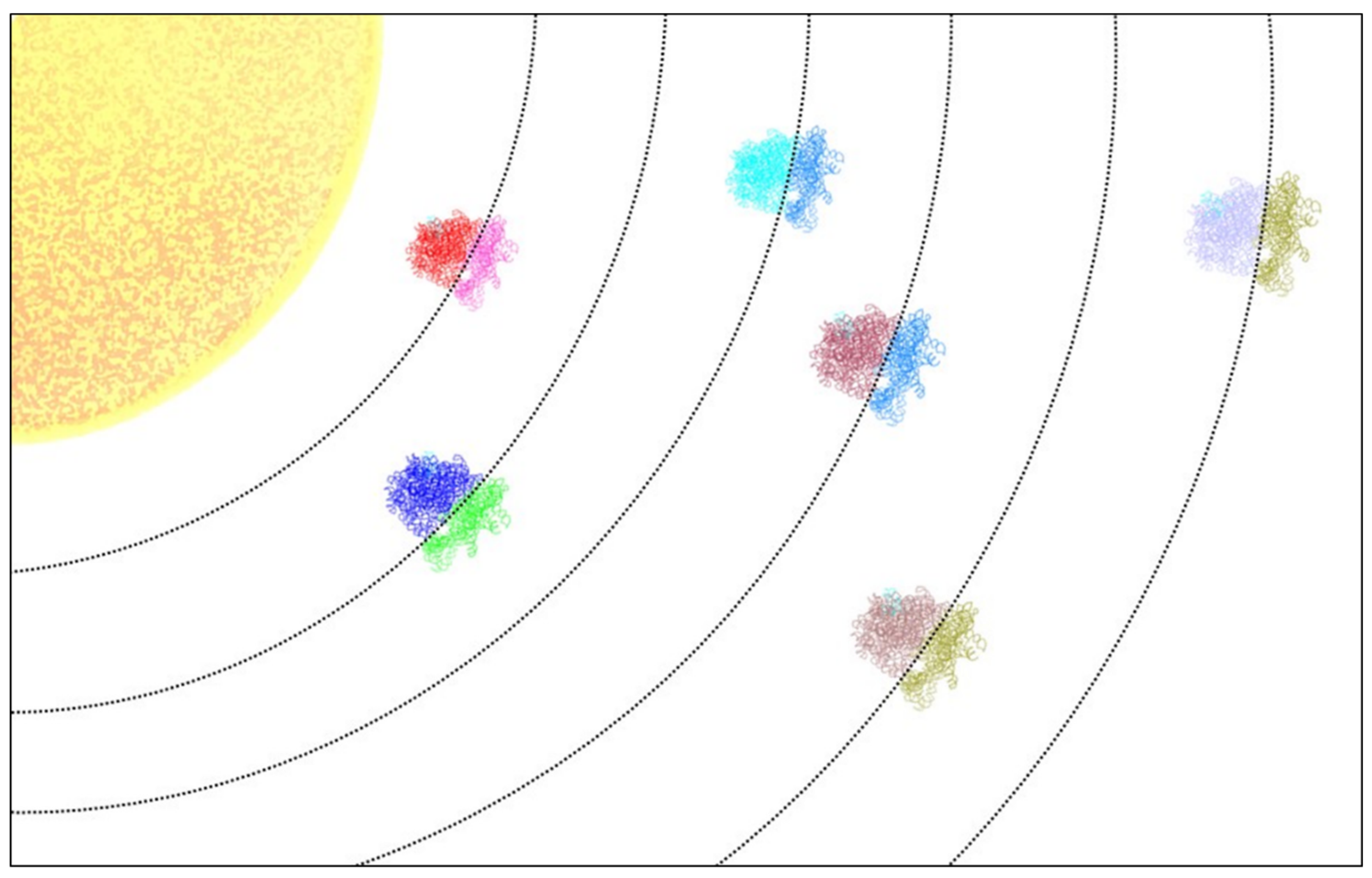
The Expanding Riboverse
Abstract
1. Introduction
2. Aliens Among Us
2.1. Mitoribosomes and Chlororibosomes
2.2. Extreme Ribosomes
2.3. Ribosomes of Parasites
3. Aliens Within Us
3.1. Ribosomal Protein Paralogs
3.2. Immunoribosomes
3.3. Onco-Ribosomes
4. The Expanding Universe of Ribosome Diversity
5. Conclusions and Perspectives
5.1. Medical Implications of the Riboverse
5.2. Designer Ribosomes
6. Final Remarks: Looking Backwards and Forwards
Funding
Acknowledgments
Conflicts of Interest
References
- Crick, F.H.C. Biochemical activities of nucleic acids. The present position of the coding problem. Brookhaven. Symp. Biol. 1959, 12, 35–39. [Google Scholar] [PubMed]
- Mikkola, R.; Kurland, C.G. Is there a unique ribosome phenotype for naturally occurring Escherichia coli? Biochimie 1991, 73, 1061–1066. [Google Scholar] [CrossRef]
- Wickner, R.B.; Leibowitz, M.J. Mak mutants of yeast: Mapping and characterization. J. Bacteriol. 1979, 140, 154–160. [Google Scholar] [PubMed]
- Komili, S.; Farny, N.G.; Roth, F.P.; Silver, P.A. Functional specificity among ribosomal proteins regulates gene expression. Cell 2007, 131, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.W.; Brandl, C.; Haubenreisser, O.; Wimmer, B.; Weber, M.; Karl, T.; Klausegger, A.; Breitenbach, M.; Hintner, H.; von der Haar, T.; et al. Specialized yeast ribosomes: A customized tool for selective mRNA translation. PLoS ONE 2013, 8, e67609. [Google Scholar] [CrossRef]
- Cheng, Z.; Mugler, C.F.; Keskin, A.; Hodapp, S.; Chan, L.Y.-L.; Weis, K.; Mertins, P.; Regev, A.; Jovanovic, M.; Brar, G.A. Small and Large Ribosomal Subunit Deficiencies Lead to Distinct Gene Expression Signatures that Reflect Cellular Growth Rate. Mol. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Genuth, N.R.; Barna, M. Heterogeneity and specialized functions of translation machinery: From genes to organisms. Nat. Rev. Genet. 2018, 19, 431–452. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- Xue, S.; Tian, S.; Fujii, K.; Kladwang, W.; Das, R.; Barna, M. RNA regulons in Hox 5′UTRs confer ribosome specificity to gene regulation and body plan formation. Nature 2015, 517, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Mageeney, C.M.; Ware, V.C. Specialized eRpL22 paralogue-specific ribosomes regulate specific mRNA translation in spermatogenesis in Drosophila melanogaster. Mol. Biol. Cell 2019, 30, 2240–2253. [Google Scholar] [CrossRef]
- Mayor, M.; Queloz, D. A jupiter-mass companion to a solar-type star. Nature 1995, 378, 355–359. [Google Scholar] [CrossRef]
- Greber, B.J.; Ban, N. Structure and Function of the Mitochondrial Ribosome. Annu. Rev. Biochem. 2016, 85, 103–132. [Google Scholar] [CrossRef]
