Differential Regulation of TLE3 in Sertoli Cells of the Testes during Postnatal Development
Abstract
1. Introduction
2. Material and Methods
2.1. Animal Management
2.2. RNA Preparation, RT-PCR, and qRT-PCR
2.3. Protein Preparation and Western Blot Analysis
2.4. Immunofluorescence Analysis
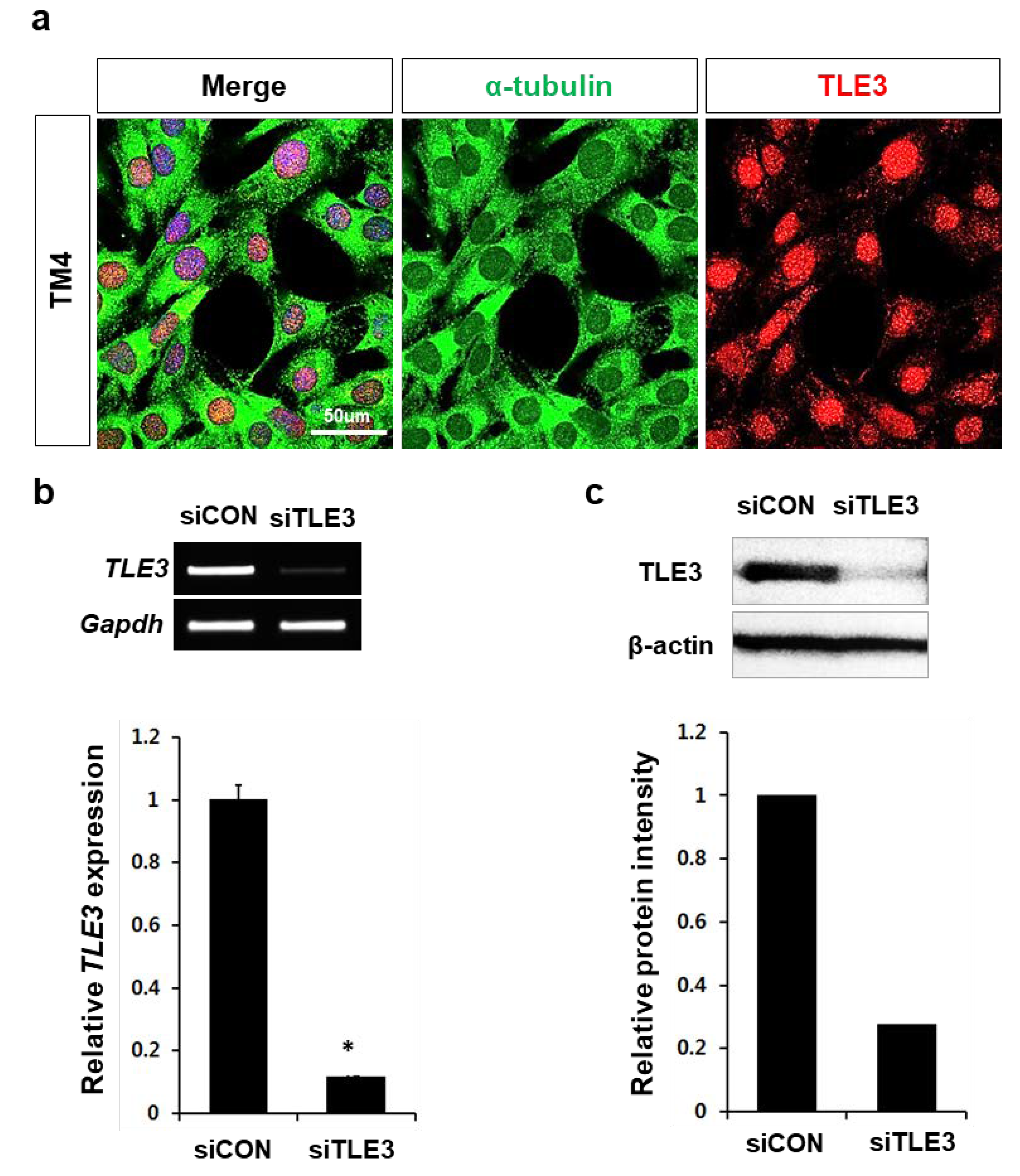
2.5. Knockdown of TLE3 and Microarray Analysis
2.6. Statistical Analysis
3. Results
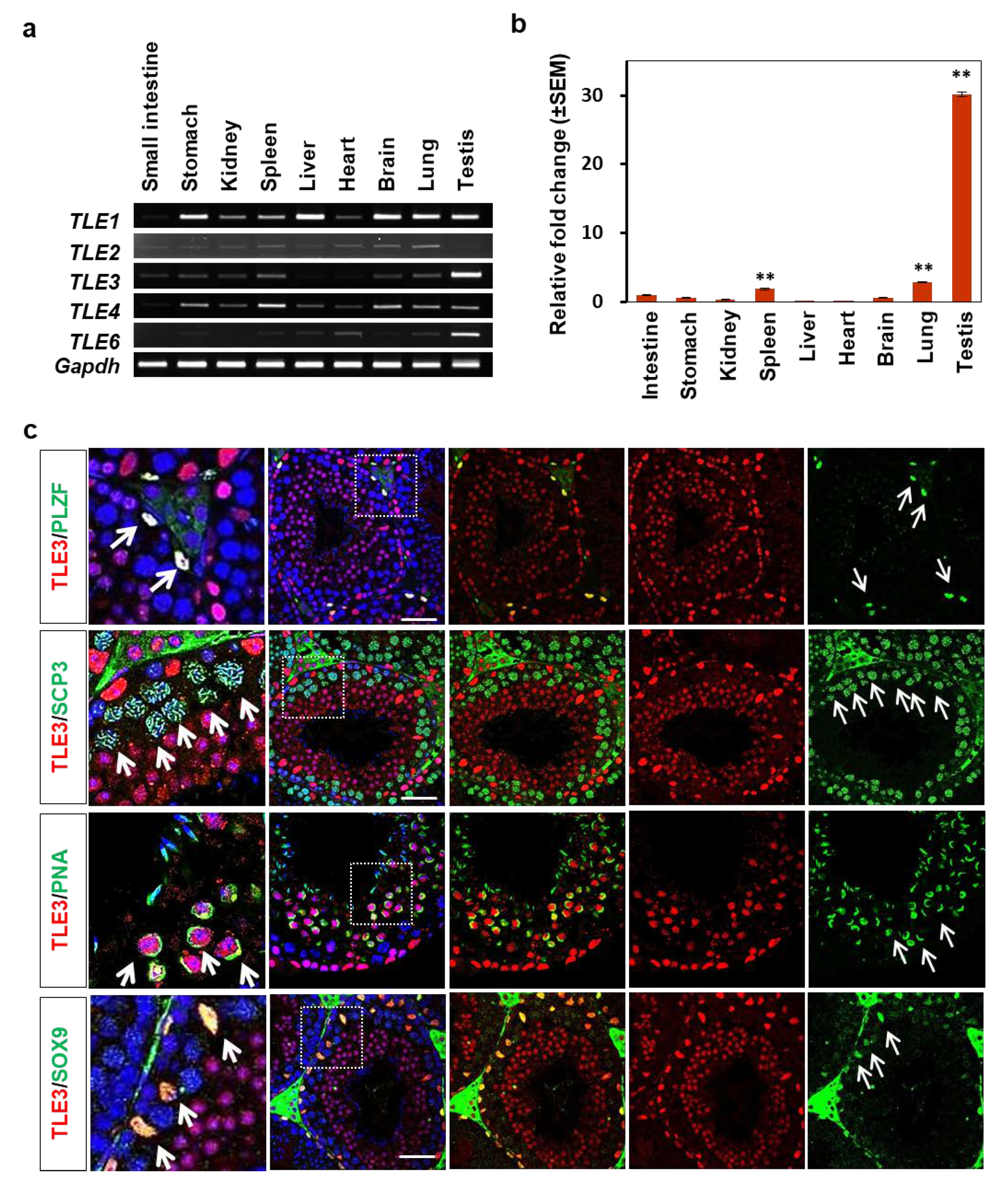
3.1. Expression of TLE3 in Mouse Tissues Including the Testes
3.2. Localization and Differential Expression of TLE3 in the Seminiferous Tubule during Testicular Development
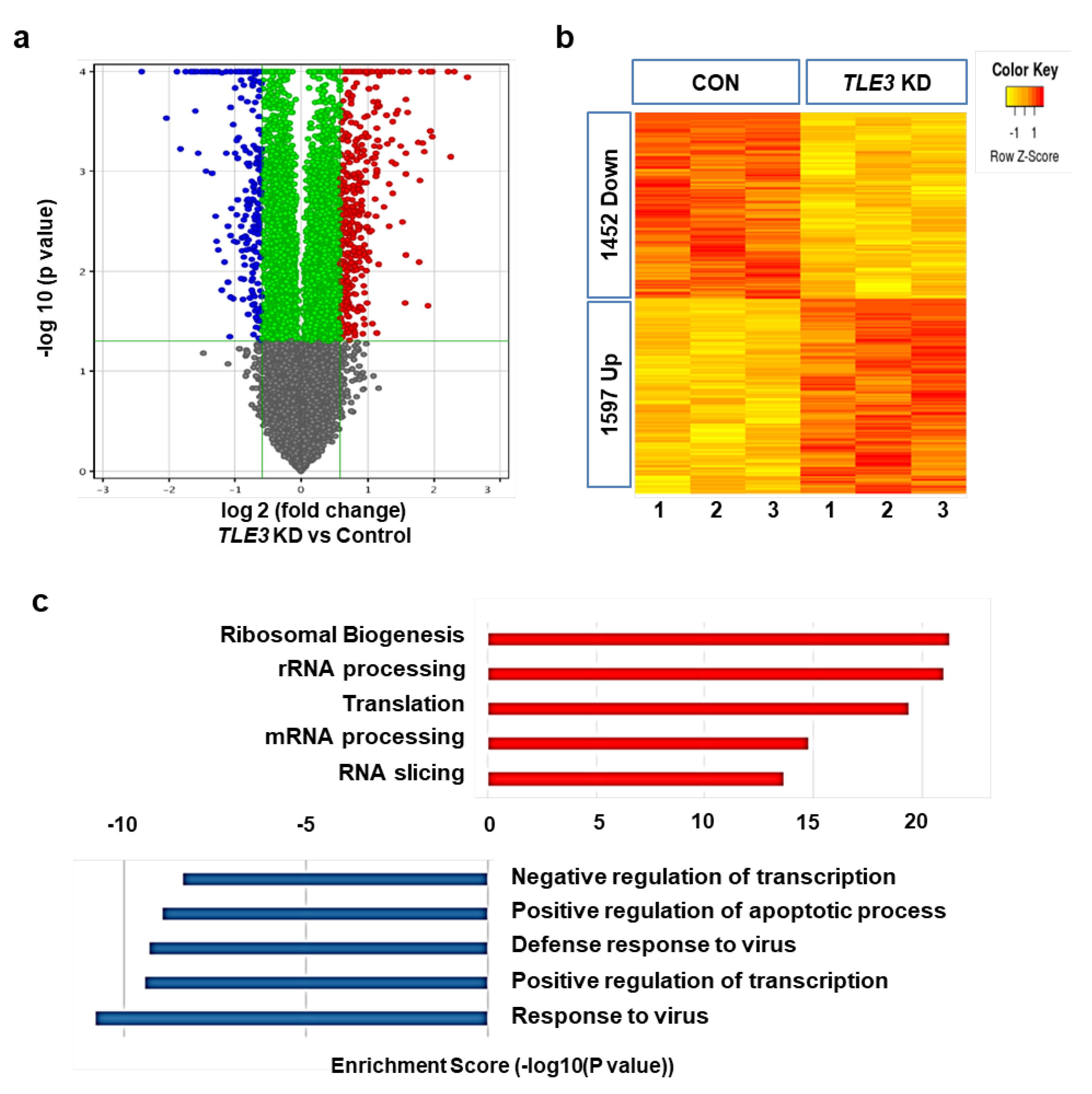
3.3. Gene profiles after TLE3 Knockdown in Sertoli Cells
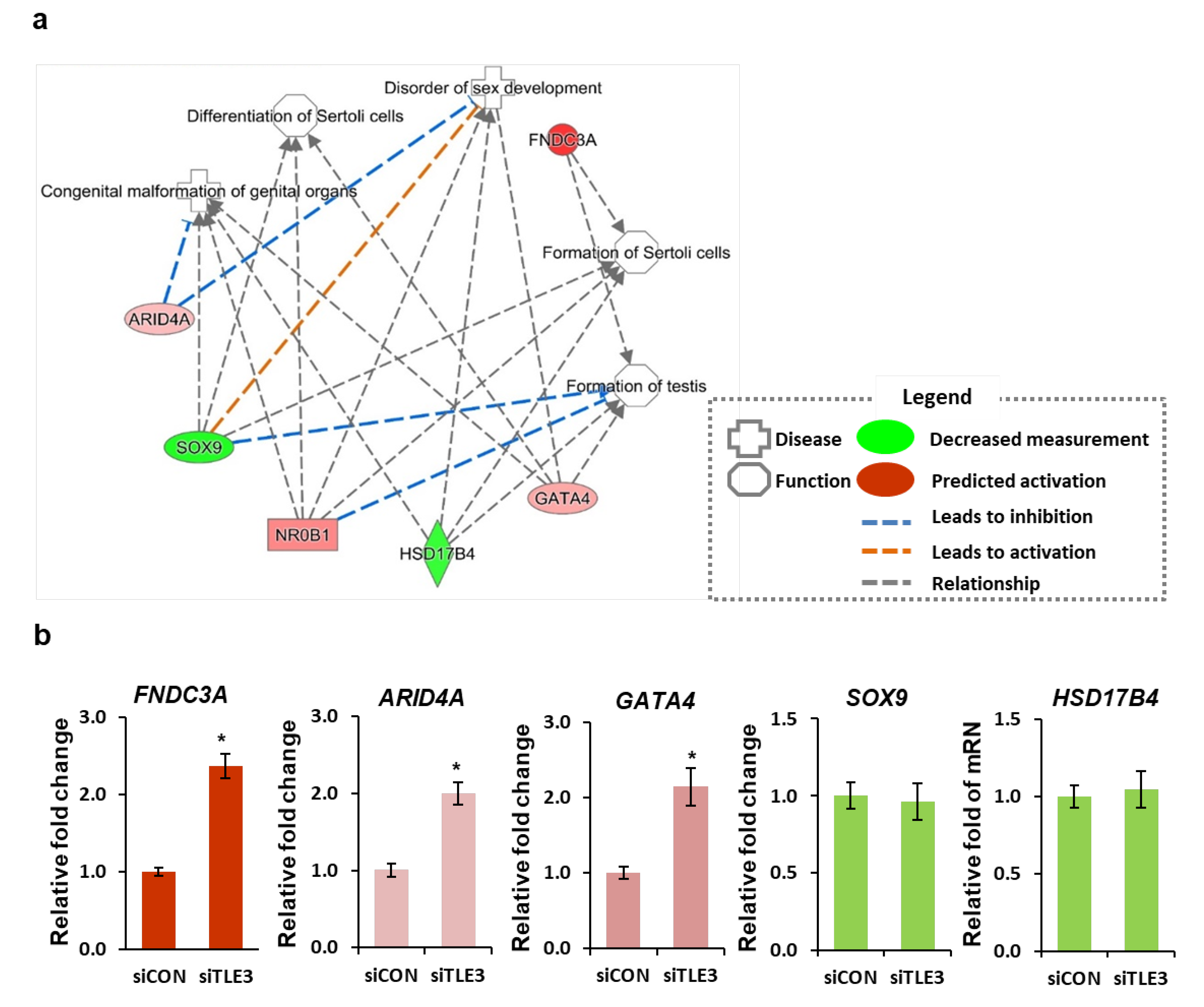
3.4. Role of TLE3 in Sertoli Cells during Spermatogenesis
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Jan, S.Z.; Hamer, G.; Repping, S.; De Rooij, D.G.; Van Pelt, A.M.; Vormer, T.L. Molecular control of rodent spermatogenesis. Biochim. et Biophys. Acta (BBA) - Mol. Basis Dis. 2012, 1822, 1838–1850. [Google Scholar] [CrossRef] [PubMed]
- Moricard, R.; Moricard, F. [Fecundation and study of spermatogenesis in human sterility]. Ann. Biol. Clin. 1950, 8, 435–452. [Google Scholar]
- Obholz, K.L.; Akopyan, A.; Waymire, K.G.; MacGregor, G.R. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev. Boil. 2006, 298, 498–513. [Google Scholar] [CrossRef] [PubMed]
- Schlatt, S.; Ehmcke, J. Regulation of spermatogenesis: An evolutionary biologist’s perspective. Semin. Cell Dev. Boil. 2014, 29, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Griswold, M.D. Interactions between germ cells and Sertoli cells in the testis. Boil. Reprod. 1995, 52, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Mruk, D.D.; Cheng, C.Y. Sertoli-Sertoli and Sertoli-Germ Cell Interactions and Their Significance in Germ Cell Movement in the Seminiferous Epithelium during Spermatogenesis. Endocr. Rev. 2004, 25, 747–806. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.; Stanton, P.G.; Bartles, J.R.; Robertson, D.M. Sertoli Cell Ectoplasmic Specializations in the Seminiferous Epithelium of the Testosterone-Suppressed Adult Rat1. Boil. Reprod. 