MiR-33a Controls hMSCS Osteoblast Commitment Modulating the Yap/Taz Expression Through EGFR Signaling Regulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Cell Transfection
2.3. Epidermal Growth Factors (EGF) Cells Treatments
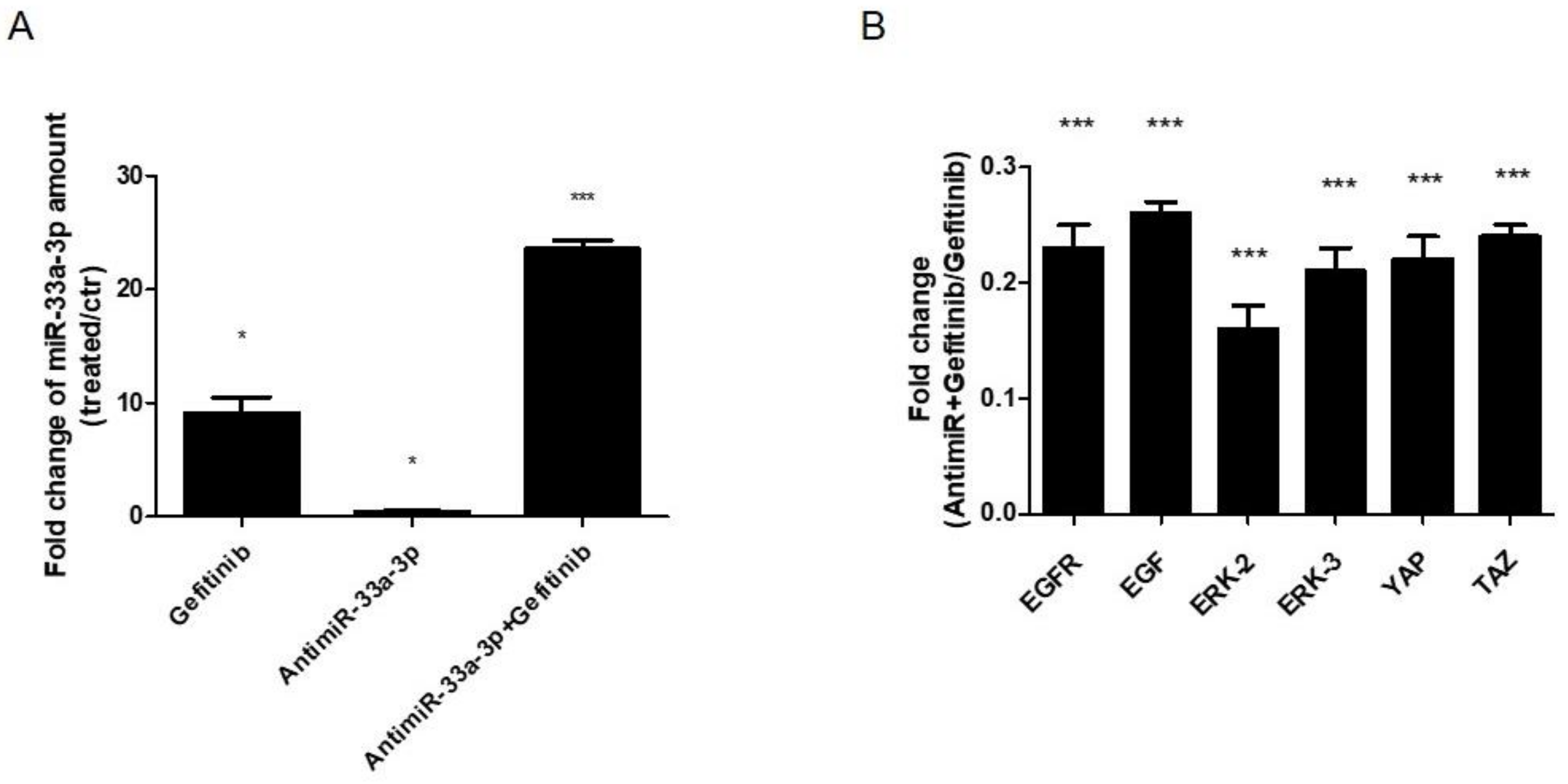
2.4. Gefitinib Cells Treatments
2.5. hMSC Viability (WST-1 Test)
2.6. RNA Extraction and Real-Time PCR
2.7. Western Blot Analysis
2.8. OsteoImage™ Bone Mineralization Assay
2.9. Statistical Analysis
3. Results
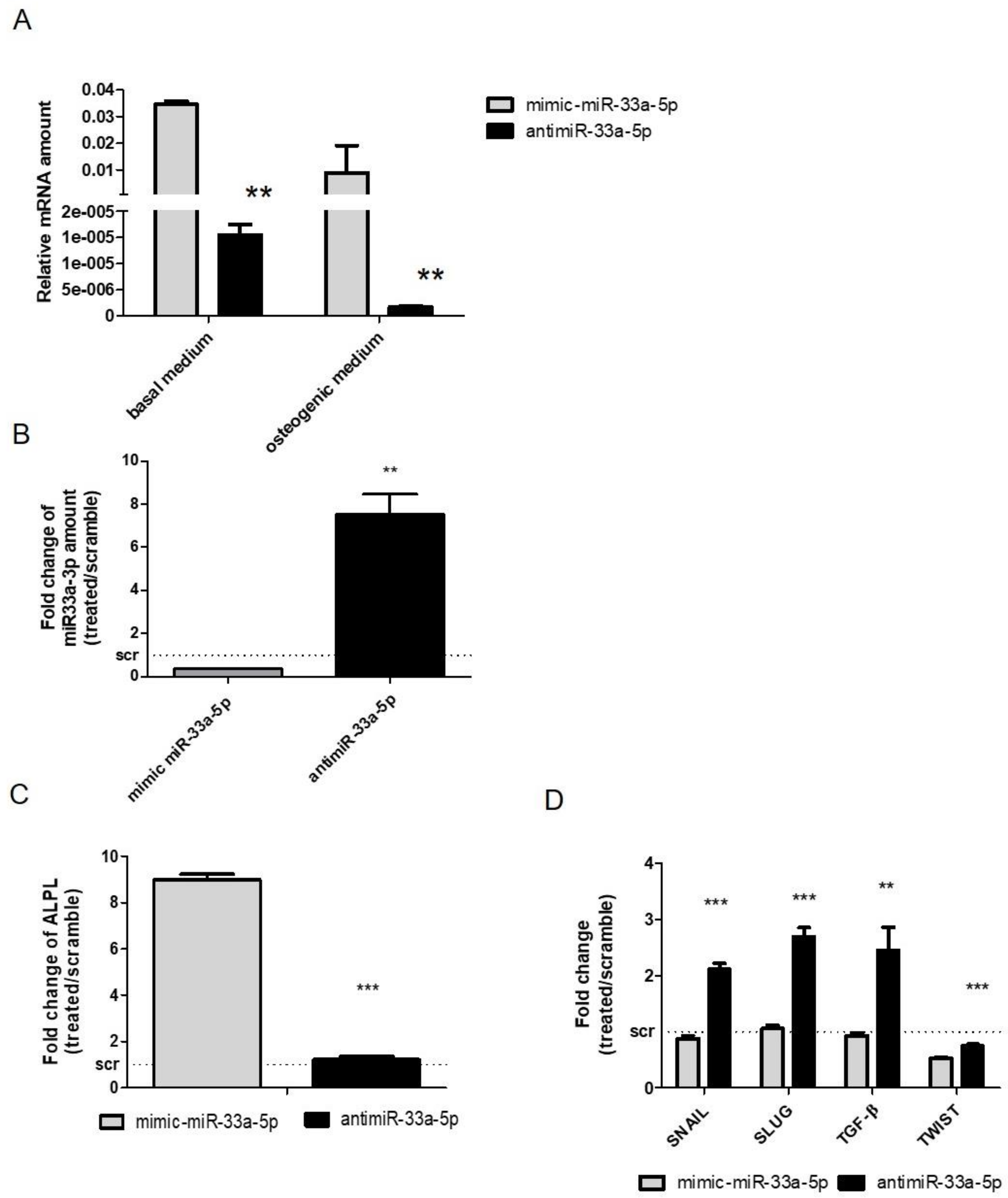
3.1. MiR-33a Family is Involved in the Maintenance of hMSCs and Osteoblast Phenotypes
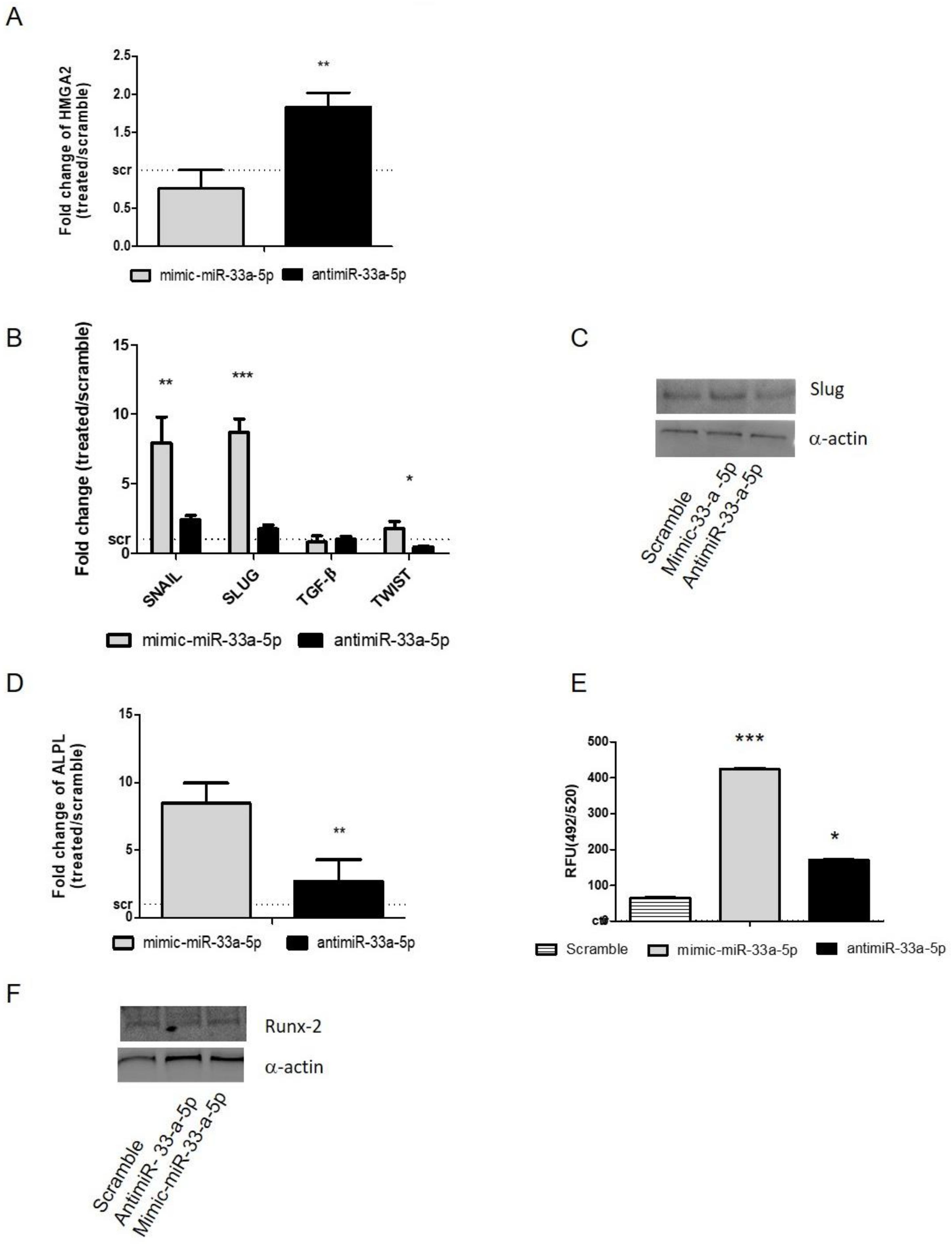
3.2. MiR-33a Family Can Promote hMSCs Osteoblast Commitments
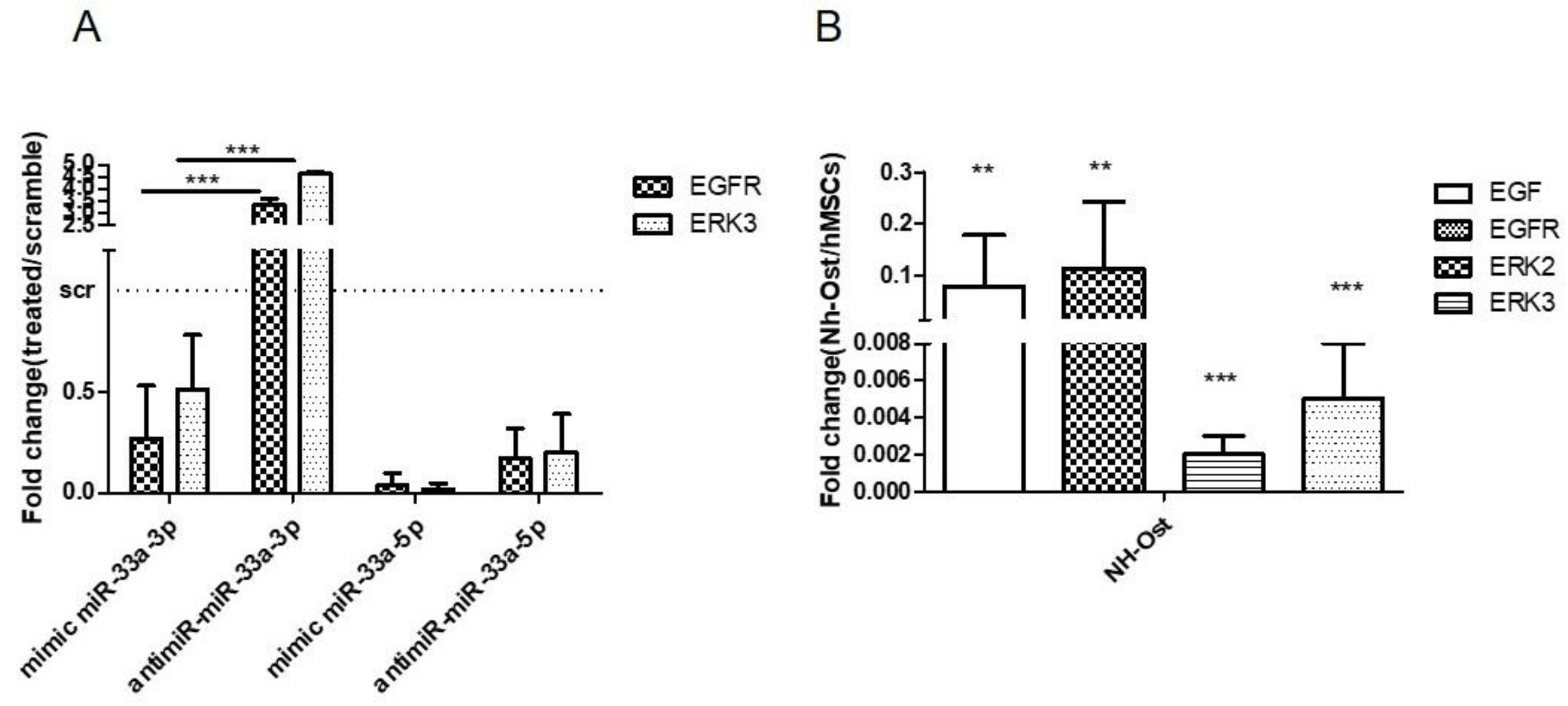
3.3. Modulation of miR-33a-5p and miR-33a-3p Expression Regulates the Activation of EMT Signaling in Pre-Osteoblast Cells
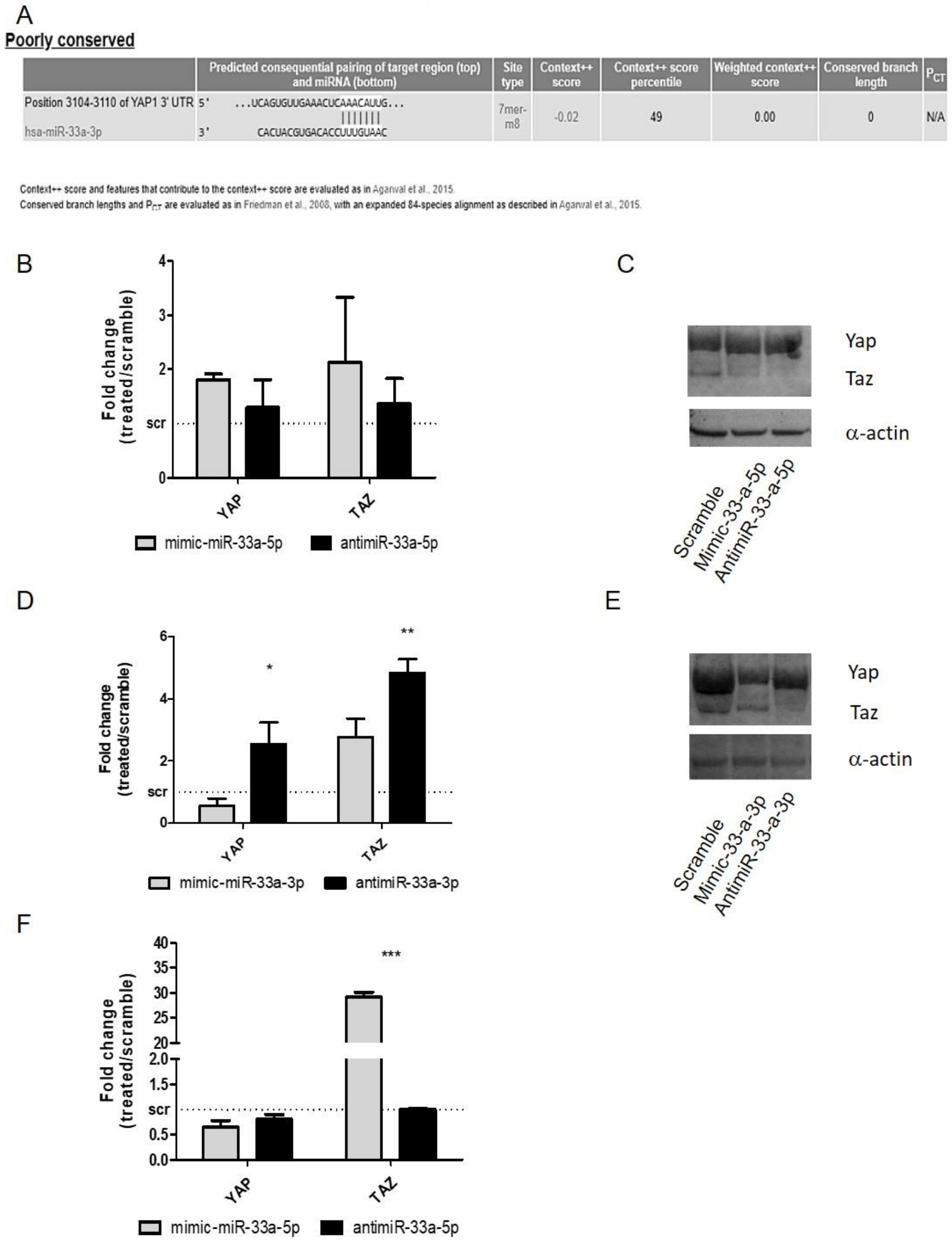
3.4. YAP/TAZ: Possible Mediators of miR-33a Effects During hMSCs Osteoblast Differentiation Process
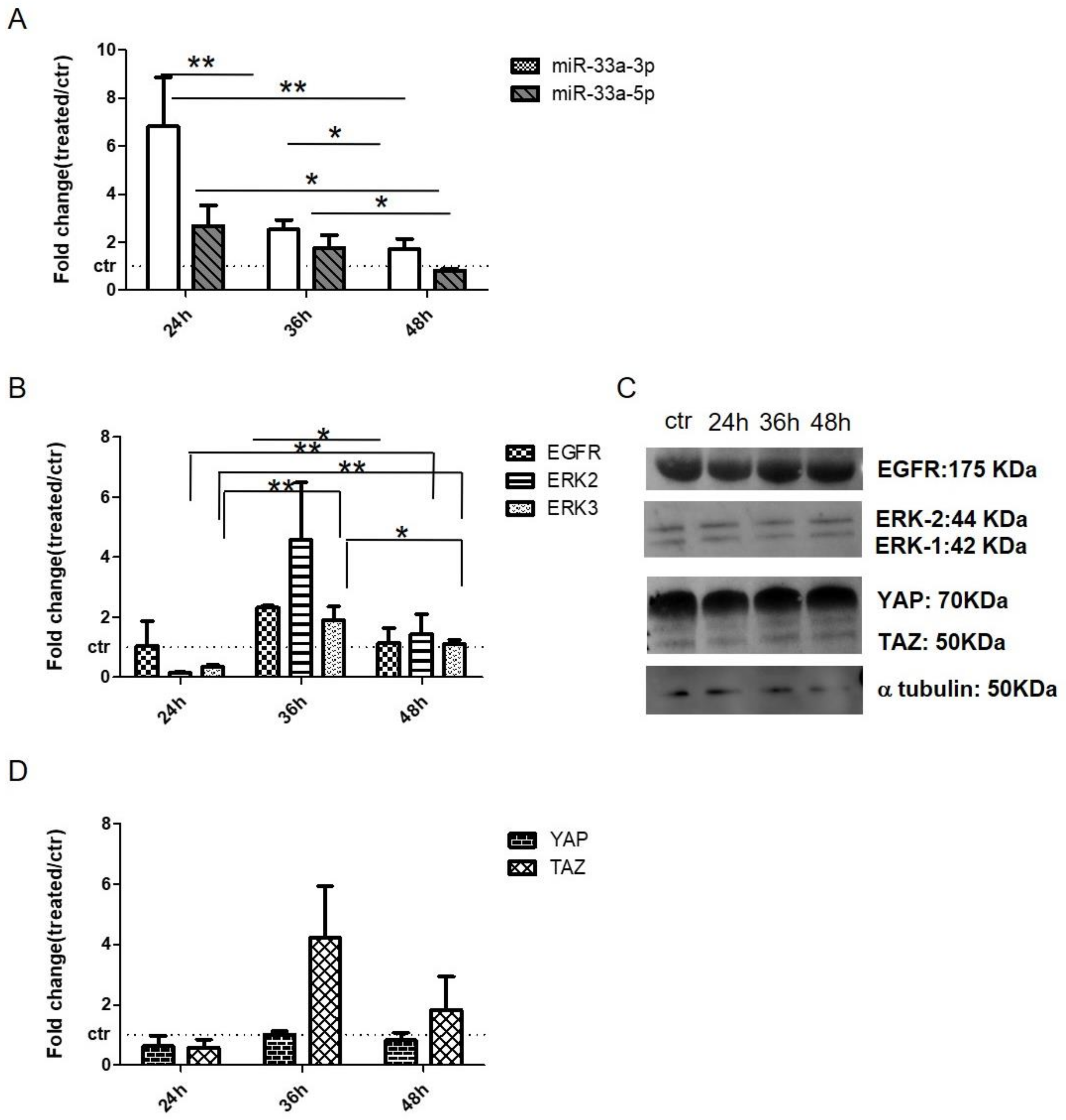
3.5. EGF Treatment Induce the Expression of miR-33a-3p and YAP/TAZ Modulation on hMSCs
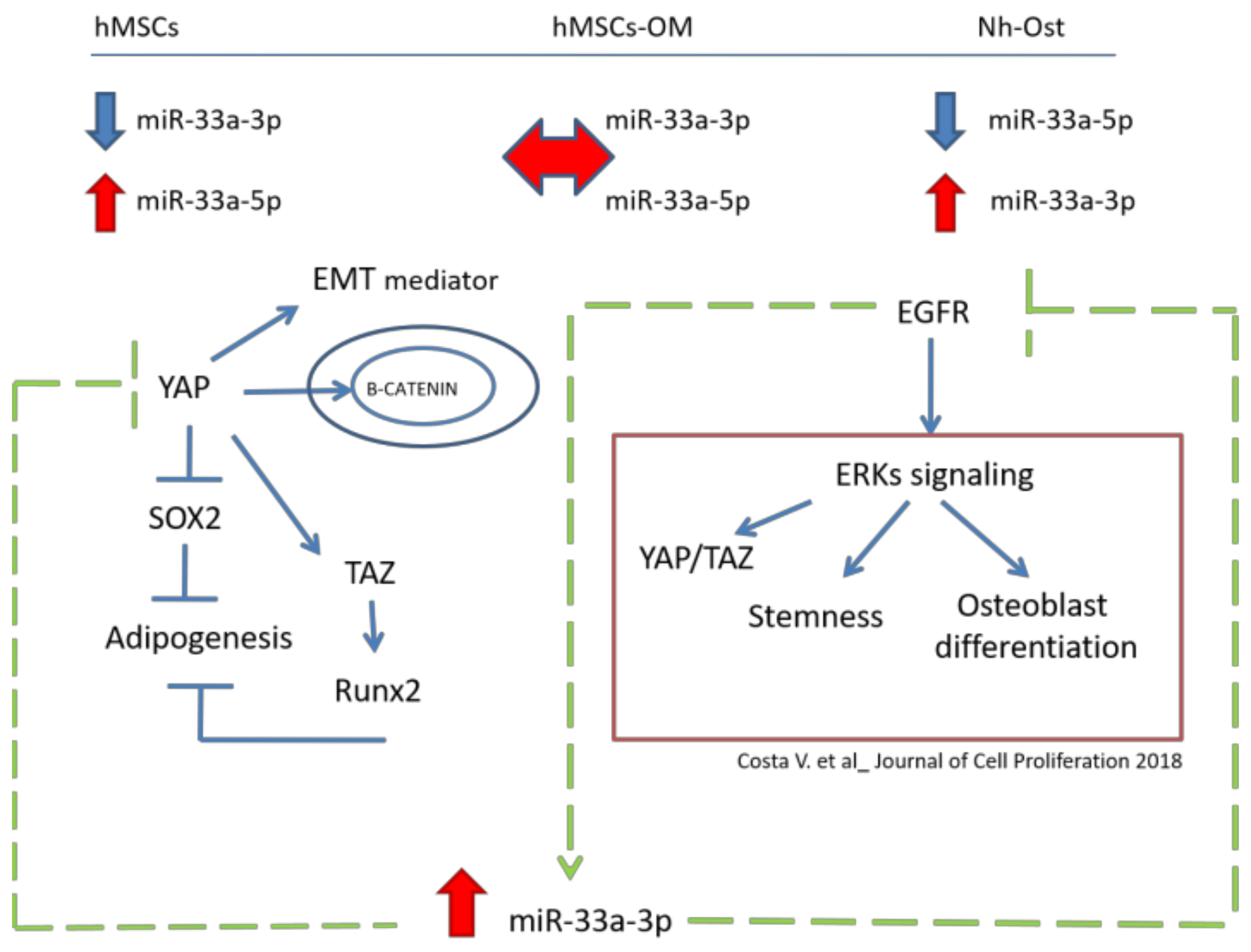
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bellavia, D.; Costa, V.; De Luca, A.; Maglio, M.; Pagani, S.; Fini, M.; Giavaresi, G. Vitamin D Level Between Calcium-Phosphorus Homeostasis and Immune System: New Perspective in Osteoporosis. Curr. Osteoporos. Rep. 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Tamama, K.; Fan, V.H.; Griffith, L.G.; Blair, H.C.; Wells, A. Epidermal growth factor as a candidate for ex vivo expansion of bone marrow-derived mesenchymal stem cells. Stem Cells 2006, 24, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Karsenty, G.; Wagner, E.F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2002, 2, 389–406. [Google Scholar] [CrossRef]