- Agrawal, R.K.; Sharma, M.R. Structural aspects of mitochondrial translational apparatus. Curr. Opin. Struct. Biol. 2012, 22, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Bieri, P.; Leibundgut, M.; Saurer, M.; Boehringer, D.; Ban, N. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017, 36, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Perez Boerema, A.; Aibara, S.; Paul, B.; Tobiasson, V.; Kimanius, D.; Forsberg, B.O.; Wallden, K.; Lindahl, E.; Amunts, A. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 2018, 4, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Clemons, W.M., Jr.; May, J.L.; Wimberly, B.T.; McCutcheon, J.P.; Capel, M.S.; Ramakrishnan, V. Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Nature 1999, 400, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Nissen, P.; Hansen, J.; Capel, M.; Moore, P.B.; Steitz, T.A. Placement of protein and RNA structures into a 5 A-resolution map of the 50S ribosomal subunit. Nature 1999, 400, 841–847. [Google Scholar] [CrossRef]
- Cate, J.H.; Yusupov, M.M.; Yusupova, G.Z.; Earnest, T.N.; Noller, H.F. X-ray crystal structures of 70S ribosome functional complexes. Science (80-.) 1999, 285, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Barandun, J.; Hunziker, M.; Vossbrinck, C.R.; Klinge, S. Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome. Nat. Microbiol. 2019, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hashem, Y.; des Georges, A.; Fu, J.; Buss, S.N.; Jossinet, F.; Jobe, A.; Zhang, Q.; Liao, H.Y.; Grassucci, R.A.; Bajaj, C.; et al. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature 2013, 494, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.P.; Syin, C.; McCutchan, T.F. Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature 1989, 342, 438–440. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.M.; Kurylo, C.M.; Batchelder, J.E.; Theresa Vincent, C.; Blanchard, S.C. Implications of sequence variation on the evolution of rRNA. Chromosom. Res. 2019, 27, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Warner, J.R. Ribosome-omics of the human ribosome. RNA 2014, 20, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, T.; Zhang, X.; McCarthy, J.J. Expression of Muscle-Specific Ribosomal Protein L3-Like Impairs Myotube Growth. J. Cell. Physiol. 2016, 231, 1894–1902. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Alimov, A.P.; McCarthy, J.J. Ribosome Biogenesis is Necessary for Skeletal Muscle Hypertrophy. Exerc. Sport Sci. Rev. 2016, 44, 110–115. [Google Scholar] [CrossRef]
- Meskauskas, A.; Dinman, J.D. Ribosomal protein L3: Gatekeeper to the A-site. Mol.Cell 2007, 25, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Antón, L.C.; Bennink, J.R. Defective ribosomal products (DRiPs): A major source of antigenic peptides for MHC class I molecules? J. Immunol. 1996, 157, 1823–1826. [Google Scholar]
- Yewdell, J.W. Plumbing the sources of endogenous MHC class I peptide ligands. Curr. Opin. Immunol. 2007, 19, 79–86. [Google Scholar] [CrossRef]
- Wei, J.; Yewdell, J.W. Immunoribosomes: Where’s there’s fire, there’s fire. Mol. Immunol. 2018. [Google Scholar] [CrossRef]
- Sulima, S.O.; Hofman, I.J.F.; De Keersmaecker, K.; Dinman, J.D. How ribosomes translate cancer. Cancer Discov. 2017, 7. [Google Scholar] [CrossRef]
- Kampen, K.R.; Sulima, S.O.; Keersmaecker, K. De Rise of the specialized onco-ribosomes. Oncotarget 2018, 9, 35205–35206. [Google Scholar] [CrossRef] [PubMed]
