The MicroRNA Centrism in the Orchestration of Neuroinflammation in Neurodegenerative Diseases
Abstract
1. Introduction
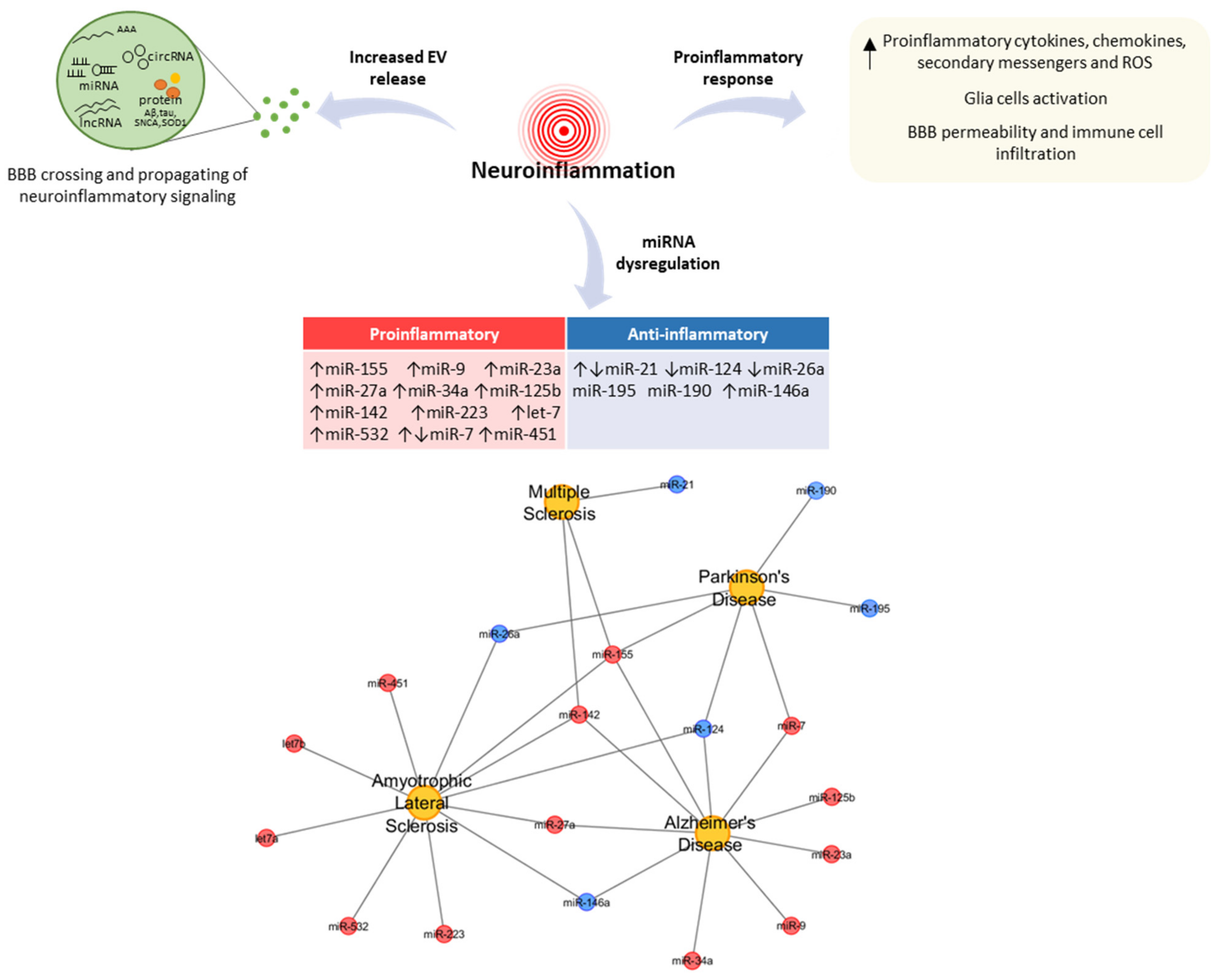
2. MiRNAs as Key Regulators of Neuroinflammation in NDDs
2.1. Proinflammatory miRNAs: Role and Function in NDDs
2.2. Anti-Anflammatory miRNAs: Role and Function in NDDs
3. miRNA-Based ceRNETs in the Orchestration of Neuroinflammation in NDDs
3.1. CircRNAs as ceRNA Regulators of Neuroinflammation in NDDs
3.2. LncRNAs as ceRNA Regulators of Neuroinflammation in NDDs
3.3. Pseudogenes as ceRNA Regulators of Neuroinflammation in NDDs
4. The Neuroinflammatory Role of EVs in NDDs
4.1. EV-Associated miRNAs as Mediators of Neuroinflammation in NDDs
4.2. The Emerging Role of Circulating ceRNETs
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pang, Y.; Mao, C.; Liu, S. Encoding activities of non-coding RNAs. Theranostics 2018, 8, 2496–2507. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Su, Y.; Wu, H.; Pavlosky, A.; Zou, L.L.; Deng, X.; Zhang, Z.X.; Jevnikar, A.M. Regulatory non-coding RNA: New instruments in the orchestration of cell death. Cell Death Dis. 2016, 7, e2333. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Giza, D.E.; Vasilescu, C.; A Calin, G. MicroRNAs and ceRNAs: Therapeutic implications of RNA networks. Expert Opin. Biol. Ther. 2014, 14, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Slota, J.A.; Booth, S.A. MicroRNAs in Neuroinflammation: Implications in Disease Pathogenesis, Biomarker Discovery and Therapeutic Applications. Non Coding RNA 2019, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Disabato, D.J.; Quan, N.; Godbout, J.P. Neuroinflammation: The devil is in the details. J. Neurochem. 2016, 139, 136–153. [Google Scholar] [CrossRef]
- Kempuraj, D.; Thangavel, R.; Natteru, P.; Selvakumar, G.; Saeed, D.; Zahoor, H.; Zaheer, S.; Iyer, S.; Zaheer, A. Neuroinflammation Induces Neurodegeneration. J. Neurol. Neurosurg. Spine 2016, 1, 1003. [Google Scholar] [PubMed]
- Aiello, M.; Cavaliere, C.; Fiorenza, D.; Duggento, A.; Passamonti, L.; Toschi, N. Neuroinflammation in Neurodegenerative Diseases: Current Multi-modal Imaging Studies and Future Opportunities for Hybrid PET/MRI. Neuroscience 2019, 403, 125–135. [Google Scholar] [CrossRef]
- Schuitemaker, A.; Kropholler, M.A.; Boellaard, R.; van der Flier, W.M.; Kloet, R.W.; van der Doef, T.F.; Knol, D.L.; Windhorst, A.D.; Luurtsema, G.; Barkhof, F.; et al. Microglial activation in Alzheimer’s disease: An (R)-[11C]PK11195 positron emission tomography study. Neurobiol. Aging 2013, 34, 128–136. [Google Scholar] [CrossRef]
- Corcia, P.; Tauber, C.; Vercoullie, J.; Arlicot, N.; Prunier, C.; Praline, J.; Nicolas, G.; Venel, Y.; Hommet, C.; Baulieu, J.L.; et al. Molecular Imaging of Microglial Activation in Amyotrophic Lateral Sclerosis. PLoS ONE 2012, 7, e52941. [Google Scholar] [CrossRef] [PubMed]
- Airas, L.; Nylund, M.; Rissanen, E. Evaluation of Microglial Activation in Multiple Sclerosis Patients Using Positron Emission Tomography. Front. Neurol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Ghadery, C.; Koshimori, Y.; Coakeley, S.; Harris, M.; Rusjan, P.; Kim, J.; Houle, S.; Strafella, A.P. Microglial activation in Parkinson’s disease using [18F]-FEPPA. J. Neuroinflamm. 2017, 14, 8. [Google Scholar] [CrossRef] [PubMed]
- Parisi, C.; Arisi, I.; D’Ambrosi, N.; Storti, A.E.; Brandi, R.; D’Onofrio, M.; Volonté, C. Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell Death Dis. 2013, 4, e959. [Google Scholar] [CrossRef] [PubMed]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132, 3342–3352. [Google Scholar] [CrossRef]
- Hutchison, E.R.; Kawamoto, E.M.; Taub, D.D.; Lal, A.; Abdelmohsen, K.; Zhang, Y.; Wood, W.H.; Lehrmann, E.; Camandola, S.; Becker, K.G.; et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 2013, 61, 1018–1028. [Google Scholar] [CrossRef]