- De Luca, A.; Raimondi, L.; Salamanna, F.; Carina, V.; Costa, V.; Bellavia, D.; Alessandro, R.; Fini, M.; Giavaresi, G. Relevance of 3d culture systems to study osteosarcoma environment. J. Exp. Clin. Cancer Res. 2018, 37, 2. [Google Scholar] [CrossRef]
- Costa, V.; Raimondi, L.; Conigliaro, A.; Salamanna, F.; Carina, V.; De Luca, A.; Bellavia, D.; Alessandro, R.; Fini, M.; Giavaresi, G. Hypoxia-inducible factor 1A may regulate the commitment of mesenchymal stromal cells toward angio-osteogenesis by miRNA-675-5P. Cytotherapy 2017, 19, 1412–1425. [Google Scholar] [CrossRef]
- Costa, V.; Carina, V.; Conigliaro, A.; Raimondi, L.; De Luca, A.; Bellavia, D.; Salamanna, F.; Setti, S.; Alessandro, R.; Fini, M.; et al. miR-31-5p Is a LIPUS-Mechanosensitive MicroRNA that Targets HIF-1α Signaling and Cytoskeletal Proteins. Int. J. Mol. Sci. 2019, 20, 1569. [Google Scholar] [CrossRef]
- Li, H.; Bai, B.; Wang, J.; Xu, Z.; Yan, S.; Liu, G. Identification of key mRNAs and microRNAs in the pathogenesis and progression of osteoarthritis using microarray analysis. Mol. Med. Rep. 2017, 16, 5659–5666. [Google Scholar] [CrossRef][Green Version]
- Chang, C.C.; Venø, M.T.; Chen, L.; Ditzel, N.; Le, D.Q.S.; Dillschneider, P.; Kassem, M.; Kjems, J. Global MicroRNA Profiling in Human Bone Marrow Skeletal-Stromal or Mesenchymal-Stem Cells Identified Candidates for Bone Regeneration. Molecular Therapy 2018, 26, 593–605. [Google Scholar] [CrossRef]
- Yu, S.; Geng, Q.; Ma, J.; Sun, F.; Yu, Y.; Pan, Q.; Hong, A. Heparin-binding EGF-like growth factor and miR-1192 exert opposite effect on Runx2-induced osteogenic differentiation. Cell Death Dis. 2013, 4, 868. [Google Scholar] [CrossRef]
