Biofabrication of Cell-Derived Nanovesicles: A Potential Alternative to Extracellular Vesicles for Regenerative Medicine
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
HEK293FT-Cre Donor Cell Line
2.2. NV Biofabrication
2.3. EV Purification
2.4. Nanoparticle Tracking Analysis
2.5. Western Blot
2.6. Transmission Electron Microscopy
2.7. Uptake Experiment
2.8. Functional Delivery of Cre by NVs and EVs
2.9. Activation of ERK Signaling by CPC-EVs and -NVs
2.10. Comparative Analysis NV and EV Production
2.11. Statistical Analysis
3. Results
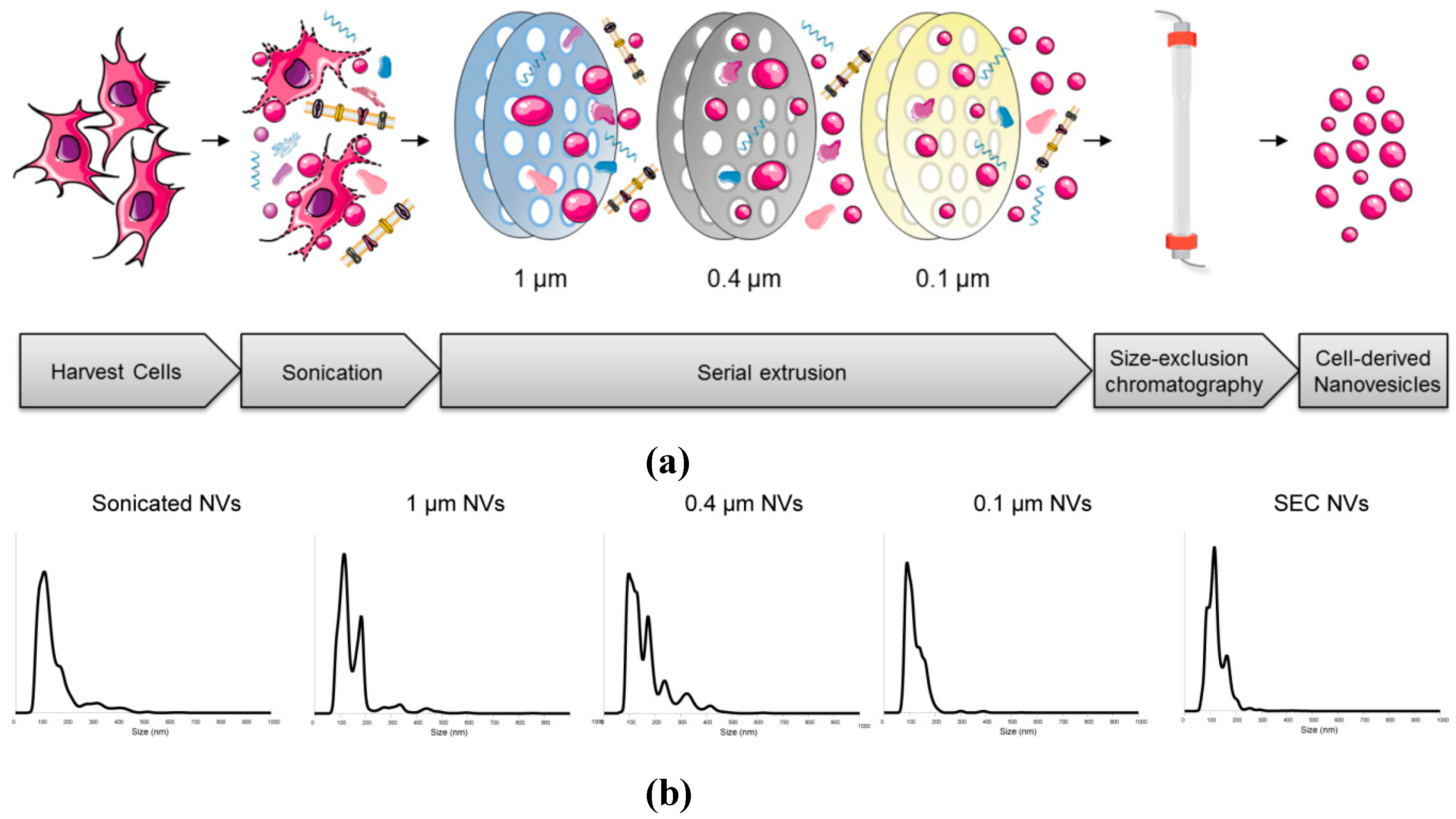
3.1. Production
3.1.1. Process Characterization
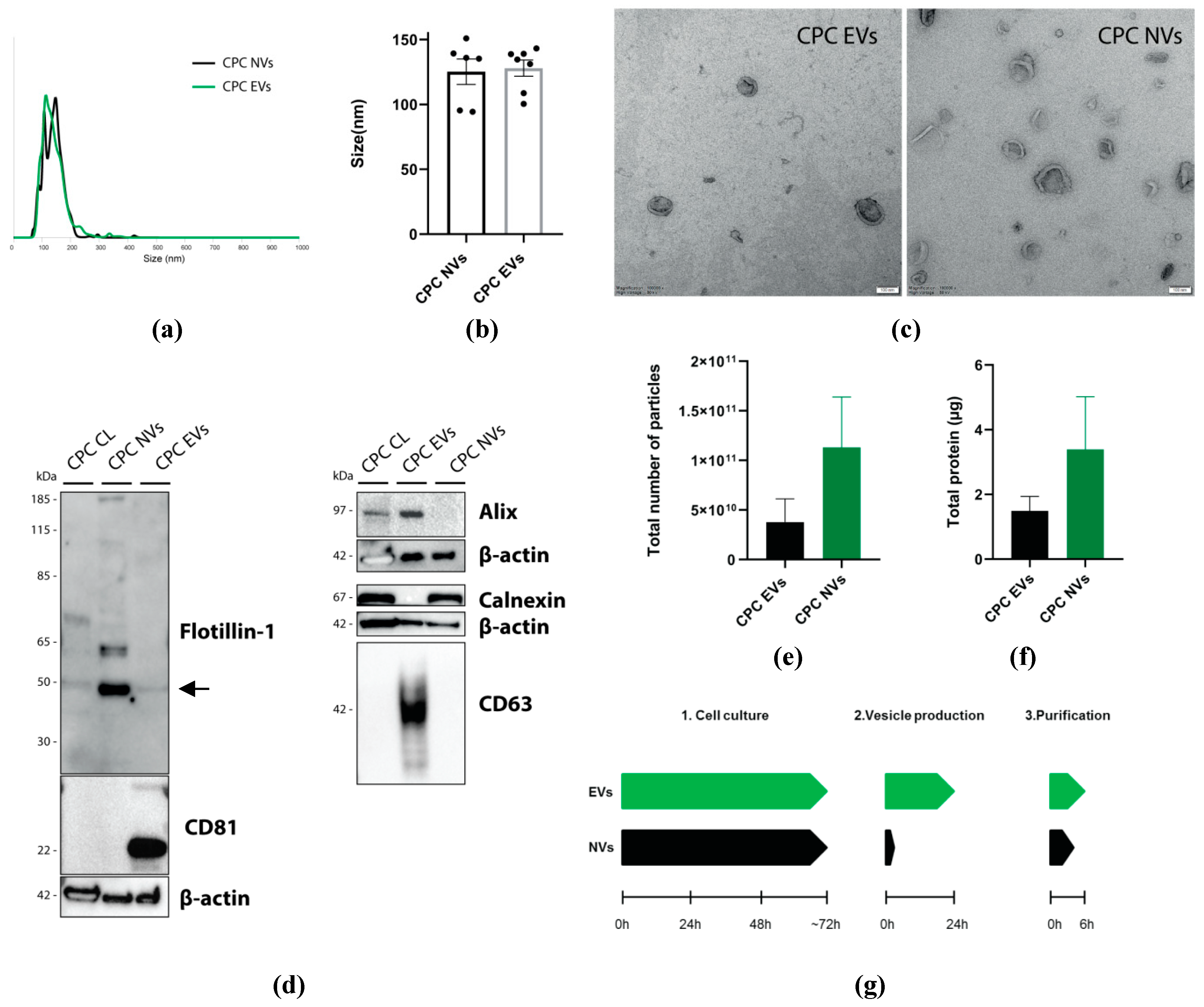
3.1.2. Characterization of CPC-NVs and CPC-EVs
3.2. Functionality
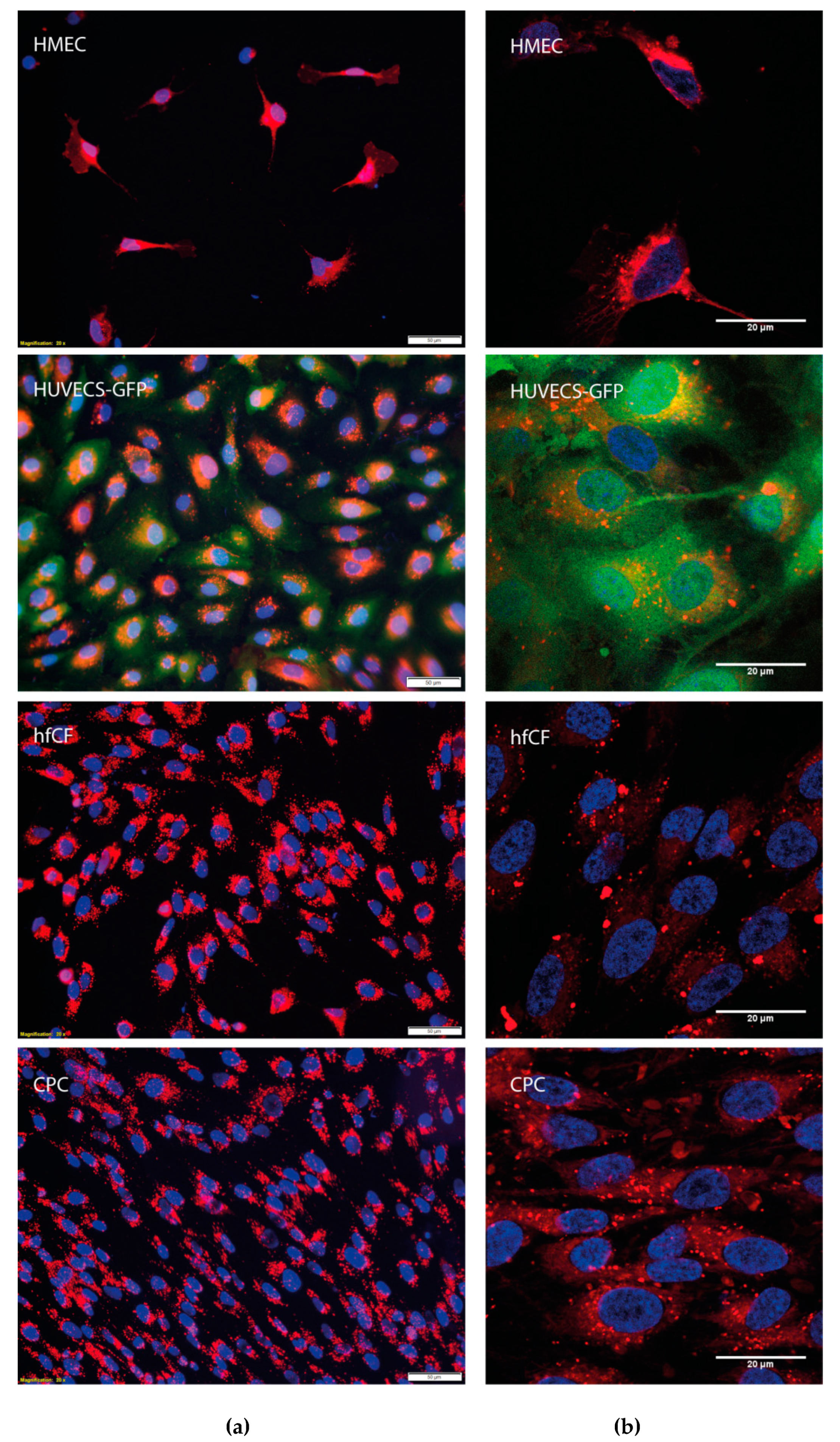
3.2.1. NVs Uptake
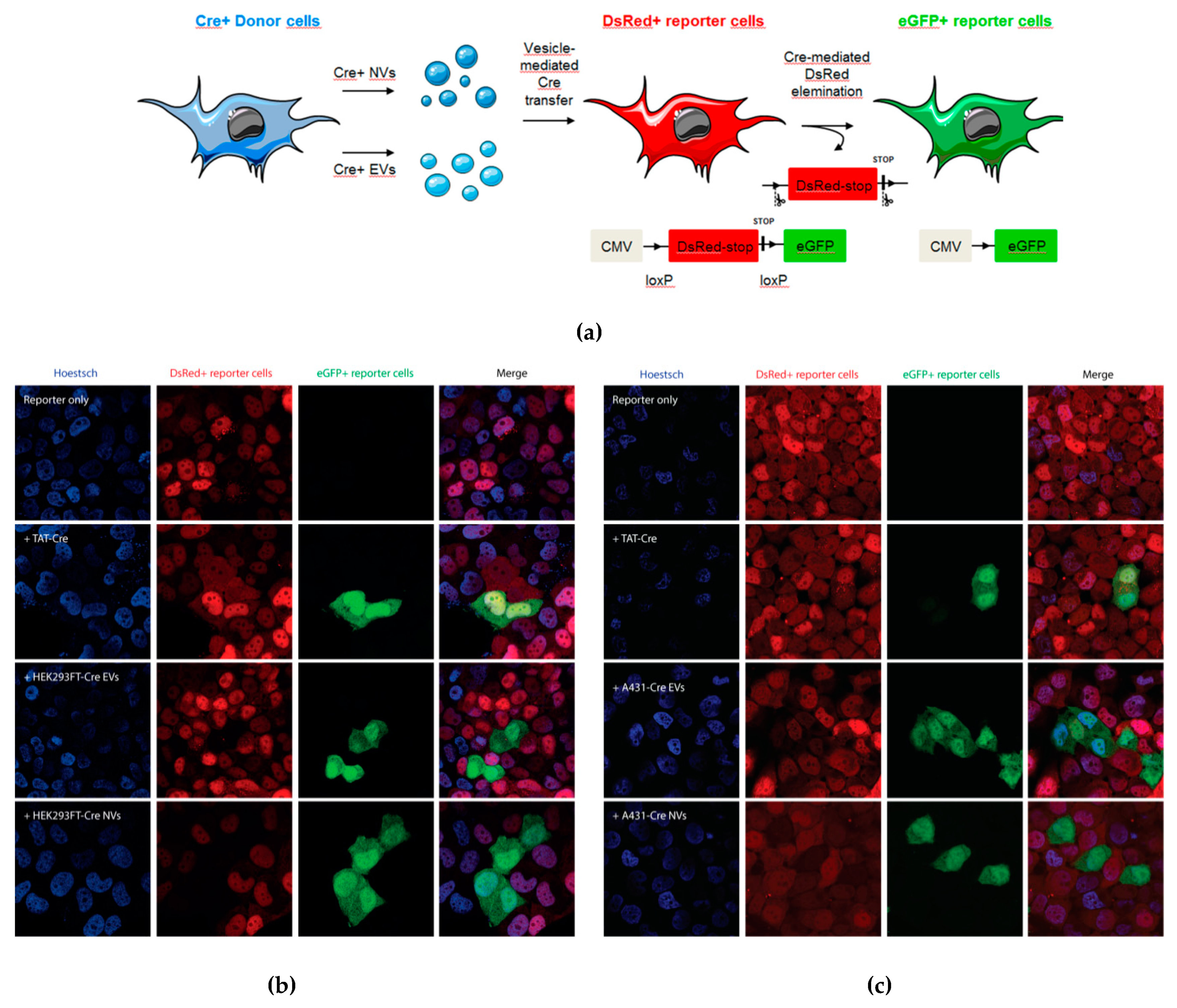
3.2.2. NVs Functionally Deliver Biomolecules to Target Cells
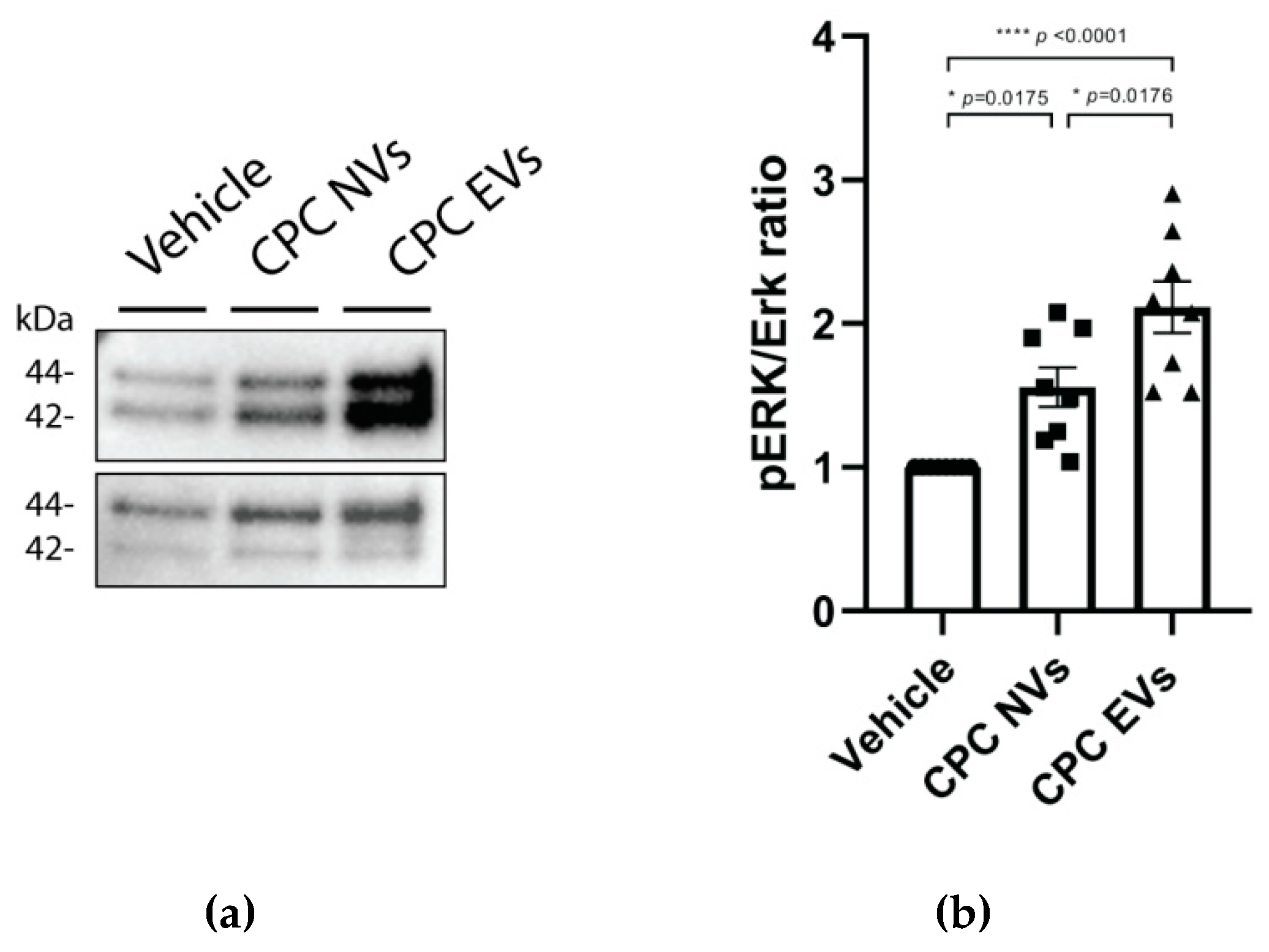
3.2.3. CPC-NVs and EVs Induce Phosphorylation of ERK1/2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wiklander, O.P.B.; Brennan, M.Á.; Lötvall, J.; Breakefield, X.O.; Andaloussi, S.E.L. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019, 11, eaav8521. [Google Scholar] [CrossRef] [PubMed]
- Vader, P.; Mol, E.A.; Pasterkamp, G.; Schiffelers, R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016, 106, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Time to deliver. Nat. Biotechnol. 2014, 32, 961. [CrossRef] [PubMed]
