Activation of Polyamine Catabolism by N1,N11-Diethylnorspermine in Hepatic HepaRG Cells Induces Dedifferentiation and Mesenchymal-Like Phenotype
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell culture
2.3. Measurement of Polyamine Levels
2.4. Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
2.5. Western Blot Analysis
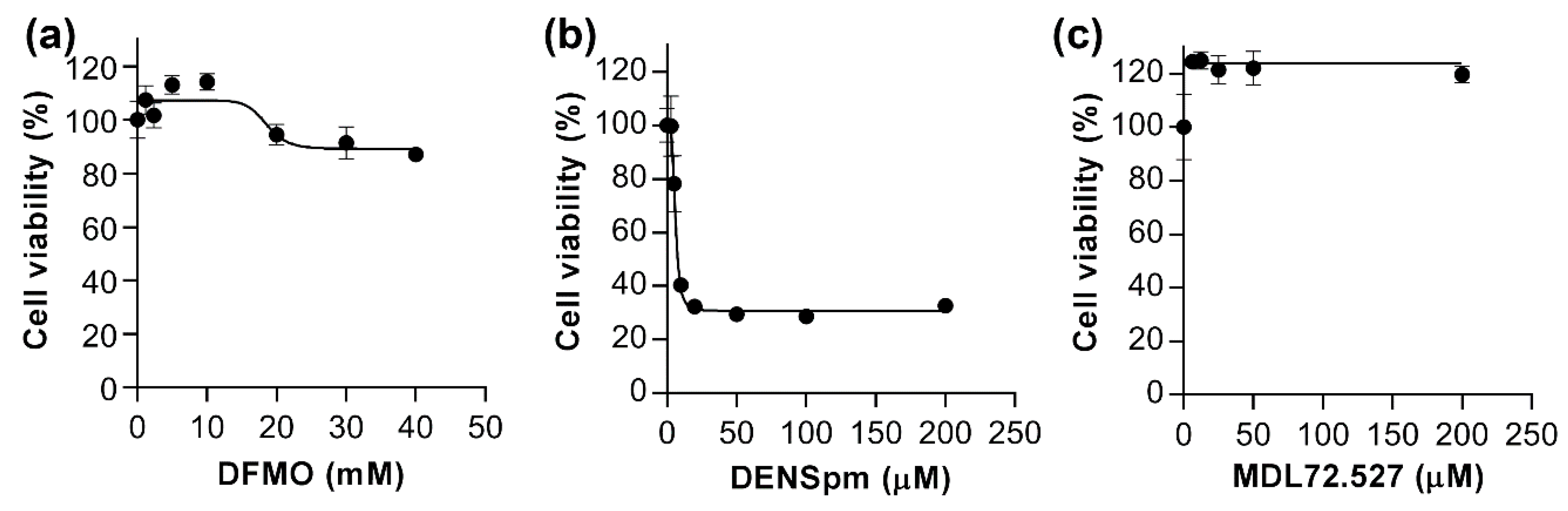
2.6. Cell Viability Assay
2.7. Isolation of RNA for NextSeq Sequencing
2.8. cDNA Library Preparation and Sequencing
2.9. Bioinformatics Analysis
2.10. Scratch Assay
2.11. Measurement of ROS Production
2.12. Statistical Analysis
3. Results
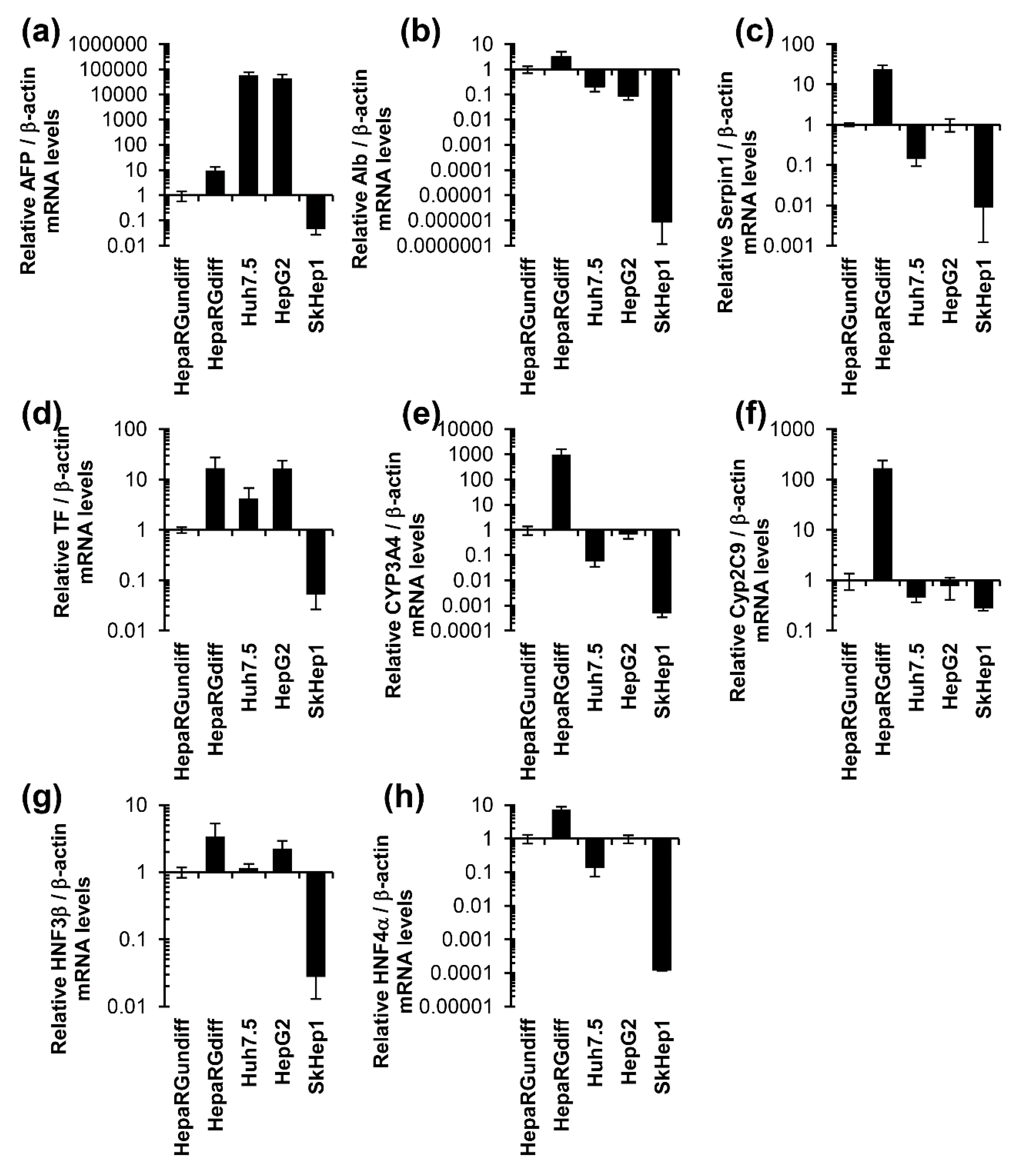
3.1. Unlike Huh7, HepG2, and SkHep1 Cells, HepaRG Cells Exhibit Elevated Expression Levels of Hepatocyte-Specific Genes
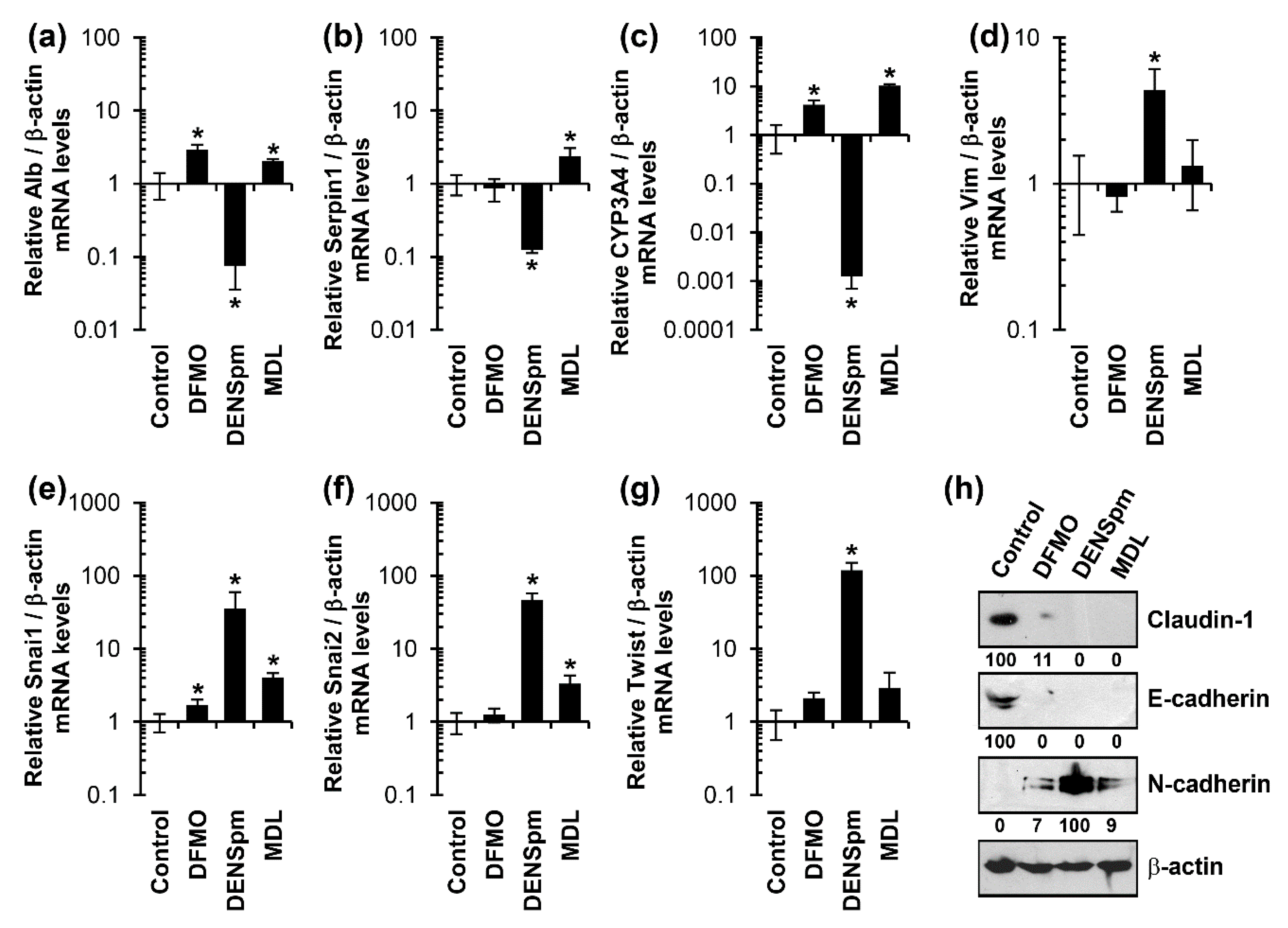
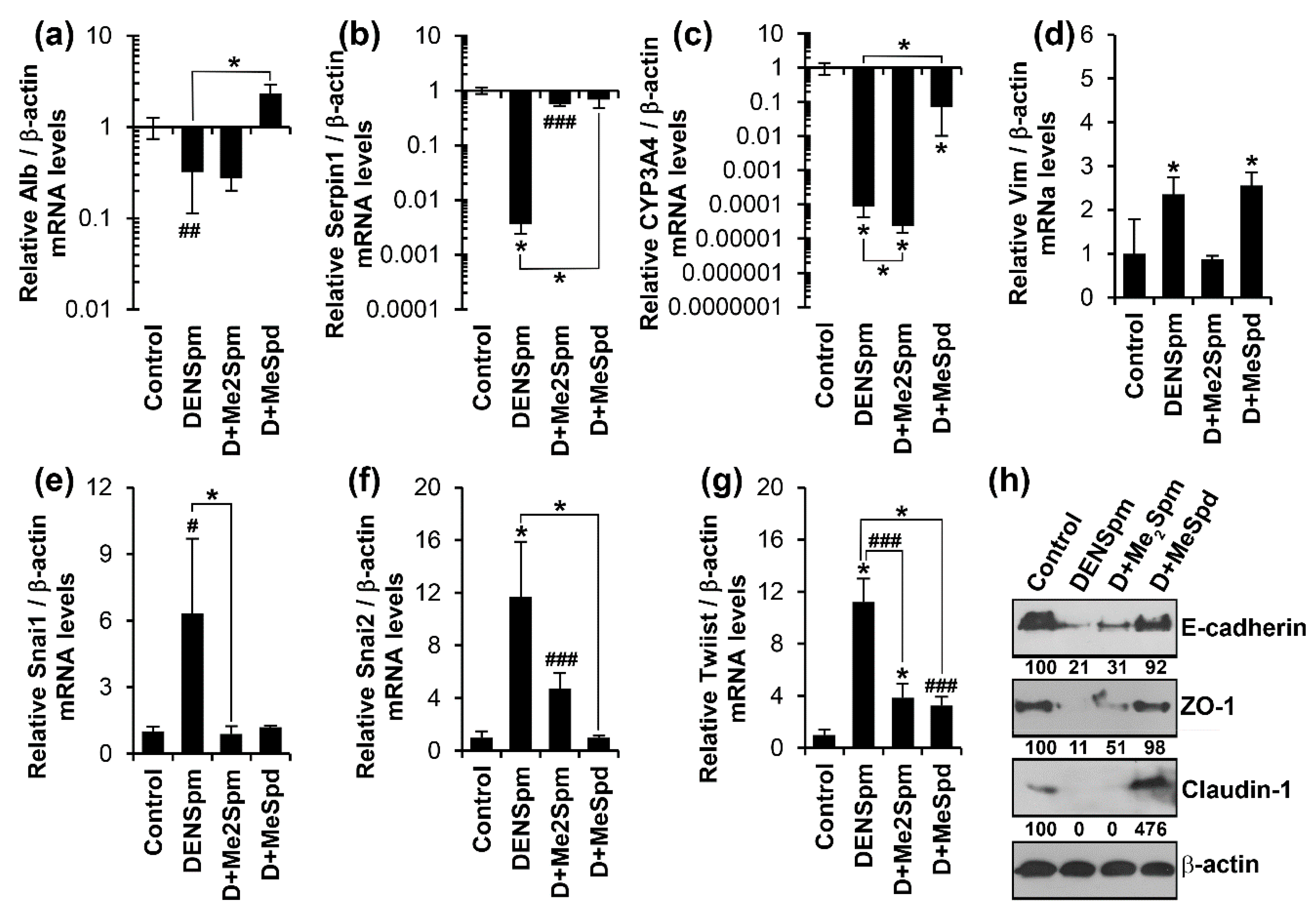
3.2. Diethylnorspermine Induces Dedifferentiation of HepaRG Cells and Mesenchymal-Like Phenotype
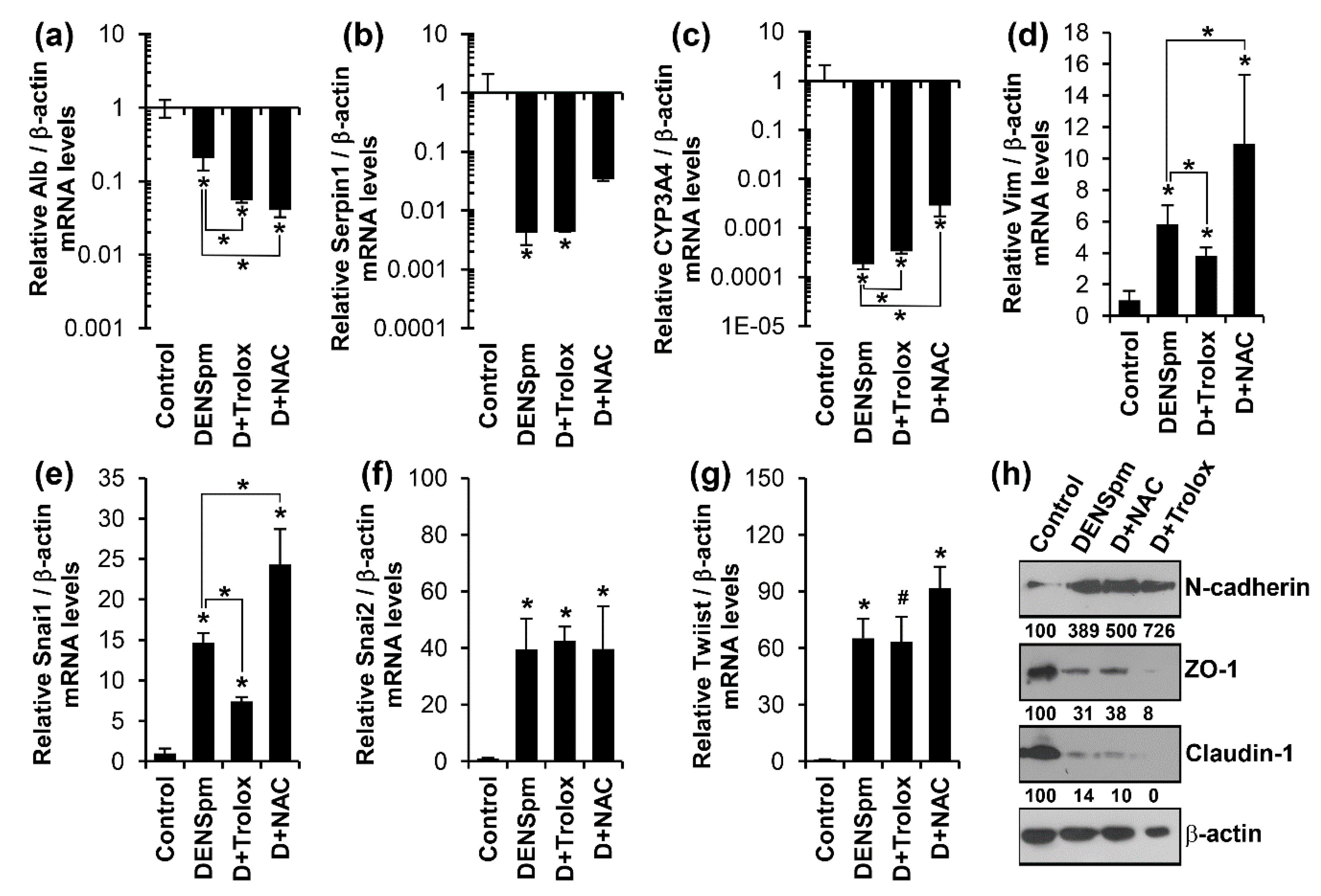
3.3. Antioxidants Trolox and N-Acetylcysteine Do Not Affect Altered Expression of Epithelial or Mesenchymal Cell-Specific Genes
3.4. DENSpm Triggers Dedifferentiation of HepaRG Cells through Exhaustion of Spermidine
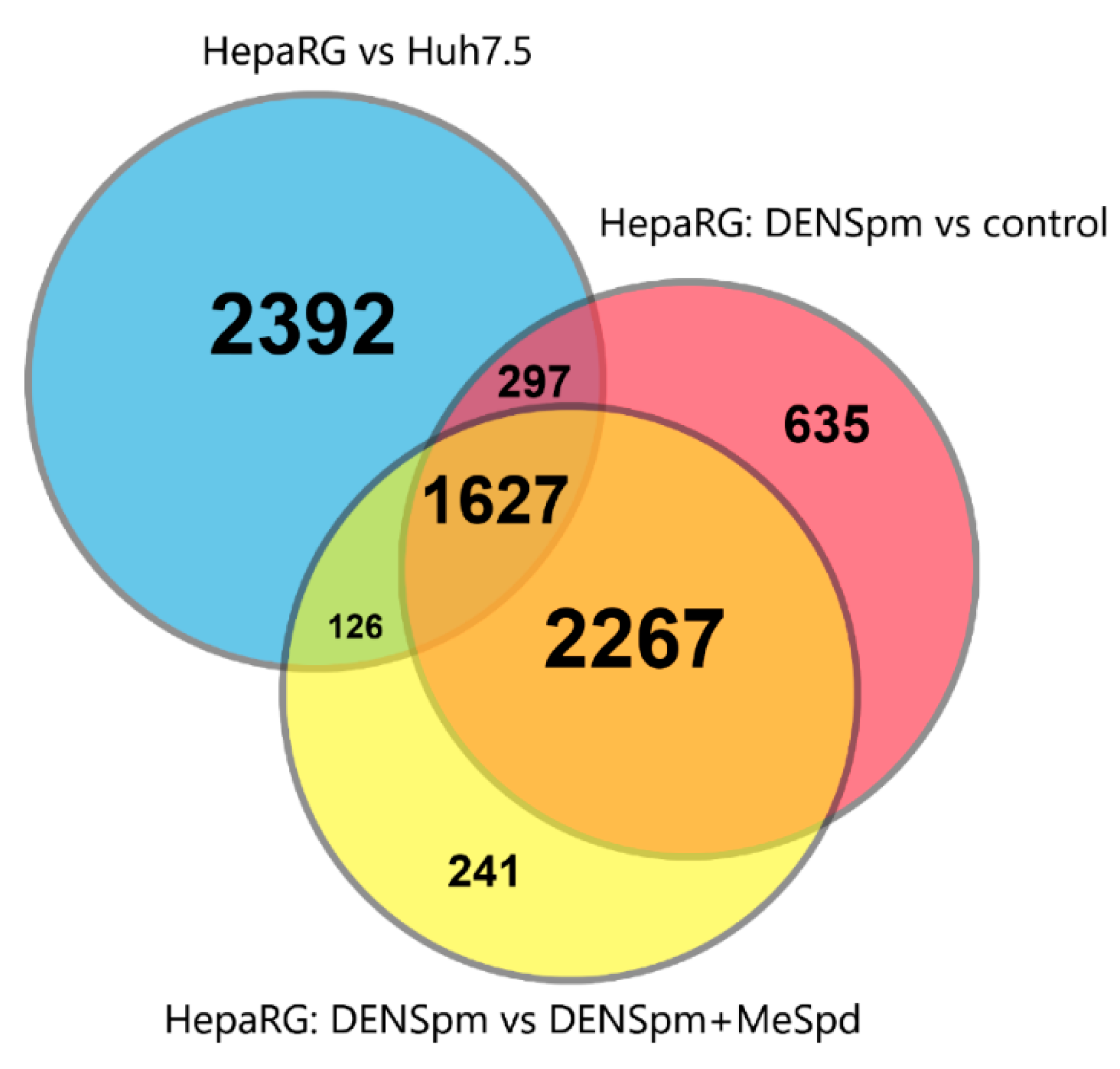
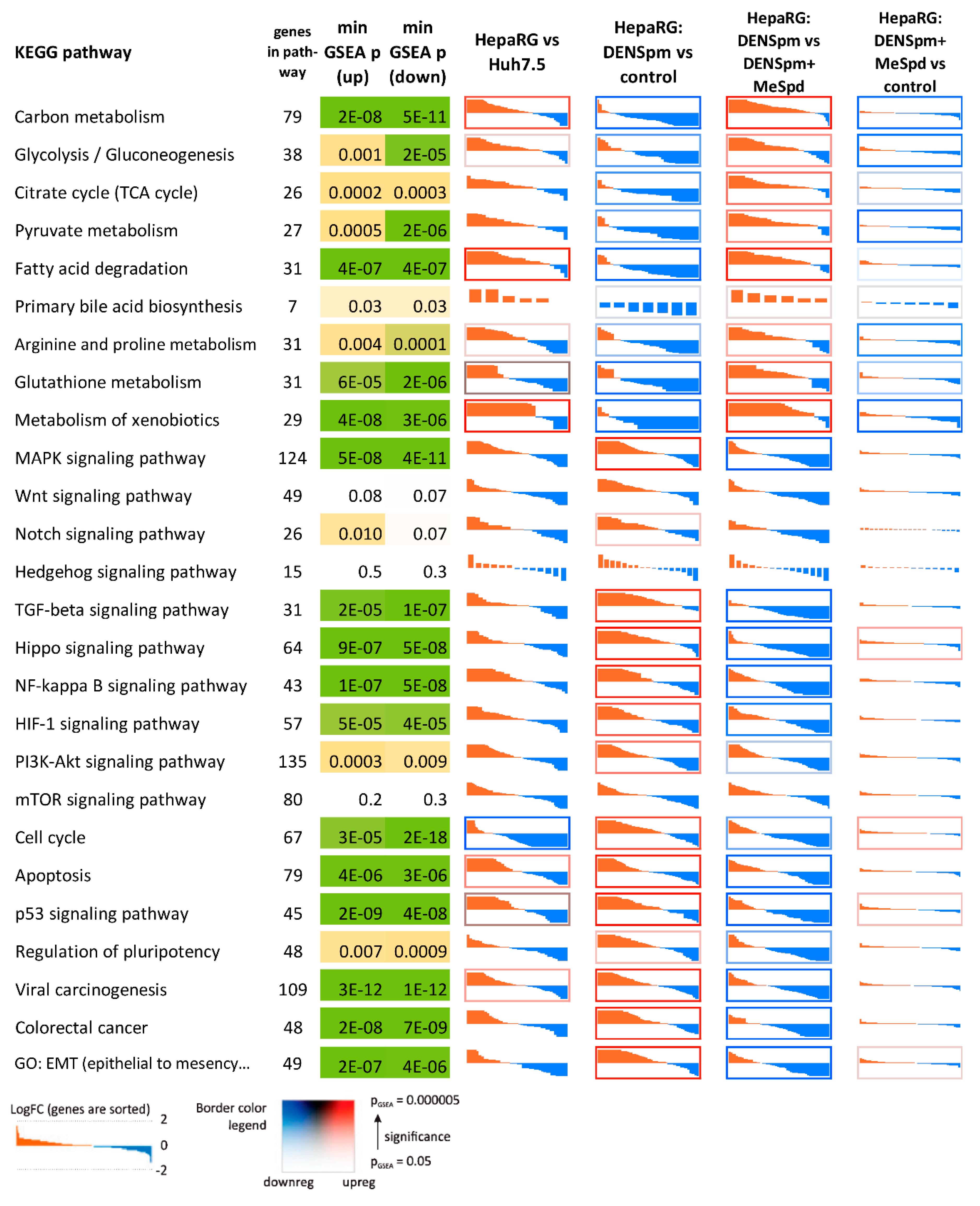
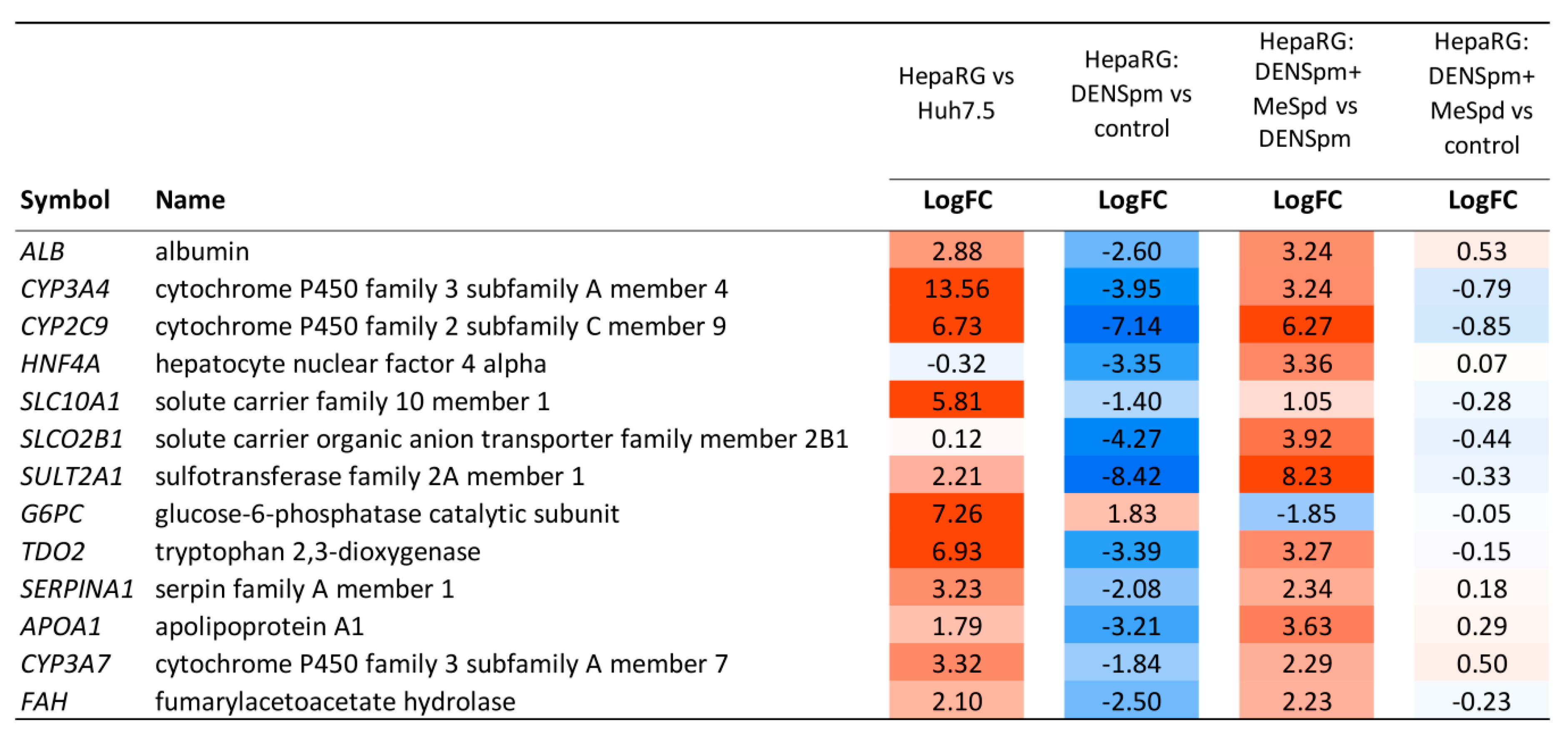
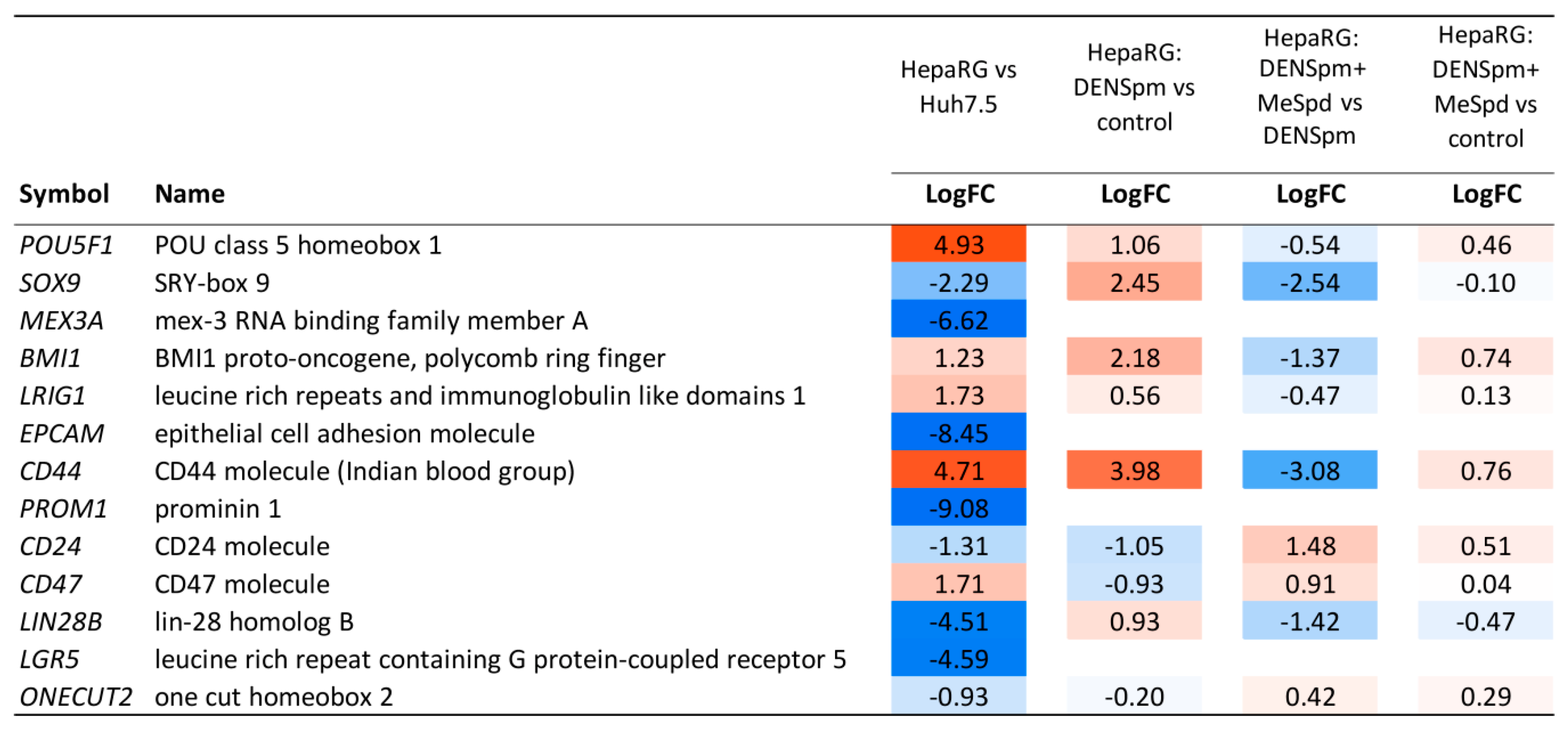
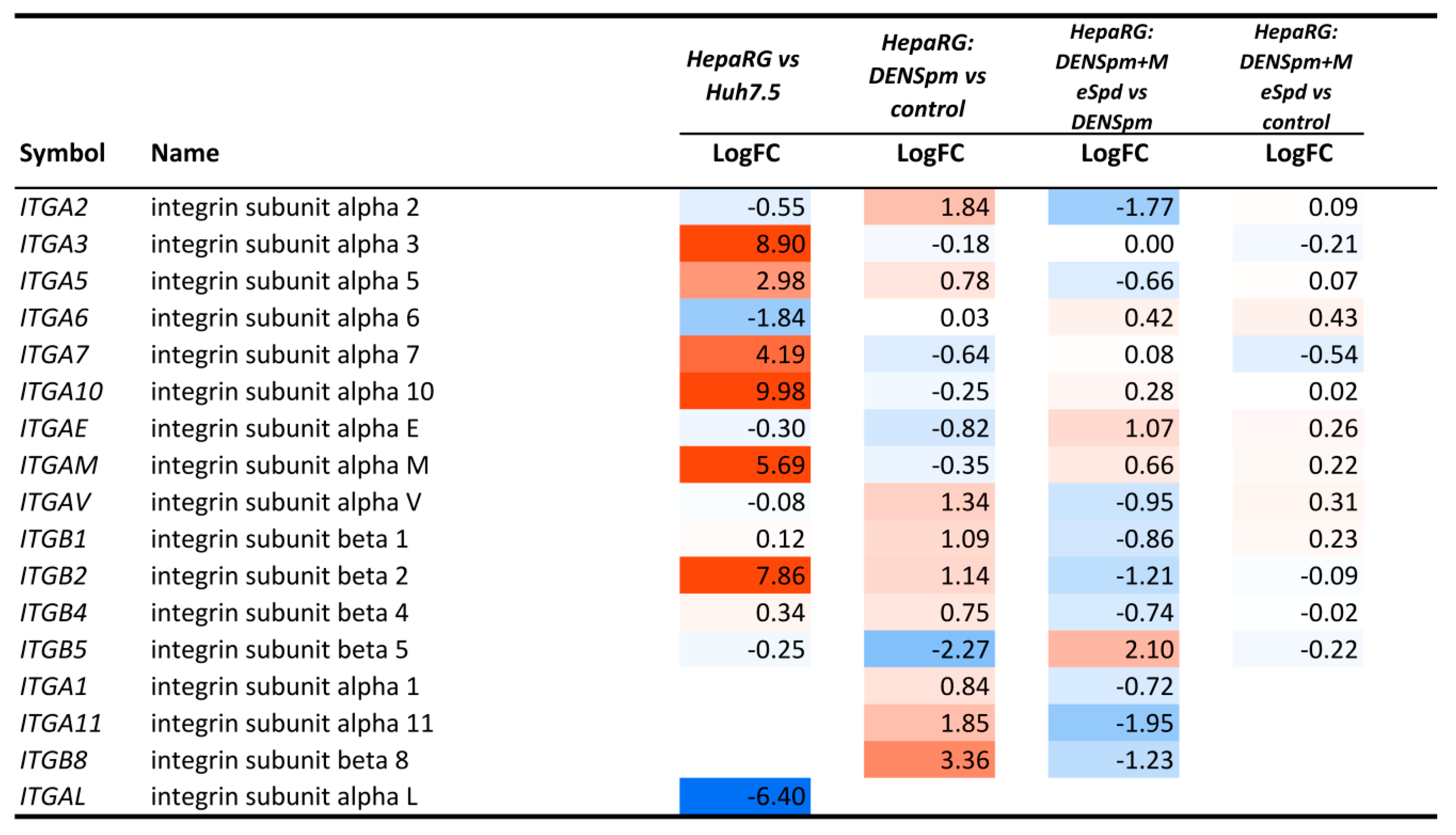
3.5. Transcriptome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.S.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Society, A.C. Cancer Facts & Figures 2018; American Cancer Society: Atlanta, GA, USA, 2018. [Google Scholar]
