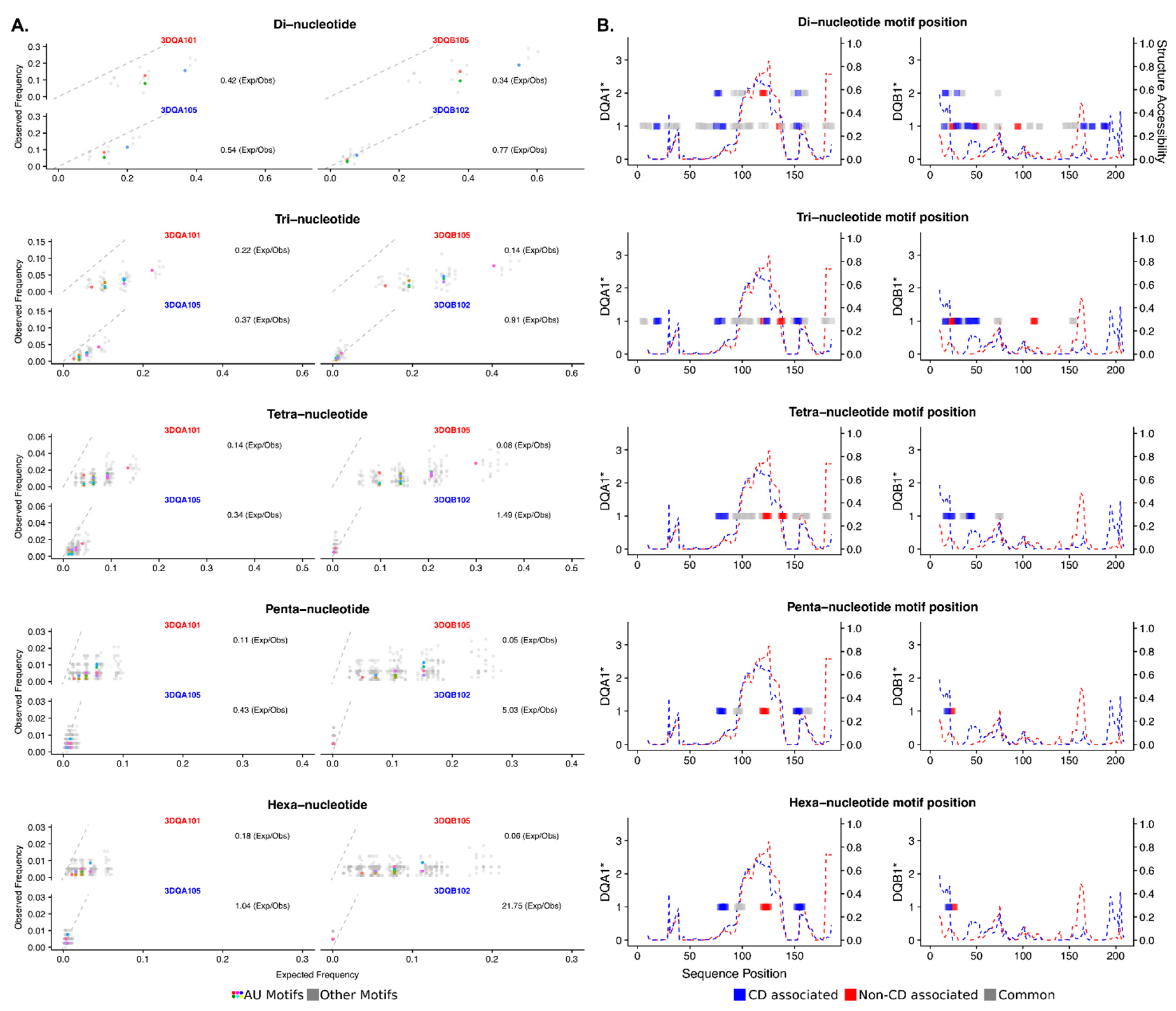
2.2. Analysis of DQA1* and DQB1* 3′UTR Sequences
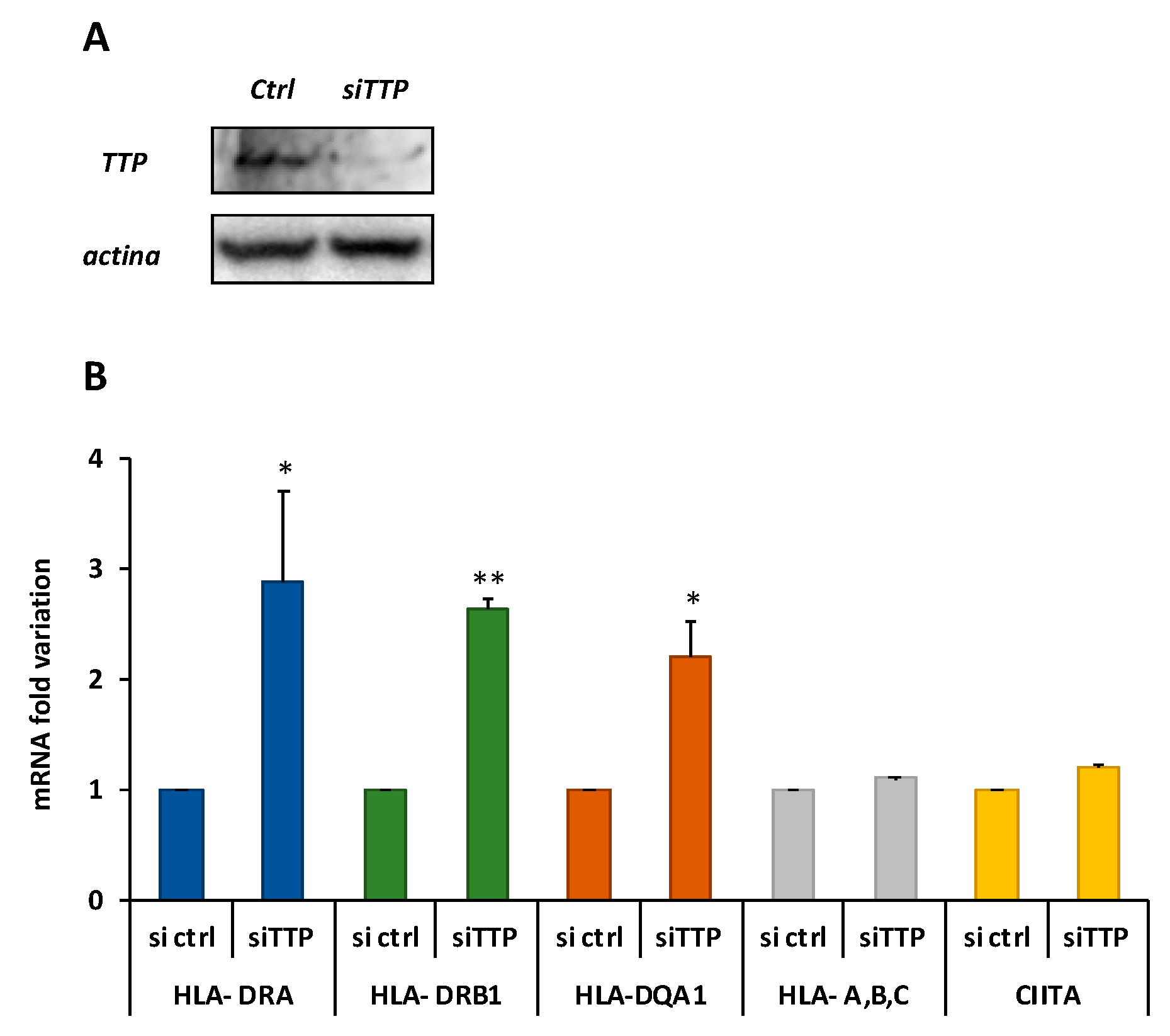
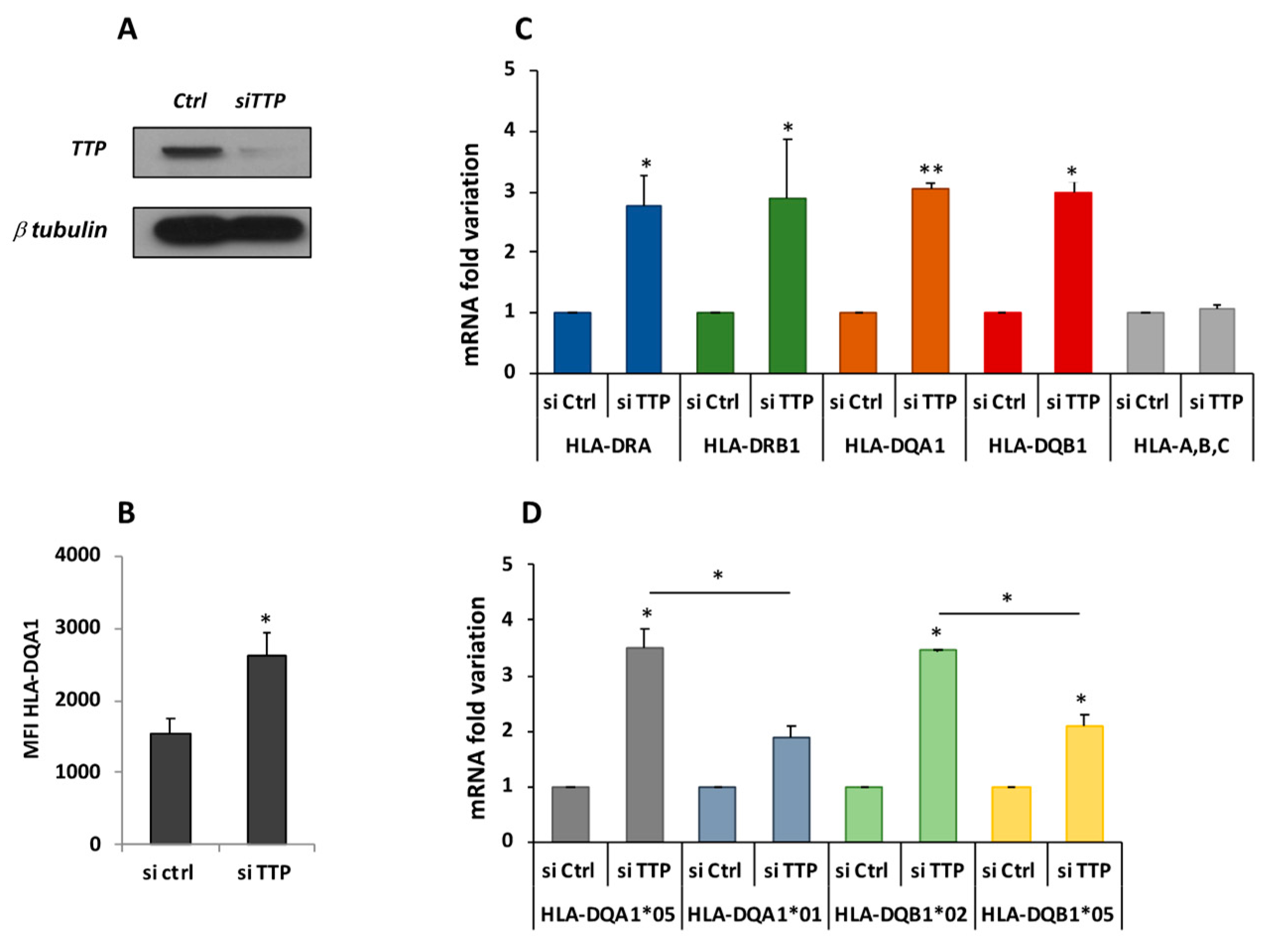
2.3. Knockdown of TTP Affects the Expression Level of DQA1 and DQB1 Transcripts
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Cell Lines
5.2. DNA Templates Synthesis and mRNA Quantification
5.3. RNA Electrophoretic Mobility Shift Assay (REMSA) and Pull-Down
5.4. TTP Silencing and Phenotype Analysis
6. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ting, J.P.Y.; Trowsdale, J. Genetic control of MHC class II expression. Cell 2002, 109, S21–S33. [Google Scholar] [CrossRef]
- Reith, W.; LeibundGut-Landmann, S.; Waldburger, J.M. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005, 5, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Corso, C.; Pisapia, L.; Citro, A.; Cicatiello, V.; Barba, P.; Cigliano, L.; Abrescia, P.; Maffei, A.; Manco, G.; Del Pozzo, G. EBP1 and DRBP76/NF90 binding proteins are included in the major histocompatibility complex class II RNA operon. Nucleic Acids Res. 2011, 39, 7263–7275. [Google Scholar] [CrossRef][Green Version]
- Pisapia, L.; Cicatiello, V.; Barba, P.; Malanga, D.; Maffei, A.; Hamilton, R.S.; Del Pozzo, G. Co-regulated expression of alpha and beta mRNAs encoding HLA-DR surface heterodimers is mediated by the MHCII RNA operon. Nucleic Acids Res. 2013, 41, 3772–3786. [Google Scholar] [CrossRef]