2000, 63, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Vogl, A.W.; Pfeiffer, D.C.; Mulholland, D.; Kimel, G.; Guttman, J. Unique and Multifunctional Adhesion Junctions in the Testis. Ectoplasmic Specializations. Arch. Histol. Cytol. 2000, 63, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mruk, D.D.; Cheng, C.Y. Cell–cell interactions at the ectoplasmic specialization in the testis. Trends Endocrinol. Metab. 2004, 15, 439–447. [Google Scholar] [CrossRef]
- Wu, R.C.; Jiang, M.; Beaudet, A.L.; Wu, M.Y. ARID4A and ARID4B regulate male fertility, a functional link to the AR and RB pathways. Proc. Natl Acad Sci U.S.A. 2013, 110, 4616–4621. [Google Scholar] [CrossRef]
- Kyronlahti, A.; Euler, R.; Bielinska, M.; Schoeller, E.L.; Moley, K.H.; Toppari, J.; Heikinheimo, M.; Wilson, D.B. GATA4 regulates Sertoli cell function and fertility in adult male mice. Mol. Cell Endocrinol. 2011, 333, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Fröjdman, K.; Harley, V.R.; Pelliniemi, L.J. Sox9 protein in rat Sertoli cells is age and stage dependent. Histochem. Cell Boil. 2000, 113, 31–36. [Google Scholar] [CrossRef]
- Beverdam, A.; Svingen, T.; Bagheri-Fam, S.; Bernard, P.; McClive, P.; Robson, M.; Khojasteh, M.B.; Salehi, M.; Sinclair, A.H.; Harley, V.R.; et al. Sox9-dependent expression of Gstm6 in Sertoli cells during testis development in mice. Reproduction 2009, 137, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-R.; Tang, J.-X.; Cheng, J.-M.; Li, J.; Jin, C.; Li, X.-Y.; Deng, S.-L.; Zhang, Y.; Wang, X.-X.; Liu, Y.-X. Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget 2015, 6, 37012–37027. [Google Scholar] [CrossRef] [PubMed]
- Huyghe, S.; Schmalbruch, H.; De Gendt, K.; Verhoeven, G.; Guillou, F.; Van Veldhoven, P.P.; Baes, M. Peroxisomal Multifunctional Protein 2 Is Essential for Lipid Homeostasis in Sertoli Cells and Male Fertility in Mice. Endocrinology 2006, 147, 2228–2236. [Google Scholar] [CrossRef]
- Gasperowicz, M.; Otto, F. Mammalian Groucho homologs: Redundancy or specificity? J. Cell. Biochem. 2005, 95, 670–687. [Google Scholar] [CrossRef] [PubMed]