- Morrot, A.; da Fonseca, L.M.; Salustiano, E.J.; Gentile, L.B.; Conde, L.; Filardy, A.A.; Franklim, T.N.; da Costa, K.M.; Freire-de-Lima, C.G.; Freire-de-Lima, L. Metabolic symbiosis and immunomodulation: How tumor cell-derived lactate may disturb innate and adaptive immune responses. Front. Oncol. 2018, 8, 81. [Google Scholar] [CrossRef]
- Pegg, A.E.; Casero, R.A., Jr. Current status of the polyamine research field. Methods Mol. Biol. 2011, 720, 3–35. [Google Scholar] [CrossRef] [PubMed]
- Janne, J.; Poso, H.; Raina, A. Polyamines in rapid growth and cancer. Biochim. Biophys. Acta 1978, 473, 241–293. [Google Scholar] [CrossRef]
- Pegg, A.E. Mammalian polyamine metabolism and function. IUBMB Life 2009, 61, 880–894. [Google Scholar] [CrossRef]
- Smirnova, O.A.; Bartosch, B.; Zakirova, N.F.; Kochetkov, S.N.; Ivanov, A.V. Polyamine metabolism and oxidative protein folding in the ER as ROS-producing systems neglected in virology. Int. J. Mol. Sci. 2018, 19, 1219. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A., Jr.; Murray Stewart, T.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Casero, R.A.; Pegg, A.E. Polyamine catabolism and disease. Biochem. J. 2009, 421, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Goyal, L.; Supko, J.G.; Berlin, J.; Blaszkowsky, L.S.; Carpenter, A.; Heuman, D.M.; Hilderbrand, S.L.; Stuart, K.E.; Cotler, S.; Senzer, N.N.; et al. Phase 1 study of n(1),n(11)diethylnorspermine (DENSPM) in patients with advanced hepatocellular carcinoma. Cancer Chemother. Pharmacol. 