- Thome, A.D.; Harms, A.S.; Volpicelli-Daley, L.A.; Standaert, D.G. microRNA-155 Regulates Alpha-Synuclein-Induced Inflammatory Responses in Models of Parkinson Disease. J. Neurosci. 2016, 36, 2383–2390. [Google Scholar] [CrossRef]
- Cai, Y.; Wan, J. Competing Endogenous RNA Regulations in Neurodegenerative Disorders: Current Challenges and Emerging Insights. Front. Mol. Neurosci. 2018, 11, 11. [Google Scholar] [CrossRef]
- Yu, S.; Yu, M.; He, X.; Wen, L.; Bu, Z.; Feng, J. KCNQ1OT1 promotes autophagy by regulating miR-200a/FOXO3/ATG7 pathway in cerebral ischemic stroke. Aging Cell 2019, 18, e12940. [Google Scholar] [CrossRef]
- Zhang, H.; Li, D.; Zhang, Y.; Li, J.; Ma, S.; Zhang, J.; Xiong, Y.; Wang, W.; Li, N.; Xia, L. Knockdown of lncRNA BDNF-AS suppresses neuronal cell apoptosis via downregulating miR-130b-5p target gene PRDM5 in acute spinal cord injury. RNA Biol. 2018, 15, 1071–1080. [Google Scholar] [CrossRef]
- Yan, H.; Rao, J.; Yuan, J.; Gao, L.; Huang, W.; Zhao, L.; Ren, J. Long Non-Coding RNA MEG3 Functions as a Competing Endogenous RNA to Regulate Ischemic Neuronal Death by Targeting miR-21/PDCD4 Signaling Pathway. Cell Death Dis. 2017, 8, 3211. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Fu, X.; Nagvekar, R.; Calin, G.A. MicroRNAs, Regulatory Messengers Inside and Outside Cancer Cells. In Exosomes, Stem Cells and MicroRNA: Aging, Cancer and Age Related Disorders, Advances in Experimental Medicine and Biology; Mettinger, K.L., Rameshwar, P., Kumar, V., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 87–108. ISBN 978-3-319-74470-4. [Google Scholar]
- Prada, I.; Gabrielli, M.; Turola, E.; Iorio, A.; D’Arrigo, G.; Parolisi, R.; De Luca, M.; Pacifici, M.; Bastoni, M.; Lombardi, M.; et al. Glia-to-neuron transfer of miRNAs via extracellular vesicles: A new mechanism underlying inflammation-induced synaptic alterations. Acta Neuropathol. 2018, 135, 529–550. [Google Scholar] [CrossRef] [PubMed]
- Brites, D.; Fernandes, A. Neuroinflammation and Depression: Microglia Activation, Extracellular Microvesicles and microRNA Dysregulation. Front. Cell. Neurosci. 2015, 9, 14645. [Google Scholar] [CrossRef] [PubMed]
- Njock, M.S.; Cheng, H.S.; Dang, L.T.H.; Nazari-Jahantigh, M.; Lau, A.C.; Boudreau, E.; Roufaiel, M.; Cybulsky, M.I.; Schober, A.; Fish, J.E. Endothelial cells suppress monocyte activation through secretion of extracellular vesicles containing antiinflammatory microRNAs. Blood 2015, 125, 3202–3212. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef]
- Becher, B.; Spath, S.; Goverman, J. Cytokine networks in neuroinflammation. Nature Reviews Immunology 2017, 17, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Strokin, M.; Sergeeva, M.; Reiser, G. Role of Ca2+-independent phospholipase A2 and n−3 polyunsaturated fatty acid docosahexaenoic acid in prostanoid production in brain: Perspectives for protection in neuroinflammation. Int. J. Dev. Neurosci. 2004, 22, 551–557. [Google Scholar] [CrossRef]
- Skuljec, J.; Sun, H.; Pul, R.; Bénardais, K.; Ragancokova, D.; Moharregh-Khiabani, D.; Kotsiari, A.; Trebst, C.; Stangel, M. CCL5 induces a pro-inflammatory profile in microglia in vitro. Cell. Immunol. 2011, 270, 164–171. [Google Scholar] [CrossRef]
- Karunakaran, I.; Alam, S.; Jayagopi, S.; Frohberger, S.J.; Hansen, J.N.; Kuehlwein, J.; Hölbling, B.V.; Schumak, B.; Hübner, M.P.; Gräler, M.H.; et al. Neural sphingosine 1-phosphate accumulation activates microglia and links impaired autophagy and inflammation. Glia 2019, 67, 1859–1872. [Google Scholar] [CrossRef]
- Zheng, Y.; Hou, X.; Yang, S. Lidocaine Potentiates SOCS3 to Attenuate Inflammation in Microglia and Suppress Neuropathic Pain. Cell. Mol. Neurobiol. 2019, 39, 1081–1092. [Google Scholar] [CrossRef]
- Burmeister, A.R.; Johnson, M.B.; Marriott, I. Murine astrocytes are responsive to the pro-inflammatory effects of IL-20. Neurosci. Lett. 2019, 708, 134334. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhu, L.; Ma, R.; Ren, J.; Wang, J.; Gao, S.; Yang, D.; Ning, K.; Ling, B.; Lu, B.; et al. Astrocytic miR-324-5p is essential for synaptic formation by suppressing the secretion of CCL5 from astrocytes. Cell Death Dis. 2019, 10, 141. [Google Scholar] [CrossRef]
