Accumulation of Ag(I) by Saccharomyces cerevisiae Cells Expressing Plant Metallothioneins
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Media
2.2. Plasmids and Yeast Transformation
2.3. Yeast Cell Growth Assay
2.3.1. Growth in Liquid Media
2.3.2. Growth on Solid Media
2.3.3. Cell Viability and LC50
2.4. Silver Accumulation by Growing Cells
2.5. myrGFP-MT Localization
2.6. Statistics
3. Results
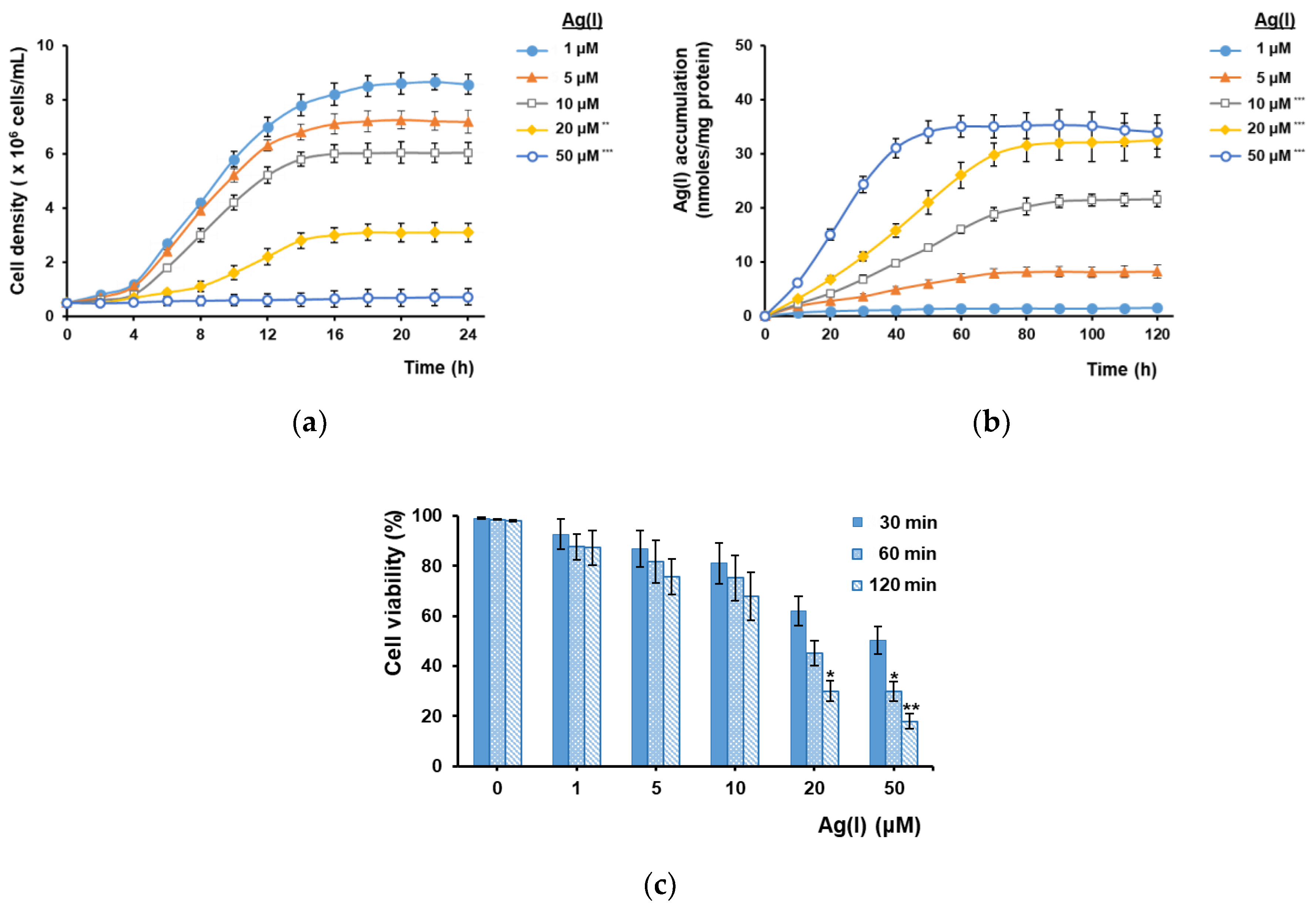
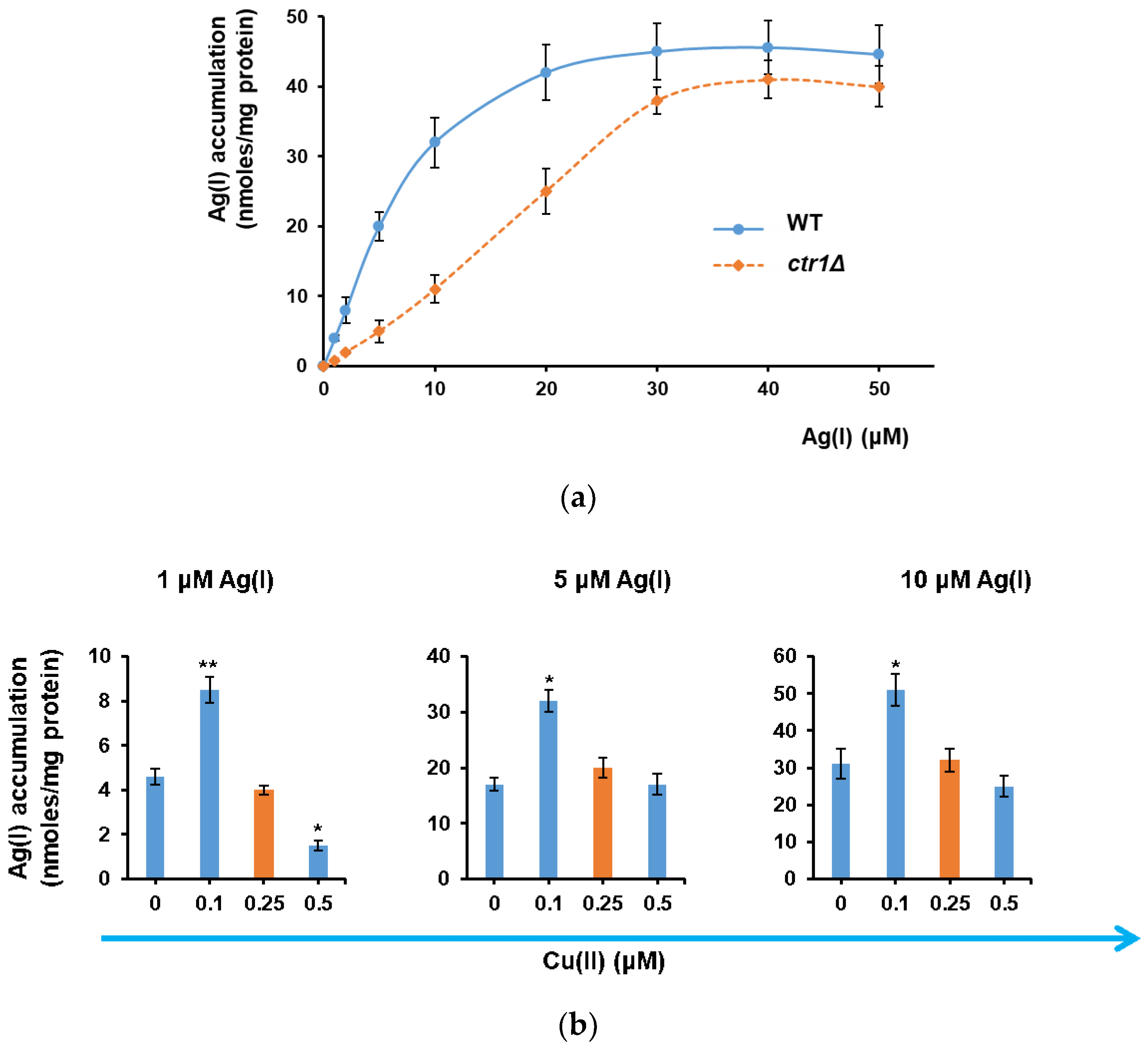
3.1. Ag(I) Uptake by Yeast Cells Depends on Ctr1 and Is Facilitated by Cu(II) Depletion
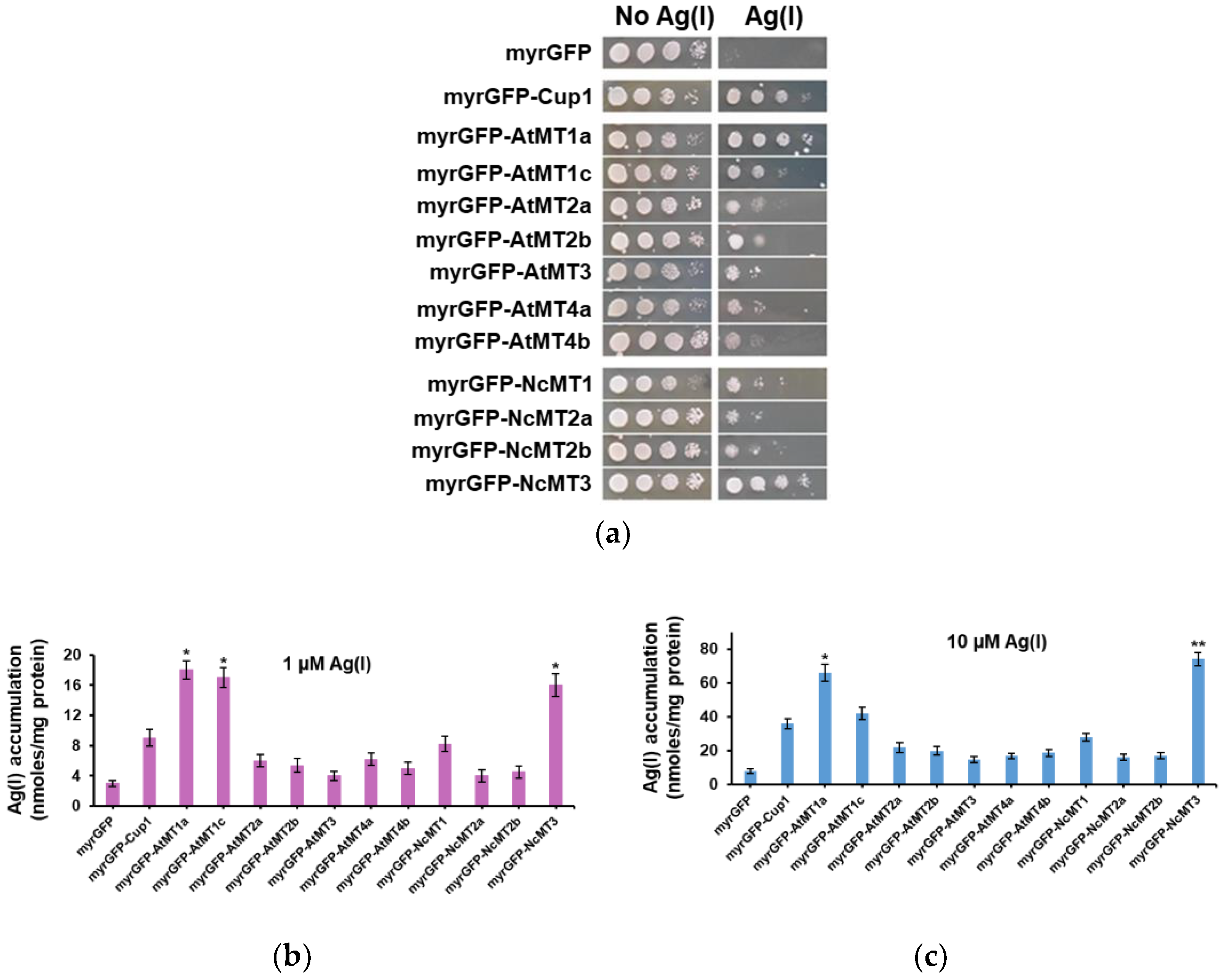
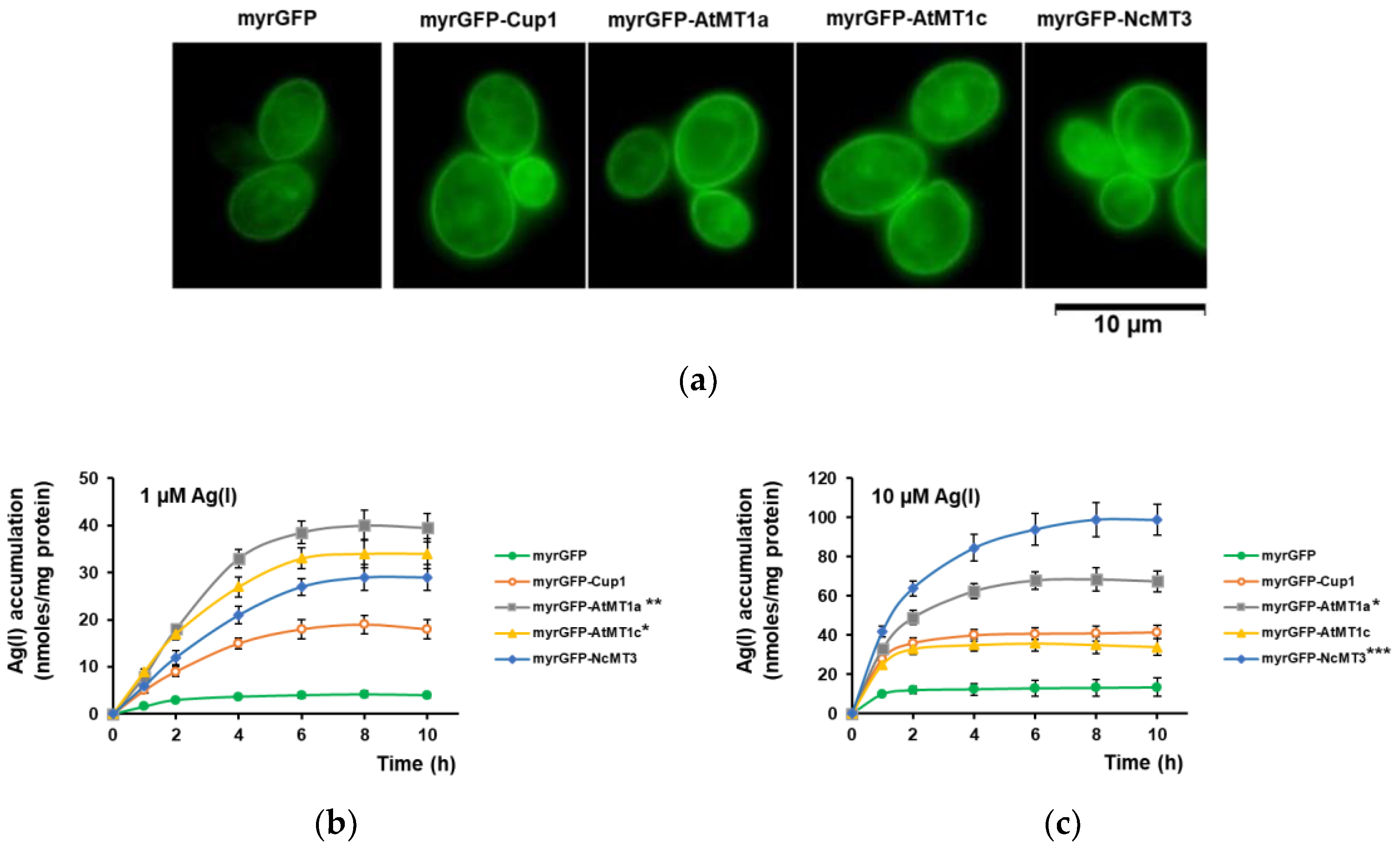
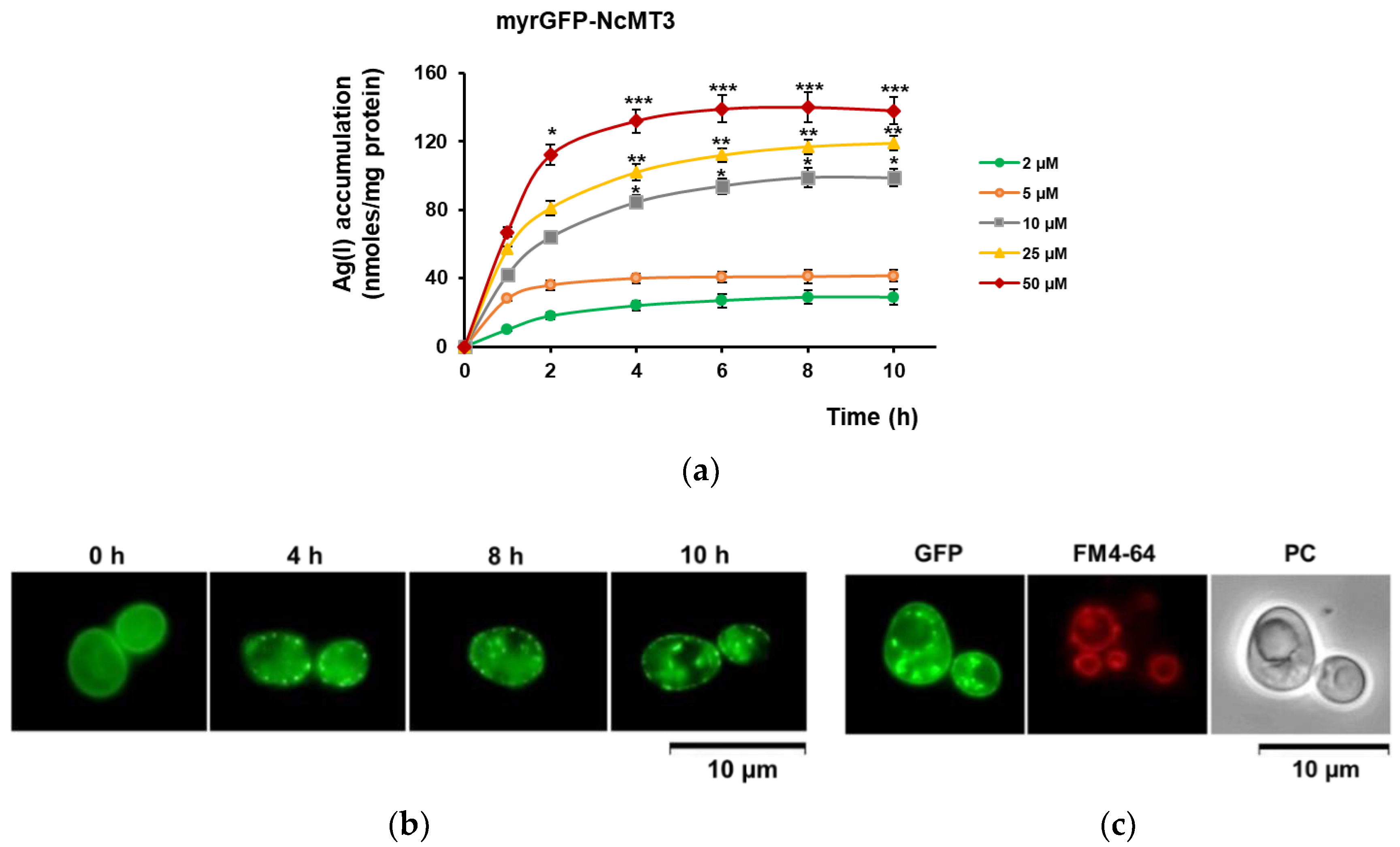
3.2. Ag(I) Accumulation by Yeast Cells Expressing Plant MTs Targeted to the Inner Face of Plasma Membrane
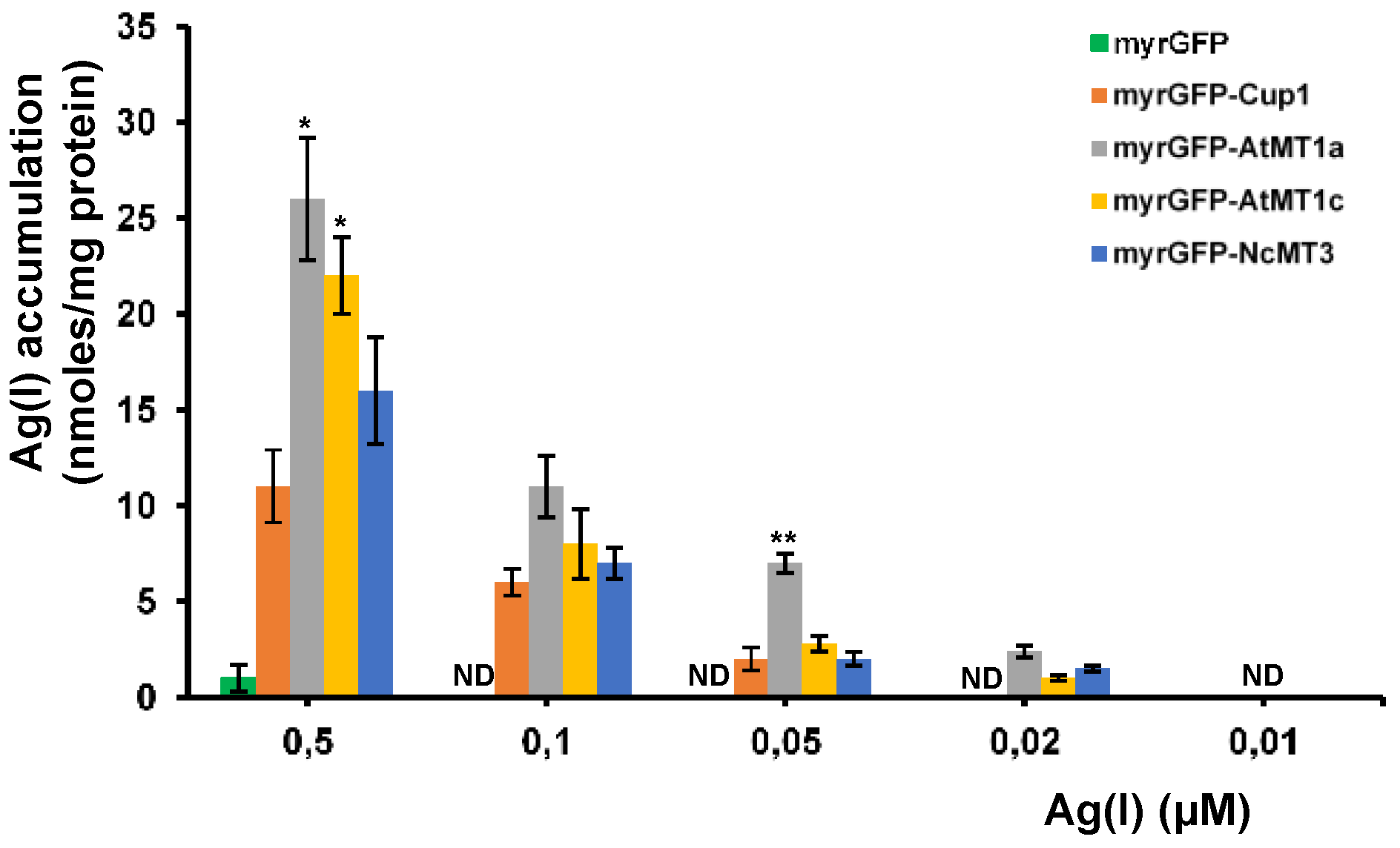
3.3. Accumulation of Ultra-Trace Ag(I)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnson, D.B. Development and application of biotechnologies in the metal mining industry. Environ. Sci. Pollut. Res. Int. 2013, 20, 7768–7776. [Google Scholar] [CrossRef] [PubMed]
- Ratte, H.T. Bioaccumulation and toxicity of silver compounds: A review. Environ. Toxicol. Chem. 1999, 18, 89–108. [Google Scholar] [CrossRef]