2013, 72, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Hahm, H.A.; Ettinger, D.S.; Bowling, K.; Hoker, B.; Chen, T.L.; Zabelina, Y.; Casero, R.A., Jr. Phase I study of n(1),n(11)-diethylnorspermine in patients with non-small cell lung cancer. Clin. Cancer Res. 2002, 8, 684–690. [Google Scholar]
- Wolff, A.C.; Armstrong, D.K.; Fetting, J.H.; Carducci, M.K.; Riley, C.D.; Bender, J.F.; Casero, R.A., Jr.; Davidson, N.E. A phase ii study of the polyamine analog n(1),n(11)-diethylnorspermine (DENSPM) daily for five days every 21 days in patients with previously treated metastatic breast cancer. Clin. Cancer Res. 2003, 9, 5922–5928. [Google Scholar] [PubMed]
- Creaven, P.J.; Perez, R.; Pendyala, L.; Meropol, N.J.; Loewen, G.; Levine, E.; Berghorn, E.; Raghavan, D. Unusual central nervous system toxicity in a phase I study of n(1),n(11)-diethylnorspermine in patients with advanced malignancy. Investig. New Drugs 1997, 15, 227–234. [Google Scholar] [CrossRef]
- Saulnier Sholler, G.L.; Gerner, E.W.; Bergendahl, G.; MacArthur, R.B.; VanderWerff, A.; Ashikaga, T.; Bond, J.P.; Ferguson, W.; Roberts, W.; Wada, R.K.; et al. A phase I trial of DFMO targeting polyamine addiction in patients with relapsed/refractory neuroblastoma. PLoS ONE 2015, 10, e0127246. [Google Scholar] [CrossRef] [PubMed]
- Samal, K.; Zhao, P.; Kendzicky, A.; Yco, L.P.; McClung, H.; Gerner, E.; Burns, M.; Bachmann, A.S.; Sholler, G. Amxt-1501, a novel polyamine transport inhibitor, synergizes with dfmo in inhibiting neuroblastoma cell proliferation by targeting both ornithine decarboxylase and polyamine