- Garibaldi, F.; Cicchini, C.; Conigliaro, A.; Santangelo, L.; Cozzolino, A.M.; Grassi, G.; Marchetti, A.; Tripodi, M.; Amicone, L. An epistatic mini-circuitry between the transcription factors Snail and HNF4α controls liver stem cell and hepatocyte features exhorting opposite regulation on stemness-inhibiting microRNAs. Cell Death Differ. 2012, 19, 937–946. [Google Scholar] [CrossRef] [PubMed]
- De Frutos, C.A.; Dacquin, R.; Vega, S.; Jurdic, P.; Machuca-Gayet, I.; Nieto, M.A. Snail1 controls bone mass by regulating Runx2 and VDR expression during osteoblast differentiation. EMBO J. 2009, 28, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Weiss, S.J. Snail/Slug-YAP/TAZ complexes cooperatively regulate mesenchymal stem cell function and bone formation. Cell Cycle 2017, 16, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Feinberg, T.; Keller, E.T.; Li, X.Y.; Weiss, S.J. Snail/Slug binding interactions with YAP/TAZ control skeletal stem cell self-renewal and differentiation. Nat. Cell Biol. 2016, 18, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Miraoui, H.; Severe, N.; Vaudin, P.; Pagès, J.C.; Marie, P.J. Molecular silencing of Twist1 enhances osteogenic differentiation of murine mesenchymal stem cells: implication of FGFR2 signaling. J. Cell Biochem. 2010, 110, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Piersma, B.; Bank, R.A.; Boersema, M. Signaling in Fibrosis: TGF-β, WNT, and YAP/TAZ Converge. Front. Med. 2015, 2, 59. [Google Scholar] [CrossRef] [PubMed]
- Varelas, X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 2014, 141, 1614–1626. [Google Scholar] [CrossRef]
- Gumbiner, B.M.; Kim, N.G. The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 2014, 127, 709–717. [Google Scholar] [CrossRef]
- Xue, P.; Wu, X.; Zhou, L.; Ma, H.; Wang, Y.; Liu, Y.; Ma, J.; Li, Y. IGF1 promotes osteogenic differentiation of mesenchymal stem cells derived from rat bone marrow by increasing TAZ expression. Biochem. Biophys. Res. Commun. 2013, 433, 226–231. [Google Scholar] [CrossRef]