- Zhang, W.; Xiao, B.; Fang, T. Chemical transformation of silver nanoparticles in aquatic environments: Mechanism, morphology and toxicity. Chemosphere 2018, 191, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Purcell, T.W.; Peters, J.J. Sources of silver in the environment. Environ. Toxicol. Chem. 1998, 17, 539–546. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Engineering of microorganisms towards recovery of rare metal ions. Appl. Microbiol. Biotechnol. 2010, 87, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Soares, E.V.; Soares, H.M. Cleanup of industrial effluents containing heavy metals: A new opportunity of valorising the biomass produced by brewing industry. Appl. Microbiol. Biotechnol. 2013, 97, 6667–6675. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ho, S.H.; Hasunuma, T.; Chang, J.S.; Ren, N.Q.; Kondo, A. Recent advances in yeast cell-surface display technologies for waste biorefineries. Bioresour. Technol. 2016, 215, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Gadd, G.M. Metal and metalloid biorecovery using fungi. Microb. Biotechnol. 2017, 10, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Ruta, L.L.; Kissen, R.; Nicolau, I.; Neagoe, A.D.; Petrescu, A.J.; Bones, A.M.; Farcasanu, I.C. Heavy metal accumulation by Saccharomyces cerevisiae cells armed with metal binding hexapeptides targeted to the inner face of the plasma membrane. Appl. Microbiol. Biotechnol. 2017, 101, 5749–5763. [Google Scholar] [CrossRef]