transport. Int. J. Cancer 2013, 133, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.R.; Li, J.; Bass, B.L.; Wang, J.Y. Expression of the transforming growth factor-beta gene during growth inhibition following polyamine depletion. Am. J. Physiol. 1998, 275, C590–C598. [Google Scholar] [CrossRef]
- Rao, J.N.; Li, L.; Bass, B.L.; Wang, J.Y. Expression of the TGF-beta receptor gene and sensitivity to growth inhibition following polyamine depletion. Am. J. Physiol. Cell Physiol. 2000, 279, C1034–C1044. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Santora, R.; Rao, J.N.; Guo, X.; Zou, T.; Zhang, H.M.; Turner, D.J.; Wang, J.Y. Activation of TGF-beta-SMAD signaling pathway following polyamine depletion in intestinal epithelial cells. Am. J. Physiol. 2003, 285, G1056–G1067. [Google Scholar] [CrossRef]
- Guo, X.; Rao, J.N.; Liu, L.; Zou, T.; Keledjian, K.M.; Boneva, D.; Marasa, B.S.; Wang, J.Y. Polyamines are necessary for synthesis and stability of occludin protein in intestinal epithelial cells. Am. J. Physiol. 2005, 288, G1159–G1169. [Google Scholar] [CrossRef]
- Prunotto, M.; Compagnone, A.; Bruschi, M.; Candiano, G.; Colombatto, S.; Bandino, A.; Petretto, A.; Moll, S.; Bochaton-Piallat, M.L.; Gabbiani, G.; et al. Endocellular polyamine availability modulates epithelial-to-mesenchymal transition and unfolded protein response in MDCK cells. Lab. Investig. 2010, 90, 929–939. [Google Scholar] [CrossRef]