- Seo, E.; Basu-Roy, U.; Gunaratne, P.H.; Coarfa, C.; Lim, D.S.; Basilico, C.; Mansukhani, A. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Rep. 2013, 3, 2075–2087. [Google Scholar] [CrossRef]
- Xia, H.; Dai, X.; Yu, H.; Zhou, S.; Fan, Z.; Wei, G.; Tang, Q.; Gong, Q.; Bi, F. EGFR-PI3K-PDK1 pathway regulates YAP signaling in hepatocellular carcinoma: the mechanism and its implications in targeted therapy. Cell Death Dis. 2018, 9, 269. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.; James, S.; Kuntin, D.; Fox, J.; Newling, K.; Hollings, S.; Pennock, R.; Genever, P. Epidermal growth factor can signal via β-catenin to control proliferation of mesenchymal stem cells independently of canonical Wntsignalling. Cell Signal. 2019, 53, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Linder, M.; Hecking, M.; Glitzner, E.; Zwerina, K.; Holcmann, M.; Bakiri, L.; Ruocco, M.G.; Tuckermann, J.; Schett, G.; Wagner, E.F.; et al. EGFR controls bone development by negatively regulating mTOR-signaling during osteoblast differentiation. Cell Death Differ. 2018, 25, 1094–1106. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Honjo, S.; Jin, J.; Chang, S.S.; Scott, A.W.; Chen, Q.; Kalhor, N.; Correa, A.M.; Hofstetter, W.L.; Albarracin, C.T.; et al. The Hippo Coactivator YAP1 Mediates EGFR Overexpression and Confers Chemoresistance in Esophageal Cancer. Clin. Cancer Res. 2015, 21, 2580–2590. [Google Scholar] [CrossRef] [PubMed]
- Watkins, D.J.; Zhou, Y.; Matthews, M.A.; Chen, L.; Besner, G.E. HB-EGF augments the ability of mesenchymal stem cells to attenuate intestinal injury. J. Pediatr. Surg. 2014, 49, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shimizu, E.; Zhang, X.; Partridge, N.C.; Qin, L. EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J. Cell Biochem. 2011, 112, 1749–1760. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, Z.; Wu, S.; Zang, X.; Liu, M.; Shi, J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J. Exp. Clin. Cancer Res. 2014, 33, 12. [Google Scholar] [CrossRef]
