lncRNA-Six1 Is a Target of miR-1611 That Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Tissues
2.3. Hematoxylin and Eosin (H&E) Staining
2.4. Cell Culture
2.5. RNA Extraction, cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR)
2.6. RNA Oligonucleotide and Plasmid Construction
2.7. Cell Transfection
2.8. Dual-Luciferase Reporter Assay
2.9. Western Blot
2.10. Flow Cytometric Analysis of the Cell Cycle
2.11. 5-Ethynyl-2’-Deoxyuridine (EdU) Assay
2.12. CCK-8 Assay
2.13. Immunofluorescence
2.14. Statistical Analysis
3. Results
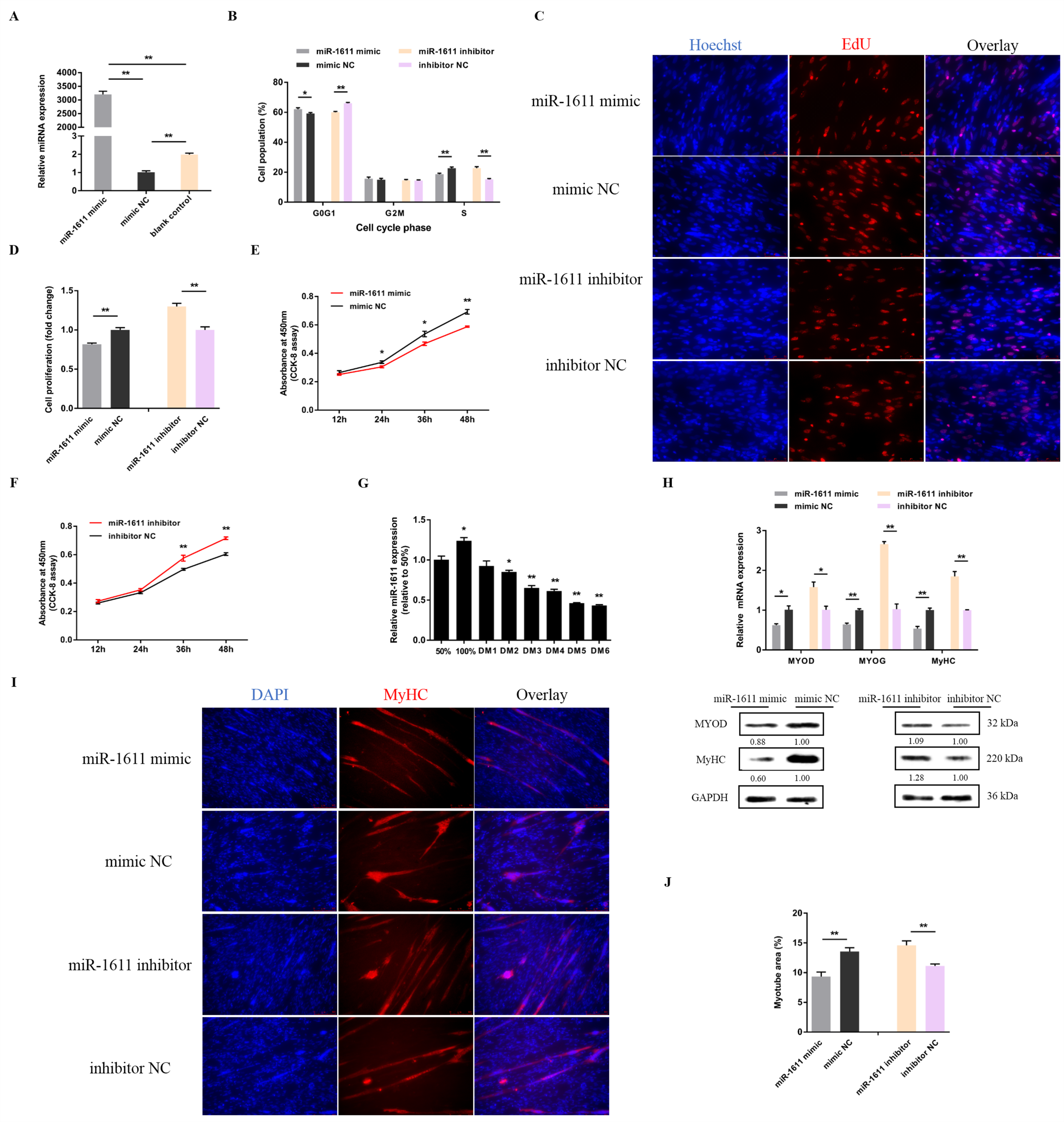
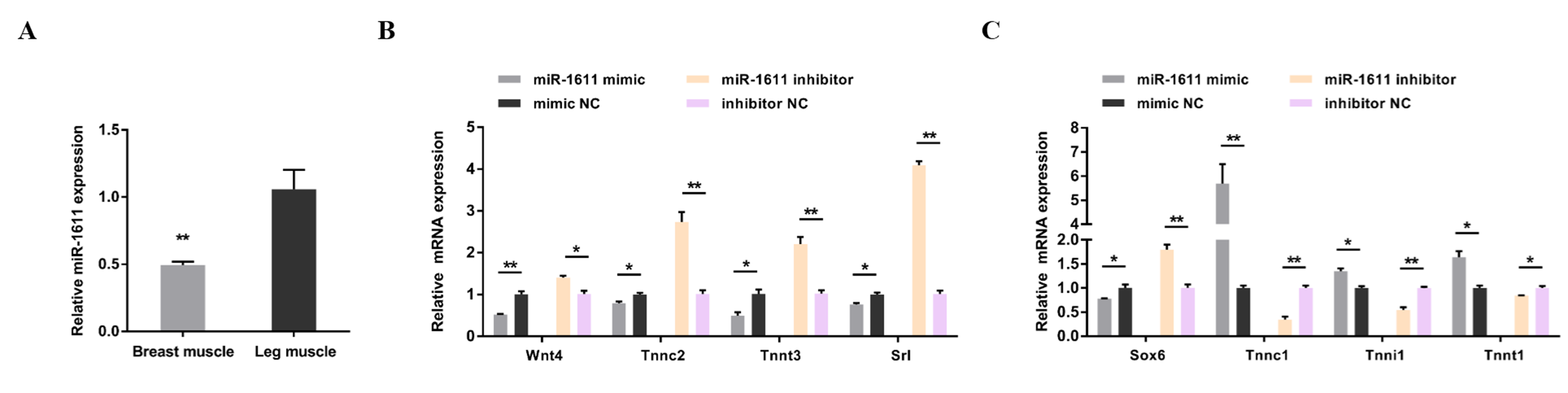