- Ruta, L.L.; Lin, Y.F.; Kissen, R.; Nicolau, I.; Neagoe, A.D.; Ghenea, S.; Bones, A.M.; Farcasanu, I.C. Anchoring plant metallothioneins to the inner face of the plasma membrane of Saccharomyces cerevisiae cells leads to heavy metal accumulation. PLoS ONE 2017, 12, e0178393. [Google Scholar] [CrossRef]
- Capdevila, M.; Atrian, S. Metallothionein protein evolution: A miniassay. J. Biol. Inorg. Chem. 2011, 16, 977–989. [Google Scholar] [CrossRef]
- Palacios, O.; Atrian, S.; Capdevila, M. Zn- and Cu-thioneins: A functional classification for metallothioneins? J. Biol. Inorg. Chem. 2011, 16, 991–1009. [Google Scholar] [CrossRef] [PubMed]
- Valls, M.; Bofill, R.; Gonzalez-Duarte, R.; Gonzalez-Duarte, P.; Capdevila, M.; Atrian, S. A new insight into metallothionein (MT) classification and evolution: The in vivo and in vitro metal binding features of Homarus americanus recombinant MT. J. Biol. Chem. 2001, 276, 32835–32843. [Google Scholar] [CrossRef] [PubMed]
- Vasak, M. Metal removal and substitution in vertebrate and invertebrate metallothioneins. In Metallobiochemistry Part. B, Metallothionein and Related Molecules, 1st ed.; Riordan, J.F., Vallee, B.L., Eds.; Academic Press: San Diego, CA, USA, 1991. [Google Scholar]
- Farcasanu, I.C.; Ruta, L.L. Metallothioneins, Saccharomyces cerevisiae, and heavy metals: A biotechnology triad? In Old Yeasts, New Questions; Lucas, C., Pais, C., Eds.; IntechOpen: London, UK, 2017; pp. 21–39. [Google Scholar]