- Compagnone, A.; Bandino, A.; Meli, F.; Bravoco, V.; Cravanzola, C.; Parola, M.; Colombatto, S. Polyamines modulate epithelial-to-mesenchymal transition. Amino Acids 2012, 42, 783–789. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis b virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y.; Kawai, K.; Yamazaki, H.; Nakamura, M.; Bree, F.; Guguen-Guillouzo, C.; Suemizu, H. The human hepatic cell line HEPARG as a possible cell source for the generation of humanized liver TK-NOG mice. Xenobiotica 2014, 44, 146–153. [Google Scholar] [CrossRef]
- Guillouzo, A.; Corlu, A.; Aninat, C.; Glaise, D.; Morel, F.; Guguen-Guillouzo, C. The human hepatoma heparg cells: A highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem. Biol. Interact. 2007, 168, 66–73. [Google Scholar] [CrossRef]
- Khomutov, M.A.; Keinanen, T.A.; Hyvonen, M.T.; Weisell, J.; Vepsalainen, J.; Alhonen, L.; Kochetkov, S.N.; Khomutov, A.R. Enantioselective synthesis of (r)- and (s)-3-methylspermidines. Russ. J. Bioorg. Chem. 2015, 41, 548–553. [Google Scholar] [CrossRef]
- Grigorenko, N.A.; Khomutov, A.R.; Keinanen, T.A.; Jarvinen, A.; Alhonen, L.; Janne, J.; Vepsalainen, J. Synthesis of novel optical isomers of alpha-methylpolyamines. Tetrahedron 2007, 63, 2257–2262. [Google Scholar] [CrossRef]
- Rehse, K.; Puchert, E.; Leissring, S. Antiaggregatory and anticoagulant effects of oligoamines. 12. Alkyl- and arylalkyl- derivatives of putrescine, spermidine and spermine. Arch. Pharm. 1990, 323, 287–294. [Google Scholar] [CrossRef]