- Zhang, L.; Gu, F.X.; Chan, J.M.; Wang, A.Z.; Langer, R.S.; Farokhzad, O.C. Nanoparticles in medicine: Therapeutic applications and developments. Clin. Pharmacol. Ther. 2008, 83, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Taheri, B.; Soleimani, M.; Fekri Aval, S.; Esmaeili, E.; Bazi, Z.; Zarghami, N. Induced pluripotent stem cell-derived extracellular vesicles: A novel approach for cell-free regenerative medicine. J. Cell. Physiol. 2019, 234, 8455–8464. [Google Scholar] [CrossRef] [PubMed]
- Vishnubhatla, I.; Corteling, R.; Stevanato, L.; Hicks, C.; Sinden, J. The development of stem cell-derived exosomes as a cell-free regenerative medicine. J. Circ. Biomarkers 2014, 3, 2–3. [Google Scholar] [CrossRef]
- Camussi, G.; Deregibus, M.C.; Cantaluppi, V. Role of stem-cell-derived microvesicles in the paracrine action of stem cells. Biochem. Soc. Trans. 2013, 41, 283–287. [Google Scholar] [CrossRef]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.E.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef]
- Timmers, L.; Lim, S.K.; Hoefer, I.E.; Arslan, F.; Lai, R.C.; van Oorschot, A.A.M.; Goumans, M.J.; Strijder, C.; Sze, S.K.; Choo, A. Human mesenchymal stem cell-conditioned medium improves cardiac function following myocardial infarction. Stem Cell Res. 2011, 6, 206–214. [Google Scholar] [CrossRef]