- Zhang, M.; Takano, T.; Liu, S.; Zhang, X. Abiotic stress response in yeast and metal-binding ability of a type 2 metallothionein-like protein (PutMT2) from Puccinellia tenuiflora. Mol. Biol Rep. 2014, 41, 5839–5849. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar]
- Guthrie, C.; Fink, G.R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991, 194, 1–863. [Google Scholar]
- Cohen, R.; Engelberg, D. Commonly used Saccharomyces cerevisiae strains (e.g. BY4741, W303) are growth sensitive on synthetic complete medium due to poor leucine uptake. FEMS Microbiol. Lett. 2007, 273, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, R.J.; Strasser, A.W.M.; Honer, C.B.; Hollenberg, C.P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 1991, 7, 691–692. [Google Scholar] [CrossRef]
- Amberg, D.C.; Burke, D.J.; Strathern, J.N. Measuring yeast cell density by spectrophotometry. In Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual; Burke, D., Dawson, D., Stearns, T., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2005; pp. 163–165. ISBN 0879697288. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Jensen, L.T.; Howard, W.R.; Strain, J.J.; Winge, D.R.; Culotta, V.C. Enhanced effectiveness of copper ion buffering by CUP1 metallothionein compared with CRS5 metallothionein in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 18514–18519. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, J.; Wintz, H.; Kim, J.H.; Poynton, H.; Fox, T.; Vulpe, C. A model organism for iron and copper metabolism studies. Biometals 2003, 16, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Stoj, C.; Romeo, A.; Kosman, D.J.; Zhu, Z. Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 50309–50315. [Google Scholar] [CrossRef] [PubMed]
- Nevitt, T.; Ohrvik, H.; Thiele, D.J. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim. Biophys. Acta 2012, 1823, 1580–1593. [Google Scholar] [CrossRef]
- Niazi, J.H.; Sang, B.I.; Kim, Y.S.; Gu, M.B. Global gene response in Saccharomyces cerevisiae exposed to silver nanoparticles. Appl. Biochem. Biotechnol. 2011, 164, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Hassett, R.; Dix, D.R.; Eide, D.J.; Kosman, D.J. The Fe(II) permease Fet4p functions as a low affinity copper transporter and supports normal copper trafficking in Saccharomyces cerevisiae. Biochem. J. 2000, 351, 477–484. [Google Scholar] [CrossRef]
- Cohen, A.; Nelson, H.; Nelson, N. The family of SMF metal ion transporters in yeast cells. J. Biol. Chem. 2000, 275, 33388–33394. [Google Scholar] [CrossRef]
- Jensen, L.T.; Ajua-Alemanji, M.; Culotta, V.C. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 2003, 278, 42036–42040. [Google Scholar] [CrossRef]
- Thiele, D.J. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol. Cell. Biol. 1988, 8, 2745–2752. [Google Scholar] [CrossRef]