3.1. miR-1611 Regulates the Proliferation and Differentiation of Myoblasts, and Is Involved in the Formation of Skeletal Muscle Fibers
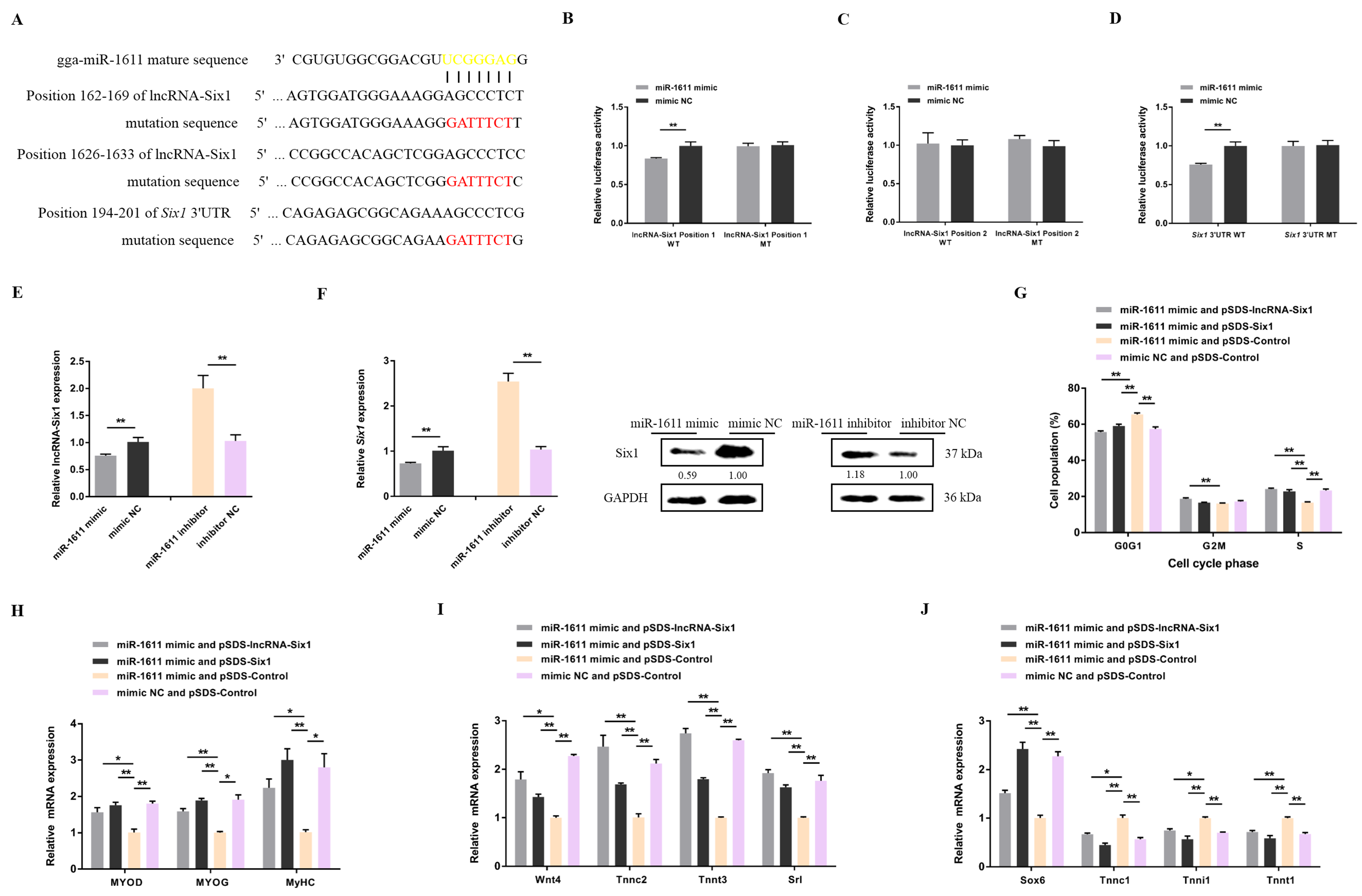