- Lai, R.C.; Chen, T.S.; Lim, S.K. Mesenchymal stem cell exosome: A novel stem cell-based therapy for cardiovascular disease. Regen. Med. 2011, 6, 481–492. [Google Scholar] [CrossRef]
- Timmers, L.; Lim, S.K.; Arslan, F.; Armstrong, J.S.; Hoefer, I.E.; Doevendans, P.A.; Piek, J.J.; El Oakley, R.M.; Choo, A.; Lee, C.N. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2008, 1, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Lionetti, V.; Cervio, E.; Matteucci, M.; Gherghiceanu, M.; Popescu, L.M.; Torre, T.; Siclari, F.; Moccetti, T.; Vassalli, G. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc. Res. 2014, 103, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Vrijsen, K.R.; Sluijter, J.P.G.; Schuchardt, M.W.L.; Van Balkom, B.W.M.; Noort, W.A.; Chamuleau, S.A.J.; Doevendans, P. Cardiomyocyte progenitor cell-derived exosomes stimulate migration of endothelial cells. J. Cell. Mol. Med. 2010, 14, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Sokic, S.; Schardt, J.S.; Raiker, R.S.; Lin, J.W.; Jay, S.M. Emerging roles for extracellular vesicles in tissue engineering and regenerative medicine. Tissue Eng. Part. B Rev. 2014, 21, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Mol, E.A.; Goumans, M.-J.; Sluijter, J.P.G. Cardiac progenitor-cell derived exosomes as cell-free therapeutic for cardiac repair. Adv. Exp. Med. Biol. 2017, 998, 207–219. [Google Scholar]