- Rice, S.J.; Lai, S.C.; Wood, L.W.; Helsley, K.R.; Runkle, E.A.; Winslow, M.M.; Mu, D. MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1). J. Biol. Chem. 2013, 288, 16348–16360. [Google Scholar] [CrossRef]
- Zuo, B.; Zhu, J.; Li, J.; Wang, C.; Zhao, X.; Cai, G.; Li, Z.; Peng, J.; Wang, P.; Shen, C.; et al. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J. Bone Min. Res. 2015, 30, 330–345. [Google Scholar] [CrossRef]
- Price, N.L.; Fernández-Hernando, C. Novel Role of miR-33 in Regulating of Mitochondrial Function. Circ. Res. 2015, 117, 225–228. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. 2018. Available online: http://www.R-project.org/ (accessed on 20 September 2019).
- Corrado, C.; Saieva, L.; Raimondo, S.; Santoro, A.; De Leo, G.; Alessandro, R. Chronic myelogenous leukaemia exosomes modulate bone marrow microenvironment through activation of epidermal growth factor receptor. J. Cell Mol. Med. 2016, 20, 1829–1839. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.C.; Chen, X.H.; Song, H.X.; Wang, H.S.; Zhang, G.; Wang, H.; Chen, D.Y.; Fang, R.; Liu, H.; Cai, S.H.; et al. Snail regulated by PKC/GSK-3β pathway is crucial for EGF-induced epithelial-mesenchymal transition (EMT) of cancer cells. Cell Tissue Res. 2014, 358, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Battula, V.L.; Evans, K.W.; Hollier, B.G.; Shi, Y.; Marini, F.C.; Ayyanan, A.; Wang, R.Y.; Brisken, C.; Guerra, R.; Andreeff, M.; et al. Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cells 2010, 28, 1435–1445. [Google Scholar] [CrossRef]
- Zou, D.; Han, W.; You, S.; Ye, D.; Wang, L.; Wang, S.; Zhao, J.; Zhang, W.; Jiang, X.; Zhang, X.; et al. In vitro study of enhanced osteogenesis induced by HIF-1α-transduced bone marrow stem cells. Cell Prolif. 2011, 44, 234–243. [Google Scholar] [CrossRef]