- Pober, J.S.; Sessa, W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 2007, 7, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, A.D.; Zarruk, J.G.; Healy, L.M.; Jesudasan, S.J.B.; Jhelum, P.; Salmon, C.K.; Formanek, A.; Russo, M.V.; Antel, J.P.; McGAVERN, D.B.; et al. Peripherally derived macrophages modulate microglial function to reduce inflammation after CNS injury. PLoS Biol. 2018, 16, e2005264. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Sætrom, P. MicroRNAs – targeting and target prediction. New Biotechnol. 2010, 27, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Rajewsky, N. microRNA target predictions in animals. Nat. Genet. 2006, 38, S8–S13. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Navarro, F.; Maher, C.A.; Maliszewski, L.E.; Yan, N.; O’Day, E.; Chowdhury, D.; Dykxhoorn, D.M.; Tsai, P.; Hofmann, O.; et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3’UTR microRNA recognition elements. Mol. Cell 2009, 35, 610–625. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, X.; Cai, Z.; Zhou, J.; Cao, R.; Zhao, Y.; Chen, Z.; Wang, D.; Ruan, W.; Zhao, Q.; et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 2018, 25, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tang, M.; Zong, P.; Liu, H.; Zhang, T.; Liu, Y.; Zhao, Y. MiRNA-155 Regulates the Th17/Treg Ratio by Targeting SOCS1 in Severe Acute Pancreatitis. Front. Physiol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.S.; Rao, V.T.; Durafourt, B.A.; Bedell, B.J.; Ludwin, S.K.; Antel, J.P.; Bar-Or, A. miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann. Neurol. 2013, 74, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, T.; Madireddy, L.; Sprenger, T.; Khankhanian, P.; Magon, S.; Naegelin, Y.; Caverzasi, E.; Lindberg, R.L.P.; Kappos, L.; Hauser, S.L.; et al. Genetic associations with brain cortical thickness in multiple sclerosis. GenesBrain Behav. 2015, 14, 217–227. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Cialic, R.; Krasemann, S.; Murugaiyan, G.; Fanek, Z.; Greco, D.J.; Wu, P.M.; Doykan, C.E.; Kiner, O.; et al. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann. Neurol. 2015, 77, 75–99. [Google Scholar] [CrossRef] [PubMed]
- Guedes, J.R.; Custódia, C.M.; Silva, R.J.; de Almeida, L.P.; Pedroso de Lima, M.C.; Cardoso, A.L. Early miR-155 upregulation contributes to neuroinflammation in Alzheimer’s disease triple transgenic mouse model. Hum. Mol. Genet. 2014, 23, 6286–6301. [Google Scholar] [CrossRef]
- Pogue, A.I.; Lukiw, W.J. Up-regulated Pro-inflammatory MicroRNAs (miRNAs) in Alzheimer’s disease (AD) and Age-Related Macular Degeneration (AMD). Cell Mol. Neurobiol. 2018, 38, 1021–1031. [Google Scholar] [CrossRef]
- Lukiw, W.J. NF-κB-regulated, proinflammatory miRNAs in Alzheimer’s disease. Alzheimer’s Res. Ther. 2012, 4, 47. [Google Scholar] [CrossRef]
- Lukiw, W.J. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. NeuroReport 2007, 18, 297–300. [Google Scholar] [CrossRef]
- Mandolesi, G.; De Vito, F.; Musella, A.; Gentile, A.; Bullitta, S.; Fresegna, D.; Sepman, H.; Di Sanza, C.; Haji, N.; Mori, F.; et al. miR-142-3p Is a Key Regulator of IL-1β-Dependent Synaptopathy in Neuroinflammation. J. Neurosci. 2017, 37, 546–561. [Google Scholar] [CrossRef] [PubMed]
- Talebi, F.; Ghorbani, S.; Chan, W.F.; Boghozian, R.; Masoumi, F.; Ghasemi, S.; Vojgani, M.; Power, C.; Noorbakhsh, F. MicroRNA-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J. Neuroinflamm. 2017, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.; Bossers, K.; Janky, R.; Salta, E.; Frigerio, C.S.; Barbash, S.; Rothman, R.; Sierksma, A.S.R.; Thathiah, A.; Greenberg, D.; et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 2013, 5, 1613–1634. [Google Scholar] [CrossRef] [PubMed]