- Zhao, Y.; Strope, P.K.; Kozmin, S.G.; McCusker, J.H.; Dietrich, F.S.; Kokoska, R.J.; Petes, T.D. Structures of naturally evolved CUP1 tandem arrays in yeast indicate that these arrays are generated by unequal nonhomologous recombination. G3 (Bethesda) 2014, 4, 2259–2269. [Google Scholar] [CrossRef]
- Welch, J.; Fogel, S.; Buchman, C.; Karin, M. The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. EMBO J. 1989, 8, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Buchman, C.; Skroch, P.; Welch, J.; Fogel, S.; Karin, M. The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol. Cell. Biol. 1989, 9, 4091–4095. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, M.; Bofilla, R.; Palaciosa, O.; Atrian, S. State-of-the-art of metallothioneins at the beginning of the 21st century. Coord Chem. Rev. 2011, 256, 46–62. [Google Scholar] [CrossRef]
- Hayden, J.; Williams, M.; Granich, A.; Ahn, H.; Tenay, B.; Lukehart, J.; Highfill, C.; Dobard, S.; Kim, K. Vps1 in the late endosome-to-vacuole traffic. J. Biosci. 2013, 38, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Orihuela, R.; Domènech, J.; Bofill, R.; You, C.; Mackay, E.A.; Kägi, J.H.; Capdevila, M.; Atrian, S. The metal-binding features of the recombinant mussel Mytilus edulis MT-10-IV metallothionein. J. Biol. Inorg. Chem. 2008, 13, 801–812. [Google Scholar] [CrossRef]