- Lo Dico, A.; Costa, V.; Martelli, C.; Diceglie, C.; Rajata, F.; Rizzo, A.; Mancone, C.; Tripodi, M.; Ottobrini, L.; Alessandro, R.; et al. MiR675-5p Acts on HIF-1α to Sustain Hypoxic Responses: A New Therapeutic Strategy for Glioma. Theranostics 2016, 6, 1105–1118. [Google Scholar] [CrossRef]
- Costa, V.; Lo Dico, A.; Rizzo, A.; Rajata, F.; Tripodi, M.; Alessandro, R.; Conigliaro, A. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget 2017, 8, 24292–24302. [Google Scholar] [CrossRef]
- Bellavia, D.; De Luca, A.; Carina, V.; Costa, V.; Raimondi, L.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. Deregulated miRNAs in bone health: Epigenetic roles in osteoporosis. Bone 2019. [Google Scholar] [CrossRef]
- Ouimet, M.; Ediriweera, H.N.; Gundra, U.M.; Sheedy, F.J.; Ramkhelawon, B.; Hutchison, S.B.; Rinehold, K.; van Solingen, C.; Fullerton, M.D.; Cecchini, K.; et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Invest. 2015, 125, 4334–4348. [Google Scholar] [CrossRef]
- Zhang, M.; Gong, W.; Zuo, B.; Chu, B.; Tang, Z.; Zhang, Y.; Yang, Y.; Zhou, D.; Weng, M.; Qin, Y.; et al. The microRNA miR-33a suppresses IL-6-induced tumor progression by binding Twist in gallbladder cancer. Oncotarget 2016, 7, 78640–78652. [Google Scholar] [CrossRef]
- Guo, X.F.; Wang, A.Y.; Liu, J. HIFs-MiR-33a-Twsit1 axis can regulate invasiveness of hepatocellular cancer cells. Eur. Rev. Med. Pharm. Sci. 2016, 20, 3011–3016. [Google Scholar]
- Wang, H.; Sun, Z.; Wang, Y.; Hu, Z.; Zhou, H.; Zhang, L.; Hong, B.; Zhang, S.; Cao, X. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci. Rep. 2016, 6, 23170. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, Z.; Shi, F.; Dong, J.; Dang, L.; Wang, Y.; Sun, Z.; Zhou, H.; Zhang, S.; Cao, X.; et al. Osteoblast-targeted delivery of miR-33-5p attenuates osteopenia development induced by mechanical unloading in mice. Cell Death Dis. 2018, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Carina, V.; Fontana, S.; De Luca, A.; Monteleone, F.; Pagani, S.; Sartori, M.; Setti, S.; Faldini, C.; Alessandro, R.; et al. Osteogenic commitment and differentiation of human mesenchymal stem cells by low-intensity pulsed ultrasound stimulation. J. Cell. Physiol. 2018, 233, 1558–1573. [Google Scholar] [CrossRef] [PubMed]
- Kitazaki, T.; Oka, M.; Nakamura, Y.; Tsurutani, J.; Doi, S.; Yasunaga, M.; Takemura, M.; Yabuuchi, H.; Soda, H.; Kohno, S. Gefitinib, an EGFR tyrosine kinase inhibitor, directly inhibits the function of P-glycoprotein in multidrug resistant cancer cells. Lung Cancer 2005, 49, 337–343. [Google Scholar] [CrossRef]


| Gene | Qiagen Primers QuantiTect Primer Assay | Catalog Number |
|---|---|---|
| RUNX-2 “Runt-related transcription factor 2” | Hs_RUNX2_1_SG | QT00020517 |
| ALPL “Alkaline phosphatase” | Hs_ALPL_1_SG | QT00012957 |
| BGLAP “bone gamma-carboxyglutamic acid-containing protein” | Hs_BGLAP_1_SG | QT00232771 |
| SPP1 “Secreted Phosphoprotein 1” | Hs_SPP1_1_SG | QT01008798 |
| Gene | Custom Made Primers Forward | Custom Made Primers Reverse |
| SNAIL “Snail family transcriptional repressor 1” | GCGAGCTGCAGGACTCTAAT | CCCGCAATGGTCCACAAAAC |
| SLUG “Snail family transcriptional repressor 2” | CATGCCTGTCATACCACAAC | GGTGTCAGATGGAGGAGGG |
| TWIST “Twist family bHLH transcription factor 1” | CGGCCAGGTACATCGACTTC | CAGAGGTGTGAGGATGGTGC |
| TGF-B “Transforming growth factor beta 1” | TGGTGGAAACCCACAACGAA | ACACAGAGATCCGCAGTCCT |
| YAP “Yes associated protein 1” | CCTCGTTTTGCCATGAACCAG | GTTCTTGCTGTTTCAGCCGCAG |
| TAZ “WW domain containing transcription regulator” | TGGACCAAGTACATGAACCACC | TCCACAGCCGACTTGTTCTC |