- Hyvonen, T.; Keinanen, T.A.; Khomutov, A.R.; Khomutov, R.M.; Eloranta, T.O. Monitoring of the uptake and metabolism of aminooxy analogues of polyamines in cultured cells by high-performance liquid chromatography. J. Chromatogr. 1992, 574, 17–21. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Smirnova, O.A.; Petrushanko, I.Y.; Ivanova, O.N.; Karpenko, I.L.; Alekseeva, E.; Sominskaya, I.; Makarov, A.A.; Bartosch, B.; Kochetkov, S.N.; et al. Hcv core protein uses multiple mechanisms to induce oxidative stress in human hepatoma huh7 cells. Viruses 2015, 7, 2745–2770. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Smirnova, O.A.; Ivanova, O.N.; Masalova, O.V.; Kochetkov, S.N.; Isaguliants, M.G. Hepatitis c virus proteins activate nrf2/are pathway by distinct ros-dependent and independent mechanisms in huh7 cells. PLoS ONE 2011, 6, e24957. [Google Scholar] [CrossRef]
- Krasnov, G.S.; Dmitriev, A.A.; Kudryavtseva, A.V.; Shargunov, A.V.; Karpov, D.S.; Uroshlev, L.A.; Melnikova, N.V.; Blinov, V.M.; Poverennaya, E.V.; Archakov, A.I.; et al. Ppline: An automated pipeline for snp, sap, and splice variant detection in the context of proteogenomics. J. Proteome Res. 2015, 14, 3729–3737. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Star: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Sigurgeirsson, B.; Emanuelsson, O.; Lundeberg, J. Sequencing degraded RNA addressed by 3’ tag counting. PLoS ONE 2014, 9, e91851. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSEQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Clusterprofiler: An r package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An r/bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Conover, W.J.; Iman, R.L. On Multiple-Comparisons Procedures; Los Alamos Scientific Laboratory: Los Alamos, NM, USA, 1979.