- Jangal, M.; Couture, J.-P.; Bianco, S.; Magnani, L.; Mohammed, H.; Gévry, N. The transcriptional co-repressor TLE3 suppresses basal signaling on a subset of estrogen receptor α target genes. Nucleic Acids Res. 2014, 42, 11339–11348. [Google Scholar] [CrossRef]
- Nakayama, H.; Liu, Y.; Stifani, S.; Cross, J.C. Developmental restriction of Mash2 expression in trophoblast correlates with potential activation of the Notch-2 pathway. Dev. Genet. 1997, 21, 21–30. [Google Scholar] [CrossRef]
- Leon, C.; Lobe, C.G. Grg3, a murineGroucho-related gene, is expressed in the developing nervous system and in mesenchyme-induced epithelial structures. Dev. Dyn. 1997, 208, 11–24. [Google Scholar] [CrossRef]
- Allen, T.; Lobe, C.G. A comparison of Notch, Hes and Grg expression during murine embryonic and post-natal development. Cell. Mol. Biol. 1999, 45, 687–708. [Google Scholar]
- Santisteban, P.; Recacha, P.; Metzger, D.E.; Zaret, K.S. Dynamic expression of Groucho-related genes Grg1 and Grg3 in foregut endoderm and antagonism of differentiation. Dev. Dyn. 2010, 239, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; He, S.; Zhang, Z.; Gao, W.; Yu, L.; Tan, Y. Foxa1 contributes to the repression of Nanog expression by recruiting Grg3 during the differentiation of pluripotent P19 embryonal carcinoma cells. Exp. Cell Res. 2014, 326, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-R.; Cho, K.-S.; Kim, E.; Lee, O.-H.; Yoon, H.; Lee, S.; Moon, S.; Park, M.; Hong, K.; Na, Y.; et al. Rapid expression of RASD1 is regulated by estrogen receptor-dependent intracellular signaling pathway in the mouse uterus. Mol. Cell. Endocrinol. 2017, 446, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Alazami, A.M.; Awad, S.M.; Coskun, S.; Al-Hassan, S.; Hijazi, H.; Abdulwahab, F.M.; Poizat, C.; Alkuraya, F.S. TLE6 mutation causes the earliest known human embryonic lethality. Genome Boil. 2015, 16, 240. [Google Scholar] [CrossRef] [PubMed]
- Bebbere, D.; Ariu, F.; Bogliolo, L.; Masala, L.; Murrone, O.; Fattorini, M.; Falchi, L.; Ledda, S. Expression of maternally derived KHDC3, NLRP5, OOEP and TLE6 is associated with oocyte developmental competence in the ovine species. BMC Dev. Boil. 2014, 14, 40. [Google Scholar]