| HMGA-2 “High mobility group AT-hook 2” | GCGCCTCAGAAGAGAGGAC | GTCTTCCCCTGGGTCTCTTAG |
| EGFR “Epidermal growth factor receptor” | CCTATGTGCAGAGGAATTATGATCTTT | CCACTGTGTTGAGGGCAATG |
| ERK-2 “Extracellular signal-regulated kinases -2” | GCGCTACACTAATCTCTCGT | CTGAGGTGCTGTGTCTTCAA |
| ERK-3 “Extracellular signal-regulated kinases -3” | GAATGGCAAATCTGCTCAATT | ACAGTCCTCCCCACCACTCA |
| Reference Gene | Forward | Reverse |
| ACTB “Beta-Actin” | ATCAAGATCATTGCTCCTCCTGA | CTGCTTGCTGATCCACATCTG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, V.; Carina, V.; Raimondi, L.; De Luca, A.; Bellavia, D.; Conigliaro, A.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. MiR-33a Controls hMSCS Osteoblast Commitment Modulating the Yap/Taz Expression Through EGFR Signaling Regulation. Cells 2019, 8, 1495. https://doi.org/10.3390/cells8121495
Costa V, Carina V, Raimondi L, De Luca A, Bellavia D, Conigliaro A, Salamanna F, Alessandro R, Fini M, Giavaresi G. MiR-33a Controls hMSCS Osteoblast Commitment Modulating the Yap/Taz Expression Through EGFR Signaling Regulation. Cells. 2019; 8(12):1495. https://doi.org/10.3390/cells8121495
Chicago/Turabian StyleCosta, Viviana, Valeria Carina, Lavinia Raimondi, Angela De Luca, Daniele Bellavia, Alice Conigliaro, Francesca Salamanna, Riccardo Alessandro, Milena Fini, and Gianluca Giavaresi. 2019. "MiR-33a Controls hMSCS Osteoblast Commitment Modulating the Yap/Taz Expression Through EGFR Signaling Regulation" Cells 8, no. 12: 1495. https://doi.org/10.3390/cells8121495
APA StyleCosta, V., Carina, V., Raimondi, L., De Luca, A., Bellavia, D., Conigliaro, A., Salamanna, F., Alessandro, R., Fini, M., & Giavaresi, G. (2019). MiR-33a Controls hMSCS Osteoblast Commitment Modulating the Yap/Taz Expression Through EGFR Signaling Regulation. Cells, 8(12), 1495. https://doi.org/10.3390/cells8121495