- Ko, H.R.; Chang, Y.S.; Park, W.S.; Ahn, J.Y. Opposing roles of the two isoforms of ErbB3 binding protein 1 in human cancer cells. Int. J. Cancer 2016, 139, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Masuda, K.; Kuwano, Y.; Nishida, K.; Rokutan, K.; Imoto, I. NF90 in posttranscriptional gene regulation and microRNA biogenesis. Int. J. Mol. Sci. 2013, 14, 17111–17121. [Google Scholar] [CrossRef]
- Abadie, V.; Sollid, L.M.; Barreiro, L.B.; Jabri, B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu. Rev. Immunol. 2011, 29, 493–525. [Google Scholar] [CrossRef]
- Noble, J.A. Immunogenetics of type 1 diabetes: A comprehensive review. J. Autoimmun. 2015, 64, 101–112. [Google Scholar] [CrossRef]
- Pisapia, L.C.A.; Picascia, S.; Bassi, V.; Barba, P.; Del Pozzo, G.; Gianfrani, C. HLA-DQ2.5 genes associated to celiac disease risk are preferentially expressed respect to non-predisposing HLA genes: Implication for anti-gluten T cell response. J. Autoimmun. 2016, 70, 63–72. [Google Scholar] [CrossRef]
- Brooks, S.A.; Blackshear, P.J. Tristetraprolin (TTP): Interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 2013, 1829, 666–679. [Google Scholar] [CrossRef]
- Mukherjee, N.; Jacobs, N.C.; Hafner, M.; Kennington, E.A.; Nusbaum, J.D.; Tuschl, T.; Blackshear, P.J.; Ohler, U. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014, 15, R12. [Google Scholar] [CrossRef] [PubMed]