- Sulima, S.O.; Kampen, K.R.; De Keersmaecker, K. Cancer Biogenesis in Ribosomopathies. Cells 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Girardi, T.; Vereecke, S.; Sulima, S.O.; Khan, Y.; Fancello, L.; Briggs, J.W.; Schwab, C.; De Beeck, J.O.; Verbeeck, J.; Royaert, J.; et al. The T-cell leukemia-associated ribosomal RPL10 R98S mutation enhances JAK-STAT signaling. Leukemia 2018, 32. [Google Scholar] [CrossRef] [PubMed]
- Kampen, K.R.; Sulima, S.O.; Verbelen, B.; Girardi, T.; Vereecke, S.; Rinaldi, G.; Verbeeck, J.; Op de Beeck, J.; Uyttebroeck, A.; Meijerink, J.P.P.; et al. The ribosomal RPL10 R98S mutation drives IRES-dependent BCL-2 translation in T-ALL. Leukemia 2019, 33, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Kampen, K.R.; Fancello, L.; Girardi, T.; Rinaldi, G.; Planque, M.; Sulima, S.O.; Loayza-Puch, F.; Verbelen, B.; Vereecke, S.; Verbeeck, J.; et al. Translatome analysis reveals altered serine and glycine metabolism in T-cell acute lymphoblastic leukemia cells. Nat. Commun. 2019, 10, 2542. [Google Scholar] [CrossRef] [PubMed]
- Morgado-Palacin, L.; Varetti, G.; Llanos, S.; Gómez-López, G.; Martinez, D.; Serrano, M. Partial Loss of Rpl11 in Adult Mice Recapitulates Diamond-Blackfan Anemia and Promotes Lymphomagenesis. Cell Rep. 2015, 13, 712–722. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Lee, S.Y.; Gutierrez, A.; Perrigoue, J.; Thapa, R.J.; Tu, Z.; Jeffers, J.R.; Rhodes, M.; Anderson, S.; Oravecz, T.; et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 2012, 120, 3764–3773. [Google Scholar] [CrossRef]
- Ludwig, L.S.; Gazda, H.T.; Eng, J.C.; Eichhorn, S.W.; Thiru, P.; Ghazvinian, R.; George, T.I.; Gotlib, J.R.; Beggs, A.H.; Sieff, C.A.; et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat. Med. 2014, 20, 748–753. [Google Scholar] [CrossRef]
- Sulima, S.O.; Kampen, K.R.; Vereecke, S.; Pepe, D.; Fancello, L.; Verbeeck, J.; Dinman, J.D.; De Keersmaecker, K. Ribosomal Lesions Promote Oncogenic Mutagenesis. Cancer Res. 2019, 79, 320–327. [Google Scholar] [CrossRef]
- Truitt, M.L.; Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 2017, 17, 332. [Google Scholar] [CrossRef]
- Kondrashov, N.; Pusic, A.; Stumpf, C.R.; Shimizu, K.; Hsieh, A.C.; Xue, S.; Ishijima, J.; Shiroishi, T.; Barna, M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 2011, 145, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.W.; Dinman, J.D. Subtractional Heterogeneity: A Crucial Step toward Defining Specialized Ribosomes. Mol. Cell 2017, 67, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Simsek, D.; Tiu, G.C.; Flynn, R.A.; Byeon, G.W.; Leppek, K.; Xu, A.F.; Chang, H.Y.; Barna, M. The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 2017, 169, 1051–1065. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Röst, H.L.; Teruel, M.N.; Barna, M. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 2017, 67, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Susanto, T.T.; Saurabh, S.; Barna, M. Decoding the Function of Expansion Segments in Ribosomes. Mol. Cell 2018, 72, 1013–1020.e6. [Google Scholar] [CrossRef]