- Paez-Colasante, X.; Figueroa-Romero, C.; Sakowski, S.A.; Goutman, S.A.; Feldman, E.L. Amyotrophic lateral sclerosis: Mechanisms and therapeutics in the epigenomic era. Nat. Rev. Neurol. 2015, 11, 266–279. [Google Scholar] [CrossRef]
- Feng, J.; Li, A.; Deng, J.; Yang, Y.; Dang, L.; Ye, Y.; Li, Y.; Zhang, W. miR-21 attenuates lipopolysaccharide-induced lipid accumulation and inflammatory response: Potential role in cerebrovascular disease. Lipids Health Dis. 2014, 13, 27. [Google Scholar] [CrossRef]
- Fenoglio, C.; Cantoni, C.; De Riz, M.; Ridolfi, E.; Cortini, F.; Serpente, M.; Villa, C.; Comi, C.; Monaco, F.; Mellesi, L.; et al. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci. Lett. 2011, 504, 9–12. [Google Scholar] [CrossRef]
- Sanders, K.A.; Benton, M.C.; Lea, R.A.; Maltby, V.E.; Agland, S.; Griffin, N.; Scott, R.J.; Tajouri, L.; Lechner-Scott, J. Next-generation sequencing reveals broad down-regulation of microRNAs in secondary progressive multiple sclerosis CD4+ T cells. Clin. Epigenetics 2016, 8, 87. [Google Scholar] [CrossRef]
- Yao, L.; Ye, Y.; Mao, H.; Lu, F.; He, X.; Lu, G.; Zhang, S. MicroRNA-124 regulates the expression of MEKK3 in the inflammatory pathogenesis of Parkinson’s disease. J. NeuroInflamm. 2018, 15, 13. [Google Scholar] [CrossRef]
- Yao, L.; Zhu, Z.; Wu, J.; Zhang, Y.; Zhang, H.; Sun, X.; Qian, C.; Wang, B.; Xie, L.; Zhang, S.; et al. MicroRNA-124 regulates the expression of p62/p38 and promotes autophagy in the inflammatory pathogenesis of Parkinson’s disease. FASEB J. 2019, 33, 8648–8665. [Google Scholar] [CrossRef]
- An, F.; Gong, G.; Wang, Y.; Bian, M.; Yu, L.; Wei, C. MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget 2017, 8, 114065–114071. [Google Scholar] [CrossRef]
- Ponomarev, E.D.; Veremeyko, T.; Barteneva, N.; Krichevsky, A.M.; Weiner, H.L. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α–PU.1 pathway. Nat. Med. 2011, 17, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, Q.; Chen, X.; Li, C.; Cao, B.; Ou, R.; Hadano, S.; Shang, H.-F. Aberration of miRNAs Expression in Leukocytes from Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2016, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- D’Erchia, A.M.; Gallo, A.; Manzari, C.; Raho, S.; Horner, D.S.; Chiara, M.; Valletti, A.; Aiello, I.; Mastropasqua, F.; Ciaccia, L.; et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci. Rep. 2017, 7, 10046. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.; Wyles, M.; Heath, P.R.; Kazoka, M.; Wollff, H.; Shaw, P.J.; Kirby, J. Small RNA Sequencing of Sporadic Amyotrophic Lateral Sclerosis Cerebrospinal Fluid Reveals Differentially Expressed miRNAs Related to Neural and Glial Activity. Front. Mol. Neurosci. 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Zhang, C.; Guan, Y.; Chen, Y.; Lu, Q.; Jie, L.; Gao, H.; Du, H.; Zhang, H.; Liu, Y.; et al. Screening the expression characteristics of several miRNAs in G93A-SOD1 transgenic mouse: Altered expression of miRNA-124 is associated with astrocyte differentiation by targeting Sox2 and Sox9. J. Neurochem. 2018, 145, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bhatia, H.S.; De Oliveira, A.C.P.; Fiebich, B.L. microRNA-26a modulates inflammatory response induced by toll-like receptor 4 stimulation in microglia. J. Neurochem. 2015, 135, 1189–1202. [Google Scholar] [CrossRef] [PubMed]
- Liguori, M.; Nuzziello, N.; Introna, A.; Consiglio, A.; Licciulli, F.; D’Errico, E.; Scarafino, A.; Distaso, E.; Simone, I.L. Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients With Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2018, 11, 1–16. [Google Scholar] [CrossRef]
- Martins, M.; Rosa, A.; Guedes, L.C.; Fonseca, B.V.; Gotovac, K.; Violante, S.; Mestre, T.; Coelho, M.; Rosa, M.M.; Martin, E.R.; et al. Convergence of miRNA Expression Profiling, α-Synuclein Interacton and GWAS in Parkinson’s Disease. PLoS ONE 2011, 6, e25443. [Google Scholar] [CrossRef]