- Segers, V.F.M.; Lee, R.T. Stem-cell therapy for cardiac disease. Nature 2008, 451, 937–942. [Google Scholar] [CrossRef]
- Steinhauser, M.L.; Lee, R.T. Regeneration of the heart. EMBO Mol. Med. 2011, 3, 701–712. [Google Scholar] [CrossRef]
- Sluijter, J.P.G.; Davidson, S.M.; Boulanger, C.M.; Buzas, E.I.; De Kleijn, D.P.V.; Engel, F.B.; Giricz, Z.; Hausenloy, D.J.; Kishore, R.; Lecour, S. Extracellular vesicles in diagnostics and therapy of the ischaemic heart: Position Paper from the Working Group on Cellular Biology of the Heart of the European Society of Cardiology. Cardiovasc. Res. 2017, 114, 19–34. [Google Scholar] [CrossRef]
- Lu, M.; Xing, H.; Yang, Z.; Sun, Y.; Yang, T.; Zhao, X.; Cai, C.; Wang, D.; Ding, P. Recent advances on extracellular vesicles in therapeutic delivery: Challenges, solutions, and opportunities. Eur. J. Pharm. Biopharm. 2017, 119, 381–395. [Google Scholar] [CrossRef]
- Jo, W.; Kim, J.; Yoon, J.; Jeong, D.; Cho, S.; Jeong, H.; Yoon, Y.J.; Kim, S.C.; Gho, Y.S.; Park, J. Large-scale generation of cell-derived nanovesicles. Nanoscale 2014, 6, 12056–12064. [Google Scholar] [CrossRef]