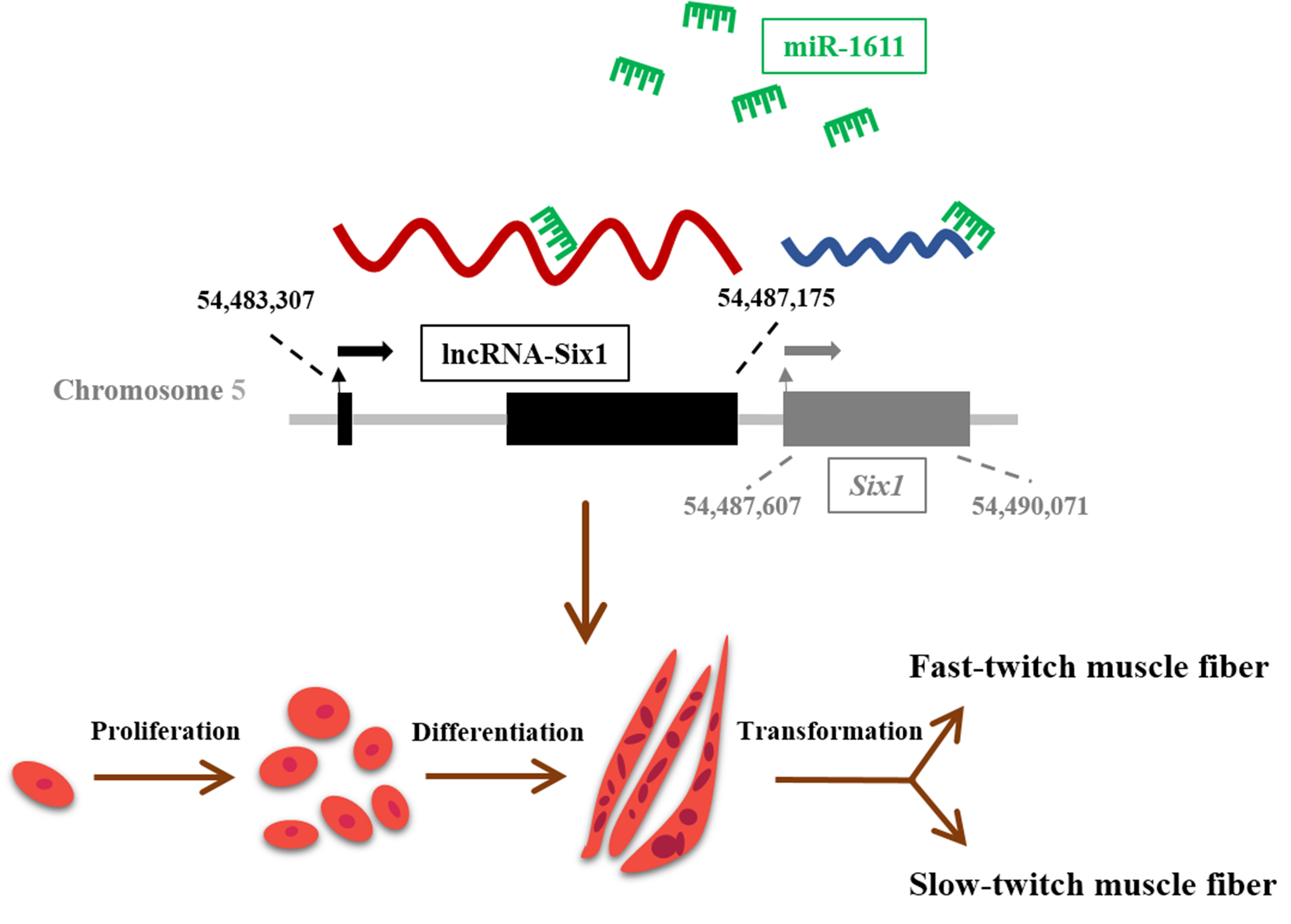
3.2. miR-1611 Is a Direct Target of the ceRNA Network between lncRNA-Six1 and Six1
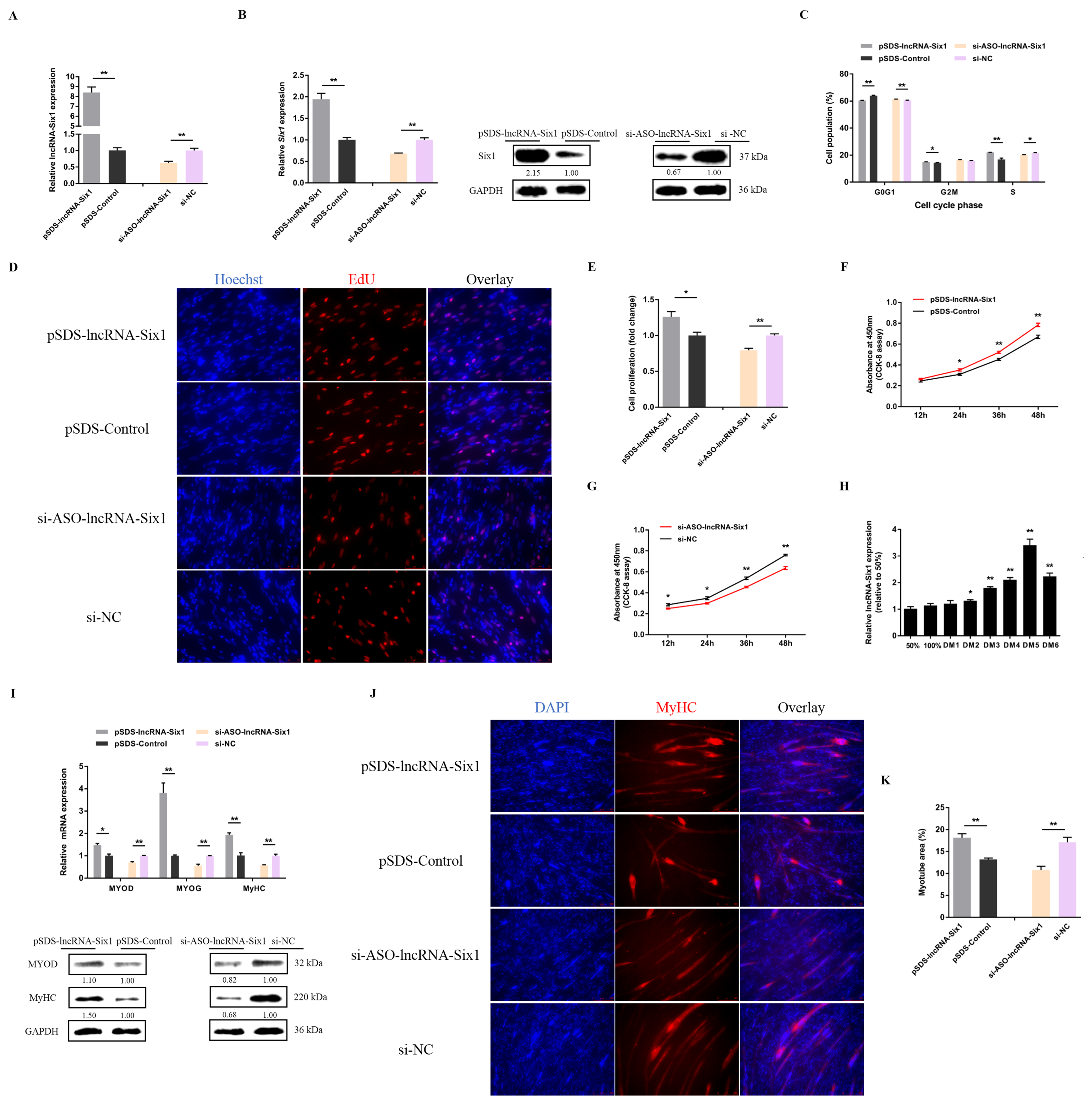
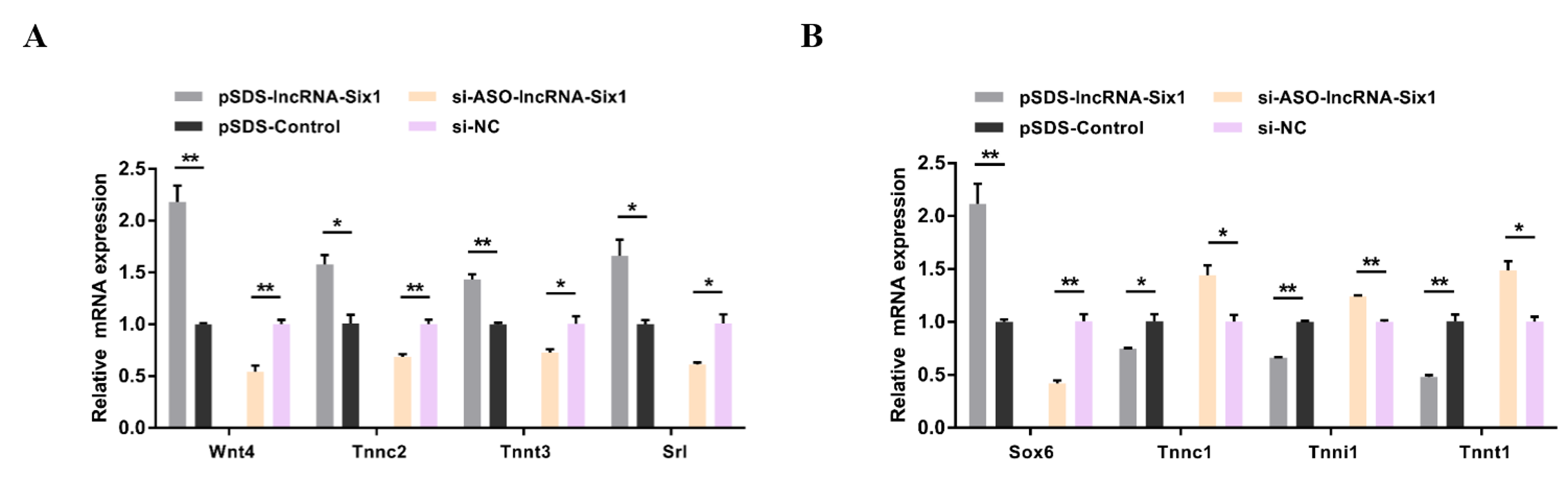