- Krzciuk, K.; Gałuszka, A. Prospecting for hyperaccumulators of trace elements: A review. Crit. Rev. Biotechnol. 2015, 35, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Pollard, A.J.; Reeves, R.D.; Baker, A.J. Facultative hyperaccumulation of heavy metals and metalloids. Plant Sci. 2014, 218, 8–17. [Google Scholar] [CrossRef]
- Miransari, M. Hyperaccumulators, arbuscular mycorrhizal fungi and stress of heavy metals. Biotechnol. Adv. 2011, 29, 645–653. [Google Scholar] [CrossRef]
- Gifford, S.; Dunstan, R.H.; O’Connor, W.; Koller, C.E.; MacFarlane, G.R. Aquatic zooremediation: Deploying animals to remediate contaminated aquatic environments. Trends Biotechnol. 2007, 25, 60–65. [Google Scholar] [CrossRef]
- Borovicka, J.; Kotrba, P.; Gryndler, M.; Mihaljevic, M.; Randa, Z.; Rohovec, J.; Cajthaml, T.; Stijve, T.; Dunn, C.E. Bioaccumulation of silver in ectomycorrhizal and saprobic macrofungi from pristine and polluted areas. Sci. Total Environ. 2010, 408, 2733–2744. [Google Scholar] [CrossRef]
- Yang, W.; Li, H.; Zhang, T.; Sen, L.; Ni, W. Classification and identification of metal-accumulating plant species by cluster analysis. Environ. Sci. Pollut. Res. Int. 2014, 21, 10626–10637. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, Y.; González, A.; Fernández, P.; Blanco, J. Interspecific variation of metal concentrations in three bivalve mollusks from Galicia. Arch. Environ. Contam. Toxicol. 