- Goh, W.J.; Zou, S.; Ong, W.Y.; Torta, F.; Alexandra, A.F.; Schiffelers, R.M.; Storm, G.; Wang, J.-W.; Czarny, B.; Pastorin, G. Bioinspired Cell-Derived Nanovesicles versus Exosomes as Drug Delivery Systems: A Cost-Effective Alternative. Sci. Rep. 2017, 7, 14322. [Google Scholar] [CrossRef] [PubMed]
- Ban, J.-J.; Lee, M.; Im, W.; Kim, M. Low pH increases the yield of exosome isolation. Biochem. Biophys. Res. Commun. 2015, 461, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.A.; Ontoria-Oviedo, I.; González-King, H.; Diez-Juan, A.; Sepúlveda, P. Glucose Starvation in Cardiomyocytes Enhances Exosome Secretion and Promotes Angiogenesis in Endothelial Cells. PLoS ONE 2015, 10, e0138849. [Google Scholar] [CrossRef] [PubMed]
- Savina, A.; Furlán, M.; Vidal, M.; Colombo, M.I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef]
- Kapustin, A.N.; Chatrou, M.L.; Drozdov, I.; Zheng, Y.; Davidson, S.M.; Soong, D.Y.H.; Furmanik, M.; Sanchis, P.; de Rosales, R.T.; Alvarez-Hernandez, D. Vascular smooth muscle cell calcification is mediated by regulated exosome secretion. Circ. Res. 2015, 116, 1312–1323. [Google Scholar] [CrossRef]
- Smits, A.M.; van Vliet, P.; Metz, C.H.; Korfage, T.; Sluijter, J.P.G.; Doevendans, P.A.; Goumans, M.-J. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: An in vitro model for studying human cardiac physiology and pathophysiology. Nat. Protoc. 2009, 4, 232–243. [Google Scholar] [CrossRef]