- Williamson, N.A.; Raliegh, J.; Morrice, N.A.; Wettenhall, R.E. Post-translational processing of rat ribosomal proteins. Ubiquitous methylation of Lys22 within the zinc-finger motif of RL40 (carboxy-terminal extension protein 52) and tissue-specific methylation of Lys4 in RL29. Eur.J.Biochem. 1997, 246, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Al-Hadid, Q.; Roy, K.; Chanfreau, G.; Clarke, S.G. Methylation of yeast ribosomal protein Rpl3 promotes translational elongation fidelity. RNA 2016, 22, 489–498. [Google Scholar] [CrossRef]
- Arnold, R.J.; Saraswat, S.; Reilly, J.P. Analysis of Methylation, Acetylation, and Other Modifications in Bacterial Ribosomal Proteins; Humana: New York, NY, USA, 2019; pp. 293–307. [Google Scholar]
- Little, R.H.; Grenga, L.; Saalbach, G.; Howat, A.M.; Pfeilmeier, S.; Trampari, E.; Malone, J.G. Adaptive Remodeling of the Bacterial Proteome by Specific Ribosomal Modification Regulates Pseudomonas Infection and Niche Colonisation. PLOS Genet. 2016, 12, e1005837. [Google Scholar] [CrossRef]
- Małecki, J.; Dahl, H.-A.; Moen, A.; Davydova, E.; Falnes, P.Ø. The METTL20 Homologue from Agrobacterium tumefaciens Is a Dual Specificity Protein-lysine Methyltransferase That Targets Ribosomal Protein L7/L12 and the β Subunit of Electron Transfer Flavoprotein (ETFβ). J. Biol. Chem. 2016, 291, 9581–9595. [Google Scholar] [CrossRef]
- Clarke, S.G. The ribosome: A hot spot for the identification of new types of protein methyltransferases. J. Biol. Chem. 2018, 293, 10438–10446. [Google Scholar] [CrossRef]
- Hamidi, T.; Singh, A.K.; Veland, N.; Vemulapalli, V.; Chen, J.; Hardikar, S.; Bao, J.; Fry, C.J.; Yang, V.; Lee, K.A.; et al. Identification of Rpl29 as a major substrate of the lysine methyltransferase Set7/9. J. Biol. Chem. 2018, 293, 12770–12780. [Google Scholar] [CrossRef] [PubMed]
- El Motiam, A.; Vidal, S.; de la Cruz-Herrera, C.F.; Da Silva-Álvarez, S.; Baz-Martínez, M.; Seoane, R.; Vidal, A.; Rodríguez, M.S.; Xirodimas, D.P.; Carvalho, A.S.; et al. Interplay between SUMOylation and NEDDylation regulates RPL11 localization and function. FASEB J. 2019, 33, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Imami, K.; Milek, M.; Bogdanow, B.; Yasuda, T.; Kastelic, N.; Zauber, H.; Ishihama, Y.; Landthaler, M.; Selbach, M. Phosphorylation of the Ribosomal Protein RPL12/uL11 Affects Translation during Mitosis. Mol. Cell 2018, 72, 84–98.e9. [Google Scholar] [CrossRef] [PubMed]
- Arif, A.; Jia, J.; Willard, B.; Li, X.; Fox, P.L. Multisite Phosphorylation of S6K1 Directs a Kinase Phospho-code that Determines Substrate Selection. Mol. Cell 2019, 73, 446–457.e6. [Google Scholar] [CrossRef] [PubMed]
- Graifer, D.; Malygin, A.; Karpova, G. Hydroxylation of protein constituents of the human translation system: Structural aspects and functional assignments. Future Med. Chem. 2019, 11, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Monaco, P.; Marcel, V.; Diaz, J.-J.; Catez, F. 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules 2018, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Krogh, N.; Jansson, M.D.; Häfner, S.J.; Tehler, D.; Birkedal, U.; Christensen-Dalsgaard, M.; Lund, A.H.; Nielsen, H. Profiling of 2′- O -Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016, 44, 7884–7895. [Google Scholar] [CrossRef]