- Villanueva, C.J.; Waki, H.; Godio, C.; Nielsen, R.; Chou, W.-L.; Vargas, L.; Wroblewski, K.; Schmedt, C.; Chao, L.C.; Boyadjian, R.; et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 2011, 13, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Kokabu, S.; Nguyen, T.; Ohte, S.; Sato, T.; Katagiri, T.; Yoda, T.; Rosen, V. TLE3, transducing-like enhancer of split 3, suppresses osteoblast differentiation of bone marrow stromal cells. Biochem. Biophys. Res. Commun. 2013, 438, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Kokabu, S.; Sato, T.; Ohte, S.; Enoki, Y.; Okubo, M.; Hayashi, N.; Nojima, J.; Tsukamoto, S.; Fukushima, Y.; Sakata, Y.; et al. Expression of TLE3 by bone marrow stromal cells is regulated by canonical Wnt signaling. FEBS Lett. 2014, 588, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Kokabu, S.; Lowery, J.W.; Jimi, E. Cell Fate and Differentiation of Bone Marrow Mesenchymal Stem Cells. Stem Cells Int. 2016, 2016, 1–7. [Google Scholar] [CrossRef]
- Doyle, M.J.; Loomis, Z.L.; Sussel, L. Nkx2.2-repressor activity is sufficient to specify alpha-cells and a small number of beta-cells in the pancreatic islet. Development 2007, 134, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.J.; Sussel, L. Nkx2.2 regulates beta-cell function in the mature islet. Diabetes 2007, 56, 1999–2007. [Google Scholar] [CrossRef] [PubMed]
- Papizan, J.B.; Singer, R.A.; Tschen, S.-I.; Dhawan, S.; Friel, J.M.; Hipkens, S.B.; Magnuson, M.A.; Bhushan, A.; Sussel, L. Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 2011, 25, 2291–2305. [Google Scholar] [CrossRef] [PubMed]
- Kokabu, S.; Nakatomi, C.; Matsubara, T.; Ono, Y.; Addison, W.N.; Lowery, J.W.; Urata, M.; Hudnall, A.M.; Hitomi, S.; Nakatomi, M.; et al. The transcriptional co-repressor TLE3 regulates myogenic differentiation by repressing the activity of the MyoD transcription factor. J. Boil. Chem. 2017, 292, 12885–12894. [Google Scholar] [CrossRef] [PubMed]