- Ren, Y.; Li, H.; Xie, W.; Wei, N.; Liu, M. MicroRNA-195 triggers neuroinflammation in Parkinson’s disease in a Rho-associated kinase 1-dependent manner. Mol. Med. Rep. 2019, 19, 5153–5161. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, X.; Dong, R.; Sun, L.; Chen, L.; Zhang, Z.; Peng, M. MicroRNA-181b-5p attenuates early postoperative cognitive dysfunction by suppressing hippocampal neuroinflammation in mice. Cytokine 2019, 120, 41–53. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, S.; Chen, J.; Cai, H.; Huang, W.; Zhang, Y.; Wang, L.; Xing, Y. MicroRNA-190 alleviates neuronal damage and inhibits neuroinflammation via Nlrp3 in MPTP-induced Parkinson’s disease mouse model. J. Cell. Physiol. 2019, 234, 23379–23387. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lu, M.; Du, R.-H.; Qiao, C.; Jiang, C.Y.; Zhang, K.Z.; Ding, J.H.; Hu, G. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol. Neurodegener. 2016, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Paik, J.H.; Jang, J.Y.; Jeon, Y.K.; Kim, W.Y.; Kim, T.M.; Heo, D.S.; Kim, C.W. MicroRNA-146a Downregulates NF B Activity via Targeting TRAF6 and Functions as a Tumor Suppressor Having Strong Prognostic Implications in NK/T Cell Lymphoma. Clin. Cancer Res. 2011, 17, 4761–4771. [Google Scholar] [CrossRef] [PubMed]
- Ye, E.A.; Steinle, J.J. miR-146a Attenuates Inflammatory Pathways Mediated by TLR4/NF-κB and TNFα to Protect Primary Human Retinal Microvascular Endothelial Cells Grown in High Glucose. Mediat. Inflamm. 2016, 2016, 3958453. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cerutti, C.; Lopez-Ramirez, M.A.; Pryce, G.; King-Robson, J.; Simpson, J.E.; van der Pol, S.M.; Hirst, M.C.; de Vries, H.E.; Sharrack, B.; et al. Brain Endothelial miR-146a Negatively Modulates T-Cell Adhesion through Repressing Multiple Targets to Inhibit NF-κB Activation. J. Cereb. Blood Flow Metab. 2015, 35, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, P.N.; Dua, P.; Lukiw, W.J. Up-Regulation of miRNA-146a in Progressive, Age-Related Inflammatory Neurodegenerative Disorders of the Human CNS. Front. Neurol. 2014, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Kartha, R.V.; Subramanian, S. Competing endogenous RNAs (ceRNAs): New entrants to the intricacies of gene regulation. Front. Genet. 2014, 5, 8. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA Regulatory Network in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Zhao, Y.; Alexandrov, P.N.; Jaber, V.; Lukiw, W.J. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes (Basel) 2016, 7, 116. [Google Scholar] [CrossRef]
- Shi, Z.; Chen, T.; Yao, Q.; Zheng, L.; Zhang, Z.; Wang, J.; Hu, Z.; Cui, H.; Han, Y.; Han, X.; et al. The circular RNA ci RS—7 promotes APP and BACE 1 degradation in an NF –κB-dependent manner. FEBS J. 2017, 284, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Chami, L.; Checler, F. BACE1 is at the crossroad of a toxic vicious cycle involving cellular stress and β-amyloid production in Alzheimer’s disease. Mol. Neurodegener. 2012, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, D.; Scarpini, E. Inflammation in Alzheimer’s Disease: A Friend or Foe? Front. Biosci. 2011, 3, 252–266. [Google Scholar] [CrossRef]
- Li, T.R.; Jia, Y.J.; Wang, Q.; Shao, X.Q.; Lv, R.J. Circular RNA: A new star in neurological diseases. Int. J. Neurosci. 2017, 127, 726–734. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, D.; Li, H.; Li, H.; Feng, C.; Zhang, W. Characterization of circRNA-Associated-ceRNA Networks in a Senescence-Accelerated Mouse Prone 8 Brain. Mol. Ther. 2017, 25, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, Y.; Faghihi, M.A.; Magistri, M.; Álvarez-García, Óscar; Lotz, M.; Wahlestedt, C. Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 2015, 11, 967–976. [Google Scholar] [CrossRef]
- May, P. The low-density lipoprotein receptor-related protein 1 in inflammation. Curr. Opin. Lipidol. 2013, 24, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, P.; Chen, B.; Zhang, Z.; Zhang, C.; Zhan, Q.; Huang, S.; Xia, Z.; Peng, W. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer’s disease-like rats using microarray analysis. Aging (Albany NY) 2018, 10, 775–788. [Google Scholar] [CrossRef]