- Tardu, M.; Jones, J.D.; Kennedy, R.T.; Lin, Q.; Koutmou, K.S. Identification and Quantification of Modified Nucleosides in Saccharomyces cerevisiae mRNAs. ACS Chem. Biol. 2019, 14, 1403–1409. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Kurylo, C.M.; Parks, M.M.; Juette, M.F.; Zinshteyn, B.; Altman, R.B.; Thibado, J.K.; Vincent, C.T.; Blanchard, S.C. Endogenous rRNA Sequence Variation Can Regulate Stress Response Gene Expression and Phenotype. Cell Rep. 2018, 25, 236–248.e6. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.D. Pathways to Specialized Ribosomes: The Brussels Lecture. J. Mol. Biol. 2016, 428, 2186–2194. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.M.; Kurylo, C.M.; Dass, R.A.; Bojmar, L.; Lyden, D.; Vincent, C.T.; Blanchard, S.C. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv. 2018, 4, eaao0665. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Zhou, D.; Steitz, T.A.; Polikanov, Y.S.; Gagnon, M.G. Ribosome-Targeting Antibiotics: Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem. 2018, 87, 451–478. [Google Scholar] [CrossRef] [PubMed]
- Myasnikov, A.G.; Kundhavai Natchiar, S.; Nebout, M.; Hazemann, I.; Imbert, V.; Khatter, H.; Peyron, J.-F.; Klaholz, B.P. Structure–function insights reveal the human ribosome as a cancer target for antibiotics. Nat. Commun. 2016, 7, 12856. [Google Scholar] [CrossRef]
- Wong, W.; Bai, X.; Brown, A.; Fernandez, I.S.; Hanssen, E.; Condron, M.; Tan, Y.H.; Baum, J.; Scheres, S.H. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Lintner, N.G.; McClure, K.F.; Petersen, D.; Londregan, A.T.; Piotrowski, D.W.; Wei, L.; Xiao, J.; Bolt, M.; Loria, P.M.; Maguire, B.; et al. Selective stalling of human translation through small-molecule engagement of the ribosome nascent chain. PLOS Biol. 2017, 15, e2001882. [Google Scholar] [CrossRef]
- Petersen, D.N.; Hawkins, J.; Ruangsiriluk, W.; Stevens, K.A.; Maguire, B.A.; O’Connell, T.N.; Rocke, B.N.; Boehm, M.; Ruggeri, R.B.; Rolph, T.; et al. A Small-Molecule Anti-secretagogue of PCSK9 Targets the 80S Ribosome to Inhibit PCSK9 Protein Translation. Cell Chem. Biol. 2016, 23, 1362–1371. [Google Scholar] [CrossRef][Green Version]
- Škrtić, M.; Sriskanthadevan, S.; Jhas, B.; Gebbia, M.; Wang, X.; Wang, Z.; Hurren, R.; Jitkova, Y.; Gronda, M.; Maclean, N.; et al. Inhibition of Mitochondrial Translation as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell 2011, 20, 674–688. [Google Scholar] [CrossRef]