- Gasperowicz, M.; Surmann-Schmitt, C.; Hamada, Y.; Otto, F.; Cross, J.C. The transcriptional co-repressor TLE3 regulates development of trophoblast giant cells lining maternal blood spaces in the mouse placenta. Dev. Boil. 2013, 382, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Laing, A.F.; Lowell, S.; Brickman, J.M. Gro/TLE enables embryonic stem cell differentiation by repressing pluripotent gene expression. Dev. Boil. 2015, 397, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Serrano, M.; Rodriguez-Cuenca, S.; Moreno-Navarrete, J.M.; Gomez-Serrano, M.; Sabater, M.; Rodriguez-Hermosa, J.I.; Xifra, G.; Ricart, W.; Peral, B.; et al. Transducin-like enhancer of split 3 (TLE3) in adipose tissue is increased in situations characterized by decreased PPARgamma gene expression. J. Mol. Med. 2015, 93, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Samimi, G.; Ring, B.Z.; Ross, D.T.; Seitz, R.S.; Sutherland, R.L.; O’Brien, P.M.; Hacker, N.F.; Huh, W.K. TLE3 expression is associated with sensitivity to taxane treatment in ovarian carcinoma. Cancer Epidemiol. Prev. Biomark. 2012, 21, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Susini, T.; Berti, B.; Carriero, C.; Tavella, K.; Nori, J.; Vanzi, E.; Molino, C.; Di Tommaso, M.; Santini, M.; Saladino, V.; et al. Topoisomerase II alpha and TLE3 as predictive markers of response to anthracycline and taxane-containing regimens for neoadjuvant chemotherapy in breast cancer. Onco Targets Ther. 2014, 7, 2111–2120. [Google Scholar] [CrossRef][Green Version]
- Sharpe, R.M.; McKinnell, C.; Kivlin, C.; Fisher, J.S. Proliferation and functional maturation of Sertoli cells, and their relevance to disorders of testis function in adulthood. Reprod. Camb. 2003, 125, 769–784. [Google Scholar] [CrossRef]
- Chen, S.-R.; Liu, Y.-X. Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 2015, 149, R159–R167. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.; Braun, R.E. Histone lysine trimethylation exhibits a distinct perinuclear distribution in Plzf-expressing spermatogonia. Dev. Boil. 2006, 293, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, Y.; Saito, K.; Shinya, M.; Kawasaki, T.; Sakai, N. Evaluation of Sycp3, Plzf and Cyclin B3 expression and suitability as spermatogonia and spermatocyte markers in zebrafish. Gene Expr. Patterns 2011, 11, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Lovelace, D.L.; Gao, Z.; Mutoji, K.; Song, Y.C.; Ruan, J.; Hermann, B.P. The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development 2016, 143, 1893–1906. [Google Scholar] [CrossRef] [PubMed]
- Oatley, M.J.; Racicot, K.E.; Oatley, J.M. Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol. Reprod. 2011, 84, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Hemendinger, R.A.; Gores, P.; Blacksten, L.; Harley, V.; Halberstadt, C. Identification of a specific Sertoli cell marker, Sox9, for use in transplantation. Cell Transplant. 2002, 11, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Sekido, R.; Bar, I.; Narváez, V.; Penny, G.; Lovell-Badge, R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev. Boil. 2004, 274, 271–279. [Google Scholar] [CrossRef] [PubMed]
- LaSala, C.; Schteingart, H.F.; Arouche, N.; Bedecarrás, P.; Grinspon, R.P.; Picard, J.-Y.; Josso, N.; Di Clemente, N.; Rey, R.A. SOX9 and SF1 are involved in cyclic AMP-mediated upregulation of anti-Müllerian gene expression in the testicular prepubertal Sertoli cell line SMAT1. Am. J. Physiol. Metab. 2011, 301, E539–E547. [Google Scholar] [CrossRef] [PubMed]
- Montoto, L.G.; Arregui, L.; Sánchez, N.M.; Gomendio, M.; Roldan, E.R.S. Postnatal testicular development in mouse species with different levels of sperm competition. Reprod. 2012, 143, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Carson, D.D.; I Rosenberg, L.; Blaner, W.S.; Kato, M.; Lennarz, W.J. Synthesis and secretion of a novel binding protein for retinol by a cell line derived from Sertoli cells. J. Boil. Chem. 1984, 259, 3117–3123. [Google Scholar]
- Agarwal, M.; Kumar, P.; Mathew, S.J.; And, P.K. The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 2015, 67, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Turki-Judeh, W.; Courey, A.J. Groucho: A corepressor with instructive roles in development. Curr. Top. Dev. Biol. 2012, 98, 65–96. [Google Scholar]