- Xia, X.; Tang, X.; Wang, S. Roles of CircRNAs in Autoimmune Diseases. Front. Immunol. 2019, 10, 639. [Google Scholar] [CrossRef]
- Friese, M.A.; Craner, M.J.; Etzensperger, R.; Vergo, S.; A Wemmie, J.; Welsh, M.J.; Vincent, A.; Fugger, L. Acid-sensing ion channel-1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nat. Med. 2007, 13, 1483–1489. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, Y.; Han, B.; Bai, Y.; Zhou, R.; Gan, G.; Chao, J.; Hu, G.; Yao, H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124–2HG. Autophagy 2017, 13, 1722–1741. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Sun, Z.; Jia, H.; Luo, H.; Ye, X.; Wu, Q.; Xiong, Y.; Zhang, W.; Wan, J. Rpph1 Upregulates CDC42 Expression and Promotes Hippocampal Neuron Dendritic Spine Formation by Competing with miR-330-5p. Front. Mol. Neurosci. 2017, 10, 332. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Shi, Y.; Ohli, J.; Schüller, U.; Dorostkar, M.M.; Herms, J. Neuroinflammation impairs adaptive structural plasticity of dendritic spines in a preclinical model of Alzheimer’s disease. Acta Neuropathol. 2016, 131, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.K.; Chen, X.F.; He, D.D.; Li, Y.; Fu, J. Dissection of functional lncRNAs in Alzheimer’s disease by construction and analysis of lncRNA-mRNA networks based on competitive endogenous RNAs. Biochem. Biophys. Res. Commun. 2017, 485, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Griffin, R.J.; Moloney, A.; Kelliher, M.; Johnston, J.A.; Ravid, R.; Dockery, P.; O’Connor, R.; O’Neill, C. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer’s disease pathology. J. Neurochem. 2005, 93, 105–117. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; Iii, G.S.L.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Spreafico, M.; Grillo, B.; Rusconi, F.; Battaglioli, E.; Venturin, M. Multiple Layers of CDK5R1 Regulation in Alzheimer’s Disease Implicate Long Non-Coding RNAs. Int. J. Mol. Sci. 2018, 19, 2022. [Google Scholar] [CrossRef]
- Santoro, M.; Nociti, V.; Lucchini, M.; De Fino, C.; Losavio, F.A.; Mirabella, M. Expression Profile of Long Non-Coding RNAs in Serum of Patients with Multiple Sclerosis. J. Mol. Neurosci. 2016, 59, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, Z. Long non-coding RNA NEAT1 mediates the toxic of Parkinson’s disease induced by MPTP/MPP+ via regulation of gene expression. Clin. Exp. Pharmacol. Physiol. 2018, 45, 841–848. [Google Scholar] [CrossRef]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; DiMasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 30. [Google Scholar] [CrossRef]
- An, H.; Williams, N.G.; Shelkovnikova, T.A. NEAT1 and paraspeckles in neurodegenerative diseases: A missing lnc found? Non Coding RNA Res. 2018, 3, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Huang, X.; Li, R. lncRNA MALAT1/miR-205-5p axis regulates MPP+-induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am. J. Transl. Res. 2018, 10, 563–572. [Google Scholar] [PubMed]
- Li, J.Q.; Tan, L.; Yu, J.T. The role of the LRRK2 gene in Parkinsonism. Mol. Neurodegener. 2014, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Codolo, G.; Plotegher, N.; Pozzobon, T.; Brucale, M.; Tessari, I.; Bubacco, L.; De Bernard, M. Triggering of Inflammasome by Aggregated α–Synuclein, an Inflammatory Response in Synucleinopathies. PLoS ONE 2013, 8, e55375. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Q.; Zhang, J.; Pan, W.; Zhao, J.; Xu, Y. Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 2017, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Q.; Zhou, H.J. LncRNA MALAT1 promotes high glucose-induced inflammatory response of microglial cells via provoking MyD88/IRAK1/TRAF6 signaling. Sci. Rep. 2018, 8, 8346. [Google Scholar] [CrossRef]