2004, 47, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Borovicka, J.; Randa, Z.; Jelínek, E.; Kotrba, P.; Dunn, C.E. Hyperaccumulation of silver by amanita strobiliformis and related species of the section Lepidella. Mycol. Res. 2007, 111, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Haverkamp, R.G.; Marshall, A.T.; van Agterveld, D. Pick your carats: Nanoparticles of gold-silver-copper alloy produced in vivo. J. Nanopart. Res. 2007, 9, 697–700. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruta, L.L.; Banu, M.A.; Neagoe, A.D.; Kissen, R.; Bones, A.M.; Farcasanu, I.C. Accumulation of Ag(I) by Saccharomyces cerevisiae Cells Expressing Plant Metallothioneins. Cells 2018, 7, 266. https://doi.org/10.3390/cells7120266
Ruta LL, Banu MA, Neagoe AD, Kissen R, Bones AM, Farcasanu IC. Accumulation of Ag(I) by Saccharomyces cerevisiae Cells Expressing Plant Metallothioneins. Cells. 2018; 7(12):266. https://doi.org/10.3390/cells7120266
Chicago/Turabian StyleRuta, Lavinia L., Melania A. Banu, Aurora D. Neagoe, Ralph Kissen, Atle M. Bones, and Ileana C. Farcasanu. 2018. "Accumulation of Ag(I) by Saccharomyces cerevisiae Cells Expressing Plant Metallothioneins" Cells 7, no. 12: 266. https://doi.org/10.3390/cells7120266
APA StyleRuta, L. L., Banu, M. A., Neagoe, A. D., Kissen, R., Bones, A. M., & Farcasanu, I. C. (2018). Accumulation of Ag(I) by Saccharomyces cerevisiae Cells Expressing Plant Metallothioneins. Cells, 7(12), 266. https://doi.org/10.3390/cells7120266