- Vendramin, R.; Verheyden, Y.; Ishikawa, H.; Goedert, L.; Nicolas, E.; Saraf, K.; Armaos, A.; Delli Ponti, R.; Izumikawa, K.; Mestdagh, P.; et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 2018, 25, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.-H.; Chen, A.W.-G.; Fan, J.-Y.; Cheng, W.-L.; Lin, T.-T.; Chen, M.-K.; Liu, C.-S. Aminoglycoside-associated nonsyndromic deafness and speech disorder in mitochondrial A1555G mutation in a family: A case report. Medicine (Baltimore) 2018, 97, e12878. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, J.C.; Zavolan, M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016, 17, 236. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, K.; Yamamoto-Mukai, H.; Shimizu, R.; Waguri, S.; Sou, Y.-S.; Sakamoto, A.; Taya, C.; Shitara, H.; Hara, T.; Chung, C.H.; et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2011, 2, 181. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.G.; Schindler, D.; Davies, J.E. Mapping of trichodermin resistence in Saccharomyces cerevisiae: A genetic locus for a component of the 60S ribosomal subunit. Genetics 1976, 83, 667–673. [Google Scholar] [PubMed]
- Wickner, R.B.; Leibowitz, M.J. Chromosomal and non-chromosomal mutations affecting the “killer character” of Saccharomyces cerevisiae. Genetics 1974, 76, 423–432. [Google Scholar]
- Wickner, R.B.; Porter-Ridley, S.; Fried, H.M.; Ball, S.G. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1982, 79, 4706–4708. [Google Scholar] [CrossRef]
- Jenner, L.; Melnikov, S.; de Loubresse, N.G.N.G.; Ben-Shem, A.; Iskakova, M.; Urzhumtsev, A.; Meskauskas, A.; Dinman, J.; Yusupova, G.; Yusupov, M. Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 2012, 22, 759–767. [Google Scholar] [CrossRef]
- Dinman, J.D.; O’Connor, M. Mutants that affect recoding. In Recoding: Expansion of Decoding Rules Enriches Gene Expression; Atkins, J.F., Gesteland, R.F., Eds.; Nucelic Acids and Molecular Biology; Springer: New York, NY, USA; Dordrecht, The Nethterlands; Heidelberg, Germany; London, UK, 2010; ISBN 978-0-387-89381-5. [Google Scholar]
- Golovina, A.Y.; Bogdanov, A.A.; Dontsova, O.A.; Sergiev, P. V Purification of 30S ribosomal subunit by streptavidin affinity chromatography. Biochimie 2010, 92, 914–917. [Google Scholar] [CrossRef]
- Orelle, C.; Carlson, E.D.; Szal, T.; Florin, T.; Jewett, M.C.; Mankin, A.S. Protein synthesis by ribosomes with tethered subunits. Nature 2015, 524, 119–124. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, G.; Luo, Z.; Lin, Y.; Wang, L.; Guo, Y.; Wang, A.; Jiang, S.; Jiang, Q.; Gong, J.; et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science 2017, 355, eaaf3981. [Google Scholar] [CrossRef] [PubMed]
- Haag, E.S.; Dinman, J.D. Still Searching for Specialized Ribosomes. Dev. Cell 2019, 48, 744–746. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, M.B.; Karbstein, K. Does Functional Specialization of Ribosomes Really Exist? RNA 2019. rna.069823.118. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulima, S.O.; Dinman, J.D. The Expanding Riboverse. Cells 2019, 8, 1205. https://doi.org/10.3390/cells8101205
Sulima SO, Dinman JD. The Expanding Riboverse. Cells. 2019; 8(10):1205. https://doi.org/10.3390/cells8101205
Chicago/Turabian StyleSulima, Sergey O., and Jonathan D. Dinman. 2019. "The Expanding Riboverse" Cells 8, no. 10: 1205. https://doi.org/10.3390/cells8101205
APA StyleSulima, S. O., & Dinman, J. D. (2019). The Expanding Riboverse. Cells, 8(10), 1205. https://doi.org/10.3390/cells8101205