- Zhou, H.J.; Wang, L.Q.; Wang, D.B.; Yu, J.B.; Zhu, Y.; Xu, Q.S.; Zheng, X.J.; Zhan, R.Y. Long noncoding RNA MALAT1 contributes to inflammatory response of microglia following spinal cord injury via the modulation of a miR-199b/IKKβ/NF-κB signaling pathway. Am. J. Physiol. Physiol. 2018, 315, C52–C61. [Google Scholar] [CrossRef]
- Soreq, L.; Guffanti, A.; Salomonis, N.; Simchovitz, A.; Israel, Z.; Bergman, H.; Soreq, H. Long Non-Coding RNA and Alternative Splicing Modulations in Parkinson’s Leukocytes Identified by RNA Sequencing. PLoS Comput. Biol. 2014, 10. [Google Scholar] [CrossRef]
- Duan, C.; Liu, Y.; Li, Y.; Chen, H.; Liu, X.; Chen, X.; Yue, J.; Zhou, X.; Yang, J. Sulfasalazine alters microglia phenotype by competing endogenous RNA effect of miR-136-5p and long non-coding RNA HOTAIR in cuprizone-induced demyelination. Biochem. Pharmacol. 2018, 155, 110–123. [Google Scholar] [CrossRef]
- Kakhki, M.P.; Nikravesh, A.; Farsani, Z.S.; Sahraian, M.A.; Behmanesh, M. HOTAIR but not ANRIL long non-coding RNA contributes to the pathogenesis of multiple sclerosis. Immunology 2018, 153, 479–487. [Google Scholar] [CrossRef]
- Straniero, L.; Rimoldi, V.; Samarani, M.; Goldwurm, S.; Di Fonzo, A.; Krüger, R.; Deleidi, M.; Aureli, M.; Soldà, G.; Duga, S.; et al. The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci. Rep. 2017, 7, 12702. [Google Scholar] [CrossRef] [PubMed]
- Migdalska-Richards, A.; Schapira, A.H.V. The relationship between glucocerebrosidase mutations and Parkinson disease. J. Neurochem. 2016, 139, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Werth, N.; Sandhoff, K.; Mizukami, H.; Wada, R.; Kono, M.; Yamashita, T.; Liu, Y.; Sandhoff, R.; Proia, R.L. Systemic inflammation in glucocerebrosidase-deficient mice with minimal glucosylceramide storage. J. Clin. Investig. 2002, 109, 1215–1221. [Google Scholar]
- Costa, V.P.; Esposito, R.; Aprile, M.; Ciccodicola, A. Non-coding RNA and pseudogenes in neurodegenerative diseases: “The (un)Usual Suspects”. Front. Genet. 2012, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Buzás, E.I.; György, B.; Nagy, G.; Falus, A.; Gay, S. Emerging role of extracellular vesicles in inflammatory diseases. Nat. Rev. Rheumatol. 2014, 10, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Verderio, C.; Muzio, L.; Turola, E.; Bergami, A.; Novellino, L.; Ruffini, F.; Riganti, L.; Corradini, I.; Francolini, M.; Garzetti, L.; et al. Myeloid microvesicles are a marker and therapeutic target for neuroinflammation. Ann. Neurol. 2012, 72, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Wang, B.; Kodali, M.C.; Chen, C.; Kim, E.; Patters, B.J.; Lan, L.; Kumar, S.; Wang, X.; Yue, J.; et al. In vivo evidence for the contribution of peripheral circulating inflammatory exosomes to neuroinflammation. J. Neuroinflammation 2018, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Selmaj, I.; Mycko, M.P.; Raine, C.S.; Selmaj, K.W. The role of exosomes in CNS inflammation and their involvement in multiple sclerosis. J. Neuroimmunol. 2017, 306, 1–10. [Google Scholar] [CrossRef]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, K.; Chen, L.; Fan, D. Increased Interleukin-6 Levels in the Astrocyte-Derived Exosomes of Sporadic Amyotrophic Lateral Sclerosis Patients. Front. Mol. Neurosci. 2019, 13, 13. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Schwartz, J.B.; Abner, E.L.; Jicha, G.A.; Kapogiannis, D. High complement levels in astrocyte-derived exosomes of Alzheimer disease. Ann. Neurol. 2018, 83, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of Extracellular miRNA in Cerebrospinal Fluid and Serum from Patients with Alzheimer’s and Parkinson’s Diseases Correlate with Disease Status and Features of Pathology. PLoS ONE 2014, 9, e94839. [Google Scholar] [CrossRef]