- Kanebratt, K.P.; Andersson, T.B. Evaluation of HEPARG cells as an in vitro model for human drug metabolism studies. Drug Metab. Dispos. 2008, 36, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Sylvester, P.W. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol. Biol. 2011, 716, 157–168. [Google Scholar] [CrossRef]
- Sliwka, L.; Wiktorska, K.; Suchocki, P.; Milczarek, M.; Mielczarek, S.; Lubelska, K.; Cierpial, T.; Lyzwa, P.; Kielbasinski, P.; Jaromin, A.; et al. The comparison of MTT and CVS assays for the assessment of anticancer agent interactions. PLoS ONE 2016, 11, e0155772. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.M.; Desai, L.P. Reciprocal regulation of TGF-beta and reactive oxygen species: A perverse cycle for fibrosis. Redox Biol. 2015, 6, 565–577. [Google Scholar] [CrossRef]
- Yoshida, M.; Tomitori, H.; Machi, Y.; Hagihara, M.; Higashi, K.; Goda, H.; Ohya, T.; Niitsu, M.; Kashiwagi, K.; Igarashi, K. Acrolein toxicity: Comparison with reactive oxygen species. Biochem. Biophys. Res. Commun. 2009, 378, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.; Nishimura, K.; Ishii, I.; Omura, T.; Okuyama, S.; Kashiwagi, K.; Igarashi, K. Intense correlation between brain infarction and protein-conjugated acrolein. Stroke 2009, 40, 3356–3361. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Wang, S.J.; Li, D.; Chu, B.; Gu, W. Activation of sat1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. USA 2016, 113, E6806–E6812. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Chang, G.D. Methylglyoxal in cells elicits a negative feedback loop entailing transglutaminase 2 and glyoxalase 1. Redox Biol. 2014, 2, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Hyvonen, M.T.; Khomutov, M.; Petit, M.; Weisell, J.; Kochetkov, S.N.; Alhonen, L.; Vepsalainen, J.; Khomutov, A.R.; Keinanen, T.A. Enantiomers of 3-methylspermidine selectively modulate deoxyhypusine synthesis and reveal important determinants for spermidine transport. ACS Chem. Biol. 2015, 10, 1417–1424. [Google Scholar] [CrossRef]
- Hyvonen, M.T.; Keinanen, T.A.; Cerrada-Gimenez, M.; Sinervirta, R.; Grigorenko, N.; Khomutov, A.R.; Vepsalainen, J.; Alhonen, L.; Janne, J. Role of hypusinated eukaryotic translation initiation factor 5a in polyamine depletion-induced cytostasis. J. Biol. Chem. 2007, 282, 34700–34706. [Google Scholar] [CrossRef] [PubMed]
- Wehrle-Haller, B. The role of integrins in cell migration. In Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2000–2013. [Google Scholar]
- Cerec, V.; Glaise, D.; Garnier, D.; Morosan, S.; Turlin, B.; Drenou, B.; Gripon, P.; Kremsdorf, D.; Guguen-Guillouzo, C.; Corlu, A. Transdifferentiation of hepatocyte-like cells from the human hepatoma HEPARG cell line through bipotent progenitor. Hepatology 2007, 45, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Borzi, R.M.; Guidotti, S.; Minguzzi, M.; Facchini, A.; Platano, D.; Trisolino, G.; Filardo, G.; Cetrullo, S.; D’Adamo, S.; Stefanelli, C.; et al. Polyamine delivery as a tool to modulate stem cell differentiation in skeletal tissue engineering. Amino Acids 2014, 46, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Chen, Y.; Huang, Y.P.; Hsu, Y.C.; Chiang, L.H.; Chen, T.Y.; Wang, G.J. Exogenous polyamines promote osteogenic differentiation by reciprocally regulating osteogenic and adipogenic gene expression. J. Cell. Biochem. 2013, 114, 2718–2728. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.H.; Lin, K.L.; Huang, Y.P.; Hsu, Y.C.; Chen, C.H.; Chen, Y.; Sie, M.H.; Wang, G.J.; Lee, M.J. Suppression of ornithine decarboxylase promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. FEBS Lett. 2015, 589, 2058–2065. [Google Scholar] [CrossRef] [PubMed]
- Pirnes-Karhu, S.; Mantymaa, P.; Sironen, R.; Makinen, P.I.; Wojciechowski, S.; Juutinen, S.; Koistinaho, J.; Horkko, S.; Jantunen, E.; Alhonen, L.; et al. Enhanced polyamine catabolism disturbs hematopoietic lineage commitment and leads to a myeloproliferative disease in mice overexpressing spermidine/spermine n(1)-acetyltransferase. Amino Acids 2014, 46, 689–700. [Google Scholar] [CrossRef]
- Pirnes-Karhu, S.; Maatta, J.; Finnila, M.; Alhonen, L.; Uimari, A. Overexpression of spermidine/spermine n1-acetyltransferase impairs osteoblastogenesis and alters mouse bone phenotype. Transgen. Res. 2015, 24, 253–265. [Google Scholar] [CrossRef]
- Cervelli, M.; Fratini, E.; Amendola, R.; Bianchi, M.; Signori, E.; Ferraro, E.; Lisi, A.; Federico, R.; Marcocci, L.; Mariottini, P. Increased spermine oxidase (SMO) activity as a novel differentiation marker of myogenic c2c12 cells. Int. J. Biochem. Cell Biol. 2009, 41, 934–944. [Google Scholar] [CrossRef]
- Quemener, V.; Blanchard, Y.; Lescoat, D.; Havouis, R.; Moulinoux, J.P. Depletion in nuclear spermine during human spermatogenesis, a natural process of cell differentiation. Am. J. Physiol. 1992, 263, C343–C347. [Google Scholar] [CrossRef]
- Pietila, M.; Pirinen, E.; Keskitalo, S.; Juutinen, S.; Pasonen-Seppanen, S.; Keinanen, T.; Alhonen, L.; Janne, J. Disturbed keratinocyte differentiation in transgenic mice and organotypic keratinocyte cultures as a result of spermidine/spermine n-acetyltransferase overexpression. J. Investig. Dermatol. 2005, 124, 596–601. [Google Scholar] [CrossRef]
- Vuohelainen, S.; Pirinen, E.; Cerrada-Gimenez, M.; Keinanen, T.A.; Uimari, A.; Pietila, M.; Khomutov, A.R.; Janne, J.; Alhonen, L. Spermidine is indispensable in differentiation of 3t3-l1 fibroblasts to adipocytes. J. Cell. Mol. Med. 2010, 14, 1683–1692. [Google Scholar] [CrossRef]