- Van Mil, A.; Grundmann, S.; Goumans, M.-J.; Lei, Z.; Oerlemans, M.I.; Jaksani, S.; Doevendans, P.A.; Sluijter, J.P.G. MicroRNA-214 inhibits angiogenesis by targeting Quaking and reducing angiogenic growth factor release. Cardiovasc. Res. 2012, 93, 655–665. [Google Scholar] [CrossRef]
- Mol, E.A.; Goumans, M.J.; Doevendans, P.A.; Sluijter, J.P.G.; Vader, P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017, 13, 2061–2065. [Google Scholar] [CrossRef]
- Huang, C.; Quinn, D.; Sadovsky, Y.; Suresh, S.; Hsia, K.J. Formation and size distribution of self-assembled vesicles. Proc. Natl. Acad. Sci. USA 2017, 114, 2910–2915. [Google Scholar] [CrossRef]
- Yoon, J.; Jo, W.; Jeong, D.; Kim, J.; Jeong, H.; Park, J. Generation of nanovesicles with sliced cellular membrane fragments for exogenous material delivery. Biomaterials 2015, 59, 12–20. [Google Scholar] [CrossRef]
- Jeyaram, A.; Jay, S.M. Preservation and storage stability of extracellular vesicles for therapeutic applications. AAPS J. 2018, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Kim, J.-Y.; Cho, R.; Shin, D.-M.; Lee, S.W.; Oh, Y.-M. Adipose stem cell-derived nanovesicles inhibit emphysema primarily via an FGF2-dependent pathway. Exp. Mol. Med. 2017, 49, e284. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-S.; Svennerholm, K.; Shelke, G.V.; Bandeira, E.; Lässer, C.; Jang, S.C.; Chandode, R.; Gribonika, I.; Lötvall, J. Mesenchymal stromal cell-derived nanovesicles ameliorate bacterial outer membrane vesicle-induced sepsis via IL-10. Stem Cell Res. Ther. 2019, 10, 231. [Google Scholar] [CrossRef]