- Huang, S.; Ge, X.; Yu, J.; Han, Z.; Yin, Z.; Li, Y.; Chen, F.; Wang, H.; Zhang, J.; Lei, P. Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2017, 32, 512–528. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.M.; Krüger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Yelamanchili, S.V.; Lamberty, B.G.; Rennard, D.A.; Morsey, B.M.; Hochfelder, C.G.; Meays, B.M.; Levy, E.; Fox, H.S. MiR-21 in Extracellular Vesicles Leads to Neurotoxicity via TLR7 Signaling in SIV Neurological Disease. PLOS Pathog. 2015, 11, e1005032. [Google Scholar]
- Paschon, V.; Takada, S.H.; Ikebara, J.M.; Sousa, E.; Raeisossadati, R.; Ulrich, H.; Kihara, A.H. Interplay Between Exosomes, microRNAs and Toll-Like Receptors in Brain Disorders. Mol. Neurobiol. 2016, 53, 2016–2028. [Google Scholar] [CrossRef]
- Pusic, K.M.; Pusic, A.D.; Kraig, R.P. Environmental Enrichment Stimulates Immune Cell Secretion of Exosomes that Promote CNS Myelination and May Regulate Inflammation. Cell. Mol. Neurobiol. 2016, 36, 313–325. [Google Scholar] [CrossRef]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. Extracellular Vesicle-Associated RNA as a Carrier of Epigenetic Information. Genes 2017, 8, 240. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Fanale, D.; Taverna, S.; Russo, A.; Bazan, V. Circular RNA in Exosomes. In Circular RNAs: Biogenesis and Functions, Advances in Experimental Medicine and Biology; Xiao, J., Ed.; Springer Singapore: Singapore, 2018; pp. 109–117. ISBN 9789811314261. [Google Scholar]
- Wu, X.; Zheng, T.; Zhang, B. Exosomes in Parkinson’s Disease. Neurosci. Bull. 2016, 33, 331–338. [Google Scholar] [CrossRef]
- Patel, N.A.; Moss, L.D.; Lee, J.Y.; Tajiri, N.; Acosta, S.; Hudson, C.; Parag, S.; Cooper, D.R.; Borlongan, C.V.; Bickford, P.C. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J. Neuroinflamm. 2018, 15, 204. [Google Scholar] [CrossRef] [PubMed]
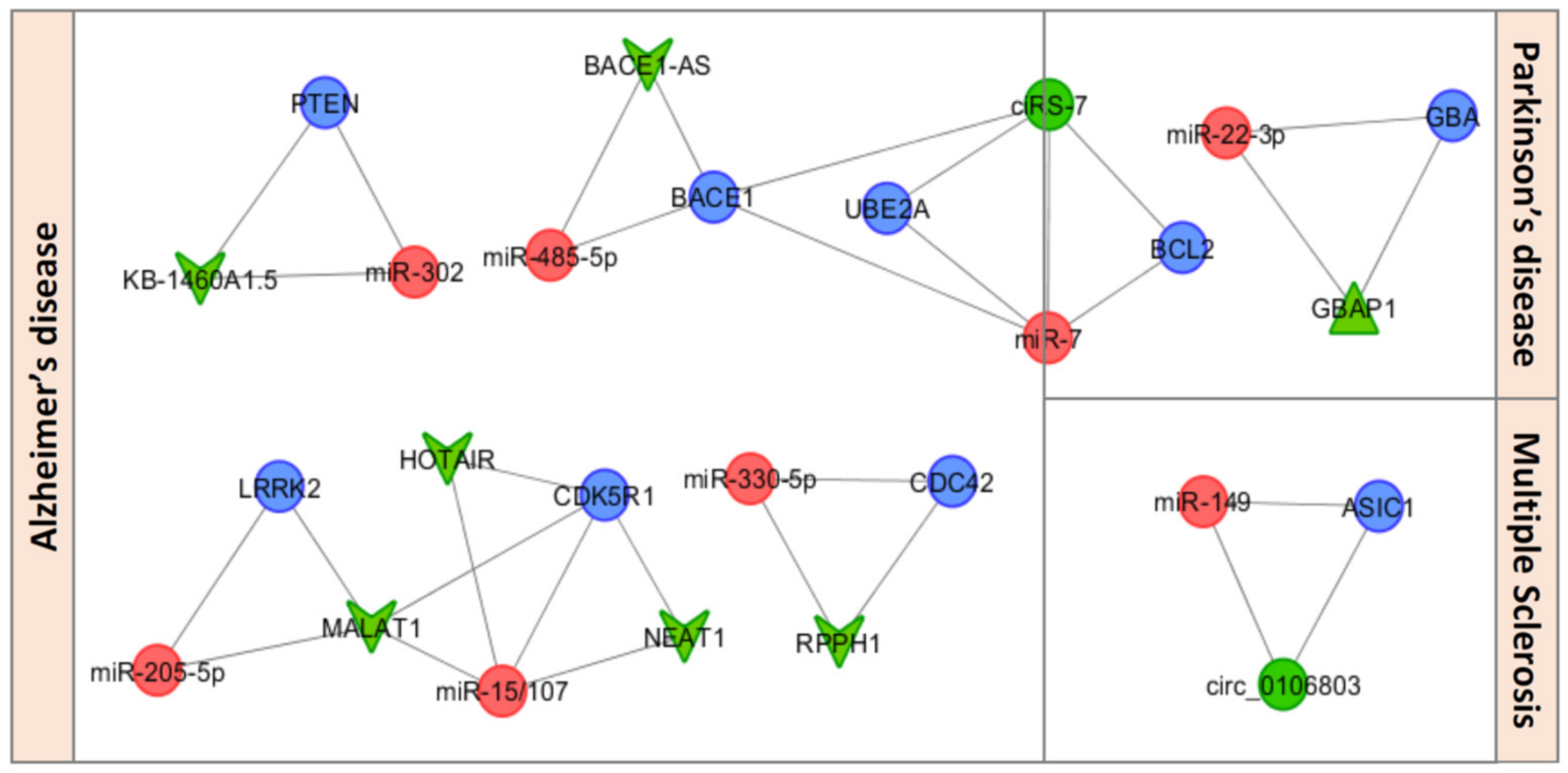
| ceRNET Type | Competing Member | Shared miRNA | Competitor (mRNA) | ceRNA Role | Ref |
|---|---|---|---|---|---|
| circRNA-miRNA-mRNA | ciRS-7 | miR-7 | UBE2A | Dysregulated in AD | [81] |
| circRNA-miRNA-mRNA | ciRS-7 | miR-7 | BACE1 | Dysregulated in AD | [82] |
| circRNA-miRNA-mRNA | ciRS-7 | miR-7 | BCL2 | Dysregulated in PD | [80,85] |
| circRNA-miRNA-mRNA | mm10_circ_0027470 mm10_circ_0011311 mm10_circ_0018430 mm10_circ_0009478 mm10_circ_0010326 mmu_circ_0001442 | let-7g-3p | HMGB2 | Dysregulated in AD | [86] |
| circRNA-miRNA-mRNA | hsa_circ_0106803 | miR-149 | ASIC1 | Progression of MS | [90] |
| circRNA-miRNA-mRNA | hsa_circ-HIPK2 | miR-124-2hg | SIGMAR1 | Astrocyte activation | [92] |
| lncRNA-miRNA-mRNA | RPPH1 | miR-330-5p | CDC42 | Dysregulated in AD | [94] |
| lncRNA-miRNA-mRNA | KB-1460A1.5 | miR-302 | PTEN | Dysregulated in AD | [95] |
| lncRNA-miRNA-mRNA | BACE1-AS | miR-485-5p | BACE1 | Dysregulated in AD | [97] |
| lncRNA-miRNA-mRNA | MALAT1 NEAT1 HOTAIR | miR-15/107 | CDK5R1 | Dysregulated in AD | [98] |
| lncRNA-miRNA-mRNA | MALAT1 | miR-205-5p | LRRK2 | Dysregulated in AD | [103] |
| lncRNA-miRNA-mRNA | MALAT1 | miR-199b | Proinflammatory cytokines | Microglia | [108] |
| lncRNA-miRNA-mRNA | HOTAIR | miR-136-5p | AKT2/NF-κB axis | cuprizone-induced demyelination | [110] |
| pseudogene-miRNA-mRNA | GBAP1 | miR-22-3p | GBA | Involved in PD | [112] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuzziello, N.; Liguori, M. The MicroRNA Centrism in the Orchestration of Neuroinflammation in Neurodegenerative Diseases. Cells 2019, 8, 1193. https://doi.org/10.3390/cells8101193
Nuzziello N, Liguori M. The MicroRNA Centrism in the Orchestration of Neuroinflammation in Neurodegenerative Diseases. Cells. 2019; 8(10):1193. https://doi.org/10.3390/cells8101193
Chicago/Turabian StyleNuzziello, Nicoletta, and Maria Liguori. 2019. "The MicroRNA Centrism in the Orchestration of Neuroinflammation in Neurodegenerative Diseases" Cells 8, no. 10: 1193. https://doi.org/10.3390/cells8101193
APA StyleNuzziello, N., & Liguori, M. (2019). The MicroRNA Centrism in the Orchestration of Neuroinflammation in Neurodegenerative Diseases. Cells, 8(10), 1193. https://doi.org/10.3390/cells8101193