- Metzger, D.E.; Liu, C.; Ziaie, A.S.; Naji, A.; Zaret, K.S. Grg3/TLE3 and Grg1/TLE1 Induce Monohormonal Pancreatic β-Cells While Repressing α-Cell Functions. Diabetes 2014, 63, 1804–1816. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, C.J.; Vergnes, L.; Wang, J.; Drew, B.G.; Hong, C.; Tu, Y.; Hu, Y.; Peng, X.; Xu, F.; Saez, E.; et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARγ specifies lipid storage versus thermogenic gene programs. Cell Metab. 2013, 17, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Kyronlahti, A.; Vetter, M.; Euler, R.; Bielinska, M.; Jay, P.Y.; Anttonen, M.; Heikinheimo, M.; Wilson, D.B. GATA4 deficiency impairs ovarian function in adult mice. Boil. Reprod. 2011, 84, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Schrade, A.; Kyrönlahti, A.; Akinrinade, O.; Pihlajoki, M.; Fischer, S.; Rodriguez, V.M.; Otte, K.; Velagapudi, V.; Toppari, J.; Wilson, D.B.; et al. GATA4 Regulates Blood-Testis Barrier Function and Lactate Metabolism in Mouse Sertoli Cells. Endocrinology 2016, 157, 2416–2431. [Google Scholar] [CrossRef] [PubMed]

| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| TLE1 | CCAGTACCTCTCACGCCTCA | GCCCACTCAGAGCACTAGAC |
| TLE2 | TGGCTGCCGTAAAGGAAGAC | CTCACTGTCATAAGGCCCTGA |
| TLE3 | TCCACCGTCTTTTACCCAAG | CAAGGCCTCATGCAGAGTTA |
| TLE4 | TTTACAGGCTCAATACCACAGTC | TGCACAGATAGCATTTAGTCGTT |
| TLE6 | ATCCAGTCGGTATTTGTCCATCG | AGGTCTGGGGTTCTACTGAAG |
| GATA4 | CCCTACCCAGCCTACATGG | ACATATCGAGATTGGGGTGTCT |
| SOX9 | AGTACCCGCATCTGCACAAC | ACGAAGGGTCTCTTCTCGCT |
| FNDC3A | GAACTCCAAAGACGTTGTCCA | GCTTGAATGCAGAACTTGTAGGA |
| ARID4A | GATGAGCCTGCCTACCTGAC | CACCAACTGTGTTGTGTTATCCT |
| HSD17B4 | GTCACGGATGACGGAGACTG | GCAAAGCCAAAGCACCAGAG |
| Gapdh | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| Gene | GenBank ID | Fold Change | Description |
|---|---|---|---|
| Mpeg1 | AK030216 | 5.7 | macrophage expressed gene 1 |
| Pydc3 | AK041099 | 4.9 | pyrin domain containing 3 |
| Gbp8 | AK041099 | 4.7 | guanylate-binding protein 8 |
| Saa3 | BC031475 | 4.6 | serum amyloid A 3 |
| Wfdc17 | BC038870 | 4.0 | WAP four-disulfide core domain 17 |