- Zomer, A.; Steenbeek, S.C.; Maynard, C.; van Rheenen, J. Studying extracellular vesicle transfer by a Cre-loxP method. Nat. Protoc. 2016, 11, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Zomer, A.; Maynard, C.; Verweij, F.J.; Kamermans, A.; Schäfer, R.; Beerling, E.; Schiffelers, R.M.; de Wit, E.; Berenguer, J.; Ellenbroek, S.I.J. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015, 161, 1046–1057. [Google Scholar] [CrossRef]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Hölttä, M.; Skantze, P.; Johansson, S.; Sundqvist, M. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Ridder, K.; Keller, S.; Dams, M.; Rupp, A.-K.; Schlaudraff, J.; Del Turco, D.; Starmann, J.; Macas, J.; Karpova, D.; Devraj, K. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014, 12, e1001874. [Google Scholar] [CrossRef]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; San Chan, Y.; Wei, L.; Chin, S.M.; Azad, A. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome–liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef]
- De Jong, O.G.; Kooijmans, S.A.A.; Murphy, D.E.; Jiang, L.; Evers, M.J.W.; Sluijter, J.P.G.; Vader, P.; Schiffelers, R.M. Drug Delivery with Extracellular Vesicles: From Imagination to Innovation. Acc. Chem. Res. 2019, 52, 1761–1770. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilahibaks, N.F.; Lei, Z.; Mol, E.A.; Deshantri, A.K.; Jiang, L.; Schiffelers, R.M.; Vader, P.; Sluijter, J.P.G. Biofabrication of Cell-Derived Nanovesicles: A Potential Alternative to Extracellular Vesicles for Regenerative Medicine. Cells 2019, 8, 1509. https://doi.org/10.3390/cells8121509
Ilahibaks NF, Lei Z, Mol EA, Deshantri AK, Jiang L, Schiffelers RM, Vader P, Sluijter JPG. Biofabrication of Cell-Derived Nanovesicles: A Potential Alternative to Extracellular Vesicles for Regenerative Medicine. Cells. 2019; 8(12):1509. https://doi.org/10.3390/cells8121509
Chicago/Turabian StyleIlahibaks, Nazma F., Zhiyong Lei, Emma A. Mol, Anil K. Deshantri, Linglei Jiang, Raymond M. Schiffelers, Pieter Vader, and Joost P.G. Sluijter. 2019. "Biofabrication of Cell-Derived Nanovesicles: A Potential Alternative to Extracellular Vesicles for Regenerative Medicine" Cells 8, no. 12: 1509. https://doi.org/10.3390/cells8121509
APA StyleIlahibaks, N. F., Lei, Z., Mol, E. A., Deshantri, A. K., Jiang, L., Schiffelers, R. M., Vader, P., & Sluijter, J. P. G. (2019). Biofabrication of Cell-Derived Nanovesicles: A Potential Alternative to Extracellular Vesicles for Regenerative Medicine. Cells, 8(12), 1509. https://doi.org/10.3390/cells8121509