- Patial, S.; Curtis, A.D., 2nd; Lai, W.S.; Stumpo, D.J.; Hill, G.D.; Flake, G.P.; Mannie, M.D.; Blackshear, P.J. Enhanced stability of tristetraprolin mRNA protects mice against immune-mediated inflammatory pathologies. Proc. Natl. Acad. Sci. USA 2016, 113, 1865–1870. [Google Scholar] [CrossRef] [PubMed]
- Sedlyarov, V.; Fallmann, J.; Ebner, F.; Huemer, J.; Sneezum, L.; Ivin, M.; Kreiner, K.; Tanzer, A.; Vogl, C.; Hofacker, I.; et al. Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol. Syst. Biol. 2016, 12, 868. [Google Scholar] [CrossRef] [PubMed]
- Bhandare, S.; Goldberg, D.S.; Dowell, R. Discriminating between HuR and TTP binding sites using the k-spectrum kernel method. PLoS ONE 2017, 12, e0174052. [Google Scholar] [CrossRef]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004, 32, W135–W141. [Google Scholar] [CrossRef]
- Sundfeld, D.; Havgaard, J.H.; de Melo, A.C.; Gorodkin, J. Foldalign 2.5: Multithreaded implementation for pairwise structural RNA alignment. Bioinformatics 2016, 32, 1238–1240. [Google Scholar] [CrossRef]
- Gianfrani, C.; Pisapia, L.; Picascia, S.; Strazzullo, M.; Del Pozzo, G. Expression level of risk genes of MHC class II is a susceptibility factor for autoimmunity: New insights. J. Autoimmun. 2018, 89, 1–10. [Google Scholar] [CrossRef]
- Patial, S.; Blackshear, P.J. Tristetraprolin as a Therapeutic Target in Inflammatory Disease. Trends Pharm. Sci. 2016, 37, 811–821. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- D’Agostino, V.G.; Adami, V.; Provenzani, A. A novel high throughput biochemical assay to evaluate the HuR protein-RNA complex formation. PLoS ONE. 2013, 8, e72426. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Havgaard, J.H.; Torarinsson, E.; Gorodkin, J. Fast pairwise structural RNA alignments by pruning of the dynamical programming matrix. PLoS Comput. Biol. 2007, 3, 1896–1908. [Google Scholar] [CrossRef] [PubMed]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [PubMed]
| Primers Used for qRT-PCR | ||
| Gene | Primers | Sequences 5′ → 3′ |
| β-Actin | ACT-F ACT-R | TCATGAAGTGTGACGTTGACA CCTAGAAGCATTTGCGGTGCAC |
| GAPDH | G-F G-R | AACGGATTTGGTCGTATTGGGC TCGCTCCTGGAAGATGGTGATG |
| HLA-DRA | DRA-F DRA-R | GGACAAAGCCAACCTGGAAA AGGACGTTGGGCTCTCTCAG |
| HLA-DRB1 | DRB1-F DRB1-R | CTCAGCATCTTGCTCTTGTGCAG CAGCATTAAAGTCAGGTGGTTCC |
| HLA-DQA1 | DQA1-F DQA1*R | GGTGTAAACTTGTACCAGT GGAGACTTGGAAAACACT |
| HLA-DQB1 | DQB1-F DQB1*R | CAGATCAAAGTCCGGTGGTTT TCTGGGCAGATTCAGACTGAGC |
| HLA-A, -B, and -C | MHCI-F MHCI-R | AGTGGGCTACGTGGACGACA ATGTAATCCTTGCCGTCGTA |
| HLA-DQA1*05 | alfa05-F alfa05-R | CGGTGGCCTGAGTTCAGCAA GGAGACTTGGAAAACACTGTGACC |
| HLA- DQA1*01 | alfa01-F alfa01-R | CGGTGGCCTGAGTTCAGCAA GGAGACTTGGAAAACACTGTGACC |
| HLA- DQB1*02 | beta02-F beta02-R | TCTTGTGAGCAGAAGCATCT CAGGATCTGGAAGGTCCAGT |
| HLA- DQB1*05 | beta05-F beta05-R | ACAACTACGAGGTGGCGTACC CAGGATCTGGAAGGTCCAGT |
| Primers used for PCR of riboprobes templates | ||
| Probe | Primers | Sequences 5′ → 3′ |
| 3DQA101 | 3DQA101T7 3DQA101R | TAATACGACTCACTATAGGCCATCCTGGAAGGGAAGTG TCAGGAGGTCAGGGAAAGAA |