- Hyvonen, M.T.; Koponen, T.; Weisell, J.; Pietila, M.; Khomutov, A.R.; Vepsalainen, J.; Alhonen, L.; Keinanen, T.A. Spermidine promotes adipogenesis of 3t3-l1 cells by preventing interaction of anp32 with HUR and pp2a. Biochem. J. 2013, 453, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Ishii, I.; Ikeguchi, Y.; Mano, H.; Wada, M.; Pegg, A.E.; Shirahata, A. Polyamine metabolism is involved in adipogenesis of 3t3-l1 cells. Amino Acids 2012, 42, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Alhonen, L.; Rasanen, T.L.; Sinervirta, R.; Parkkinen, J.J.; Korhonen, V.P.; Pietila, M.; Janne, J. Polyamines are required for the initiation of rat liver regeneration. Biochem. J. 2002, 362, 149–153. [Google Scholar] [CrossRef]
- Oh, S.H.; Swiderska-Syn, M.; Jewell, M.L.; Premont, R.T.; Diehl, A.M. Liver regeneration requires Yap1-TGFbeta-dependent epithelial-mesenchymal transition in hepatocytes. J. Hepatol. 2018, 69, 359–367. [Google Scholar] [CrossRef]
- Lu, W.Y.; Bird, T.G.; Boulter, L.; Tsuchiya, A.; Cole, A.M.; Hay, T.; Guest, R.V.; Wojtacha, D.; Man, T.Y.; Mackinnon, A.; et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 2015, 17, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Conigliaro, A.; Amicone, L.; Costa, V.; De Santis Puzzonia, M.; Mancone, C.; Sacchetti, B.; Cicchini, C.; Garibaldi, F.; Brenner, D.A.; Kisseleva, T.; et al. Evidence for a common progenitor of epithelial and mesenchymal components of the liver. Cell Death Differ. 2013, 20, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhao, L.; Fish, M.; Logan, C.Y.; Nusse, R. Self-renewing diploid axin2(+) cells fuel homeostatic renewal of the liver. Nature 2015, 524, 180–185. [Google Scholar] [CrossRef]
- Gutierrez, E.; Shin, B.S.; Woolstenhulme, C.J.; Kim, J.R.; Saini, P.; Buskirk, A.R.; Dever, T.E. Eif5a promotes translation of polyproline motifs. Mol. Cell 2013, 51, 35–45. [Google Scholar] [CrossRef]
- Li, S.S. Specificity and versatility of sh3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem. J. 2005, 390, 641–653. [Google Scholar] [CrossRef]
- Burgio, G.; Corona, D.F.; Nicotra, C.M.; Carruba, G.; Taibi, G. P/caf-mediated spermidine acetylation regulates histone acetyltransferase activity. J. Enzyme Inhib. Med. Chem. 2016, 31, 75–82. [Google Scholar] [CrossRef]
- McCormack, S.A.; Johnson, L.R. Polyamines and cell migration. J. Physiol. Pharmacol. 2001, 52, 327–349. [Google Scholar] [PubMed]
- Chen, C.; Young, B.A.; Coleman, C.S.; Pegg, A.E.; Sheppard, D. Spermidine/spermine n1-acetyltransferase specifically binds to the integrin alpha9 subunit cytoplasmic domain and enhances cell migration. J. Cell Biol. 2004, 167, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Kohn-Gaone, J.; Gogoi-Tiwari, J.; Ramm, G.A.; Olynyk, J.K.; Tirnitz-Parker, J.E. The role of liver progenitor cells during liver regeneration, fibrogenesis, and carcinogenesis. Am. J. Physiol. 2016, 310, G143–G154. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Wang, X.W. Cancer stem cells in the development of liver cancer. J. Clin. Investing. 2013, 123, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; et al. EPCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Tyurina, D.A.; Ivanova, O.N.; Kochetkov, S.N.; Bartosch, B.; Isaguliants, M.G. Oxidative stress, a trigger of hepatitis c and b virus-induced liver carcinogenesis. Oncotarget 2017, 8, 3895–3932. [Google Scholar] [CrossRef]
- Smirnova, O.A.; Keinanen, T.A.; Ivanova, O.N.; Hyvonen, M.T.; Khomutov, A.R.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Hepatitis C virus alters metabolism of biogenic polyamines by affecting expression of key enzymes of their metabolism. Biochem. Biophys. Res. Commun. 2017, 483, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Gaude, E.; Frezza, C. Tissue-specific and convergent metabolic transformation of cancer correlates with metastatic potential and patient survival. Nat. Commun. 2016, 7, 13041. [Google Scholar] [CrossRef]
- Liu, M.; Quek, L.E.; Sultani, G.; Turner, N. Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma. Cancer Metab. 2016, 4, 19. [Google Scholar] [CrossRef] [PubMed]
| Cell Line | Put (nmol/mg DNA) | Spd (nmol/mg DNA) | Spm (nmol/mg DNA) | N1-AcSpd (nmol/mg DNA) |
|---|---|---|---|---|
| Huh7.5 1 | N.D. 2 | 17.8 ± 5.9 | 63.2 ± 15.0 | N.D. 2 |
| HepG2 1 | N.D. 2 | 20.7 ± 5.8 | 54.2 ± 16.5 | 8.5 ± 1.5 |
| HepaRGundiff 1 | 10.4 ± 1.0 | 54.8 ± 6.3 | 53.9 ± 3.7 | N.D. 2 |
| HepaRGdiff 3 | N.D. 2 | N.D. 2 | 2.6 ± 0.3 | N.D. 2 |
| SkHep1 1 | 9.9 ± 4.3 | 25.0 ± 7.7 | 16.5 ± 3.4 | N.D. 2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, O.N.; Snezhkina, A.V.; Krasnov, G.S.; Valuev-Elliston, V.T.; Khomich, O.A.; Khomutov, A.R.; Keinanen, T.A.; Alhonen, L.; Bartosch, B.; Kudryavtseva, A.V.; et al. Activation of Polyamine Catabolism by N1,N11-Diethylnorspermine in Hepatic HepaRG Cells Induces Dedifferentiation and Mesenchymal-Like Phenotype. Cells 2018, 7, 275. https://doi.org/10.3390/cells7120275
Ivanova ON, Snezhkina AV, Krasnov GS, Valuev-Elliston VT, Khomich OA, Khomutov AR, Keinanen TA, Alhonen L, Bartosch B, Kudryavtseva AV, et al. Activation of Polyamine Catabolism by N1,N11-Diethylnorspermine in Hepatic HepaRG Cells Induces Dedifferentiation and Mesenchymal-Like Phenotype. Cells. 2018; 7(12):275. https://doi.org/10.3390/cells7120275
Chicago/Turabian StyleIvanova, Olga N., Anastasiya V. Snezhkina, George S. Krasnov, Vladimir T. Valuev-Elliston, Olga A. Khomich, Alexey R. Khomutov, Tuomo A. Keinanen, Leena Alhonen, Birke Bartosch, Anna V. Kudryavtseva, and et al. 2018. "Activation of Polyamine Catabolism by N1,N11-Diethylnorspermine in Hepatic HepaRG Cells Induces Dedifferentiation and Mesenchymal-Like Phenotype" Cells 7, no. 12: 275. https://doi.org/10.3390/cells7120275
APA StyleIvanova, O. N., Snezhkina, A. V., Krasnov, G. S., Valuev-Elliston, V. T., Khomich, O. A., Khomutov, A. R., Keinanen, T. A., Alhonen, L., Bartosch, B., Kudryavtseva, A. V., Kochetkov, S. N., & Ivanov, A. V. (2018). Activation of Polyamine Catabolism by N1,N11-Diethylnorspermine in Hepatic HepaRG Cells Induces Dedifferentiation and Mesenchymal-Like Phenotype. Cells, 7(12), 275. https://doi.org/10.3390/cells7120275