| Apod | AK135046 | 4.0 | apolipoprotein D |
| Gm12250 | DQ508486 | 3.9 | predicted gene 12250 |
| Serping1 | AK144292 | 3.8 | serine (or cysteine) peptidase inhibitor, clade G, member 1 |
| Gm5431 | DQ508486 | 3.8 | predicted gene 5431 |
| Angptl4 | AK00916 | 3.5 | angiopoietin-like 4 |
| Gene | GenBank ID | Fold Change | Description |
|---|---|---|---|
| Crabp1 | AK045283 | −5.3 | cellular retinoic acid binding protein I |
| Serpinb2 | AK081487 | −4.1 | serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| Idi1 | AK029302 | −3.6 | isopentenyl-diphosphate delta isomerase |
| Fdps | AK077979 | −3.4 | farnesyl diphosphate synthetase |
| Cyp51 | AK002827 | −3.1 | cytochrome P450, family 51 |
| Msmo1 | AK005090 | −3.1 | methylsterol monoxygenase 1 |
| Sqle | AK041198 | −3.1 | squalene epoxidase |
| Lss | AK012813 | −3.0 | lanosterol synthase |
| Msln | AK144391 | −3.0 | mesothelin |
| Dhcr24 | AK010192 | −3.0 | 24-dehydrocholesterol reductase |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Jang, H.; Moon, S.; Lee, O.-H.; Lee, S.; Lee, J.; Park, C.; Seol, D.W.; Song, H.; Hong, K.; et al. Differential Regulation of TLE3 in Sertoli Cells of the Testes during Postnatal Development. Cells 2019, 8, 1156. https://doi.org/10.3390/cells8101156
Lee S, Jang H, Moon S, Lee O-H, Lee S, Lee J, Park C, Seol DW, Song H, Hong K, et al. Differential Regulation of TLE3 in Sertoli Cells of the Testes during Postnatal Development. Cells. 2019; 8(10):1156. https://doi.org/10.3390/cells8101156
Chicago/Turabian StyleLee, Sangho, Hoon Jang, Sohyeon Moon, Ok-Hee Lee, Sujin Lee, Jihyun Lee, Chanhyeok Park, Dong Won Seol, Hyuk Song, Kwonho Hong, and et al. 2019. "Differential Regulation of TLE3 in Sertoli Cells of the Testes during Postnatal Development" Cells 8, no. 10: 1156. https://doi.org/10.3390/cells8101156
APA StyleLee, S., Jang, H., Moon, S., Lee, O.-H., Lee, S., Lee, J., Park, C., Seol, D. W., Song, H., Hong, K., Kim, J.-H., Uhm, S. J., Lee, D. R., Lee, J.-W., & Choi, Y. (2019). Differential Regulation of TLE3 in Sertoli Cells of the Testes during Postnatal Development. Cells, 8(10), 1156. https://doi.org/10.3390/cells8101156