| 3DQA105 | 3DQA105T7 3DQA105R | TAATACGACTCACTATAGGATCCCATCCTGGAATGGAAGTG AAAGGCATTTGTGAGCTCTGAGCAG |
| 3DQB102 | 3DQB102T7 3DQB102R | TAATACGACTCACTATAGGGGCACTGACTCCTGAGACT GCTGTGGGTCAGTGCAG |
| 3DQB105 | 3DQB105T7 3DQB105R | TAATACGACTCACTATAGGGGCACTGACTCCTGAGACTGT GGCTGTGGGTCAGTGCAG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pisapia, L.; Hamilton, R.S.; Farina, F.; D’Agostino, V.; Barba, P.; Strazzullo, M.; Provenzani, A.; Gianfrani, C.; Del Pozzo, G. Tristetraprolin/ZFP36 Regulates the Turnover of Autoimmune-Associated HLA-DQ mRNAs. Cells 2019, 8, 1570. https://doi.org/10.3390/cells8121570
Pisapia L, Hamilton RS, Farina F, D’Agostino V, Barba P, Strazzullo M, Provenzani A, Gianfrani C, Del Pozzo G. Tristetraprolin/ZFP36 Regulates the Turnover of Autoimmune-Associated HLA-DQ mRNAs. Cells. 2019; 8(12):1570. https://doi.org/10.3390/cells8121570
Chicago/Turabian StylePisapia, Laura, Russell S. Hamilton, Federica Farina, Vito D’Agostino, Pasquale Barba, Maria Strazzullo, Alessandro Provenzani, Carmen Gianfrani, and Giovanna Del Pozzo. 2019. "Tristetraprolin/ZFP36 Regulates the Turnover of Autoimmune-Associated HLA-DQ mRNAs" Cells 8, no. 12: 1570. https://doi.org/10.3390/cells8121570
APA StylePisapia, L., Hamilton, R. S., Farina, F., D’Agostino, V., Barba, P., Strazzullo, M., Provenzani, A., Gianfrani, C., & Del Pozzo, G. (2019). Tristetraprolin/ZFP36 Regulates the Turnover of Autoimmune-Associated HLA-DQ mRNAs. Cells, 8(12), 1570. https://doi.org/10.3390/cells8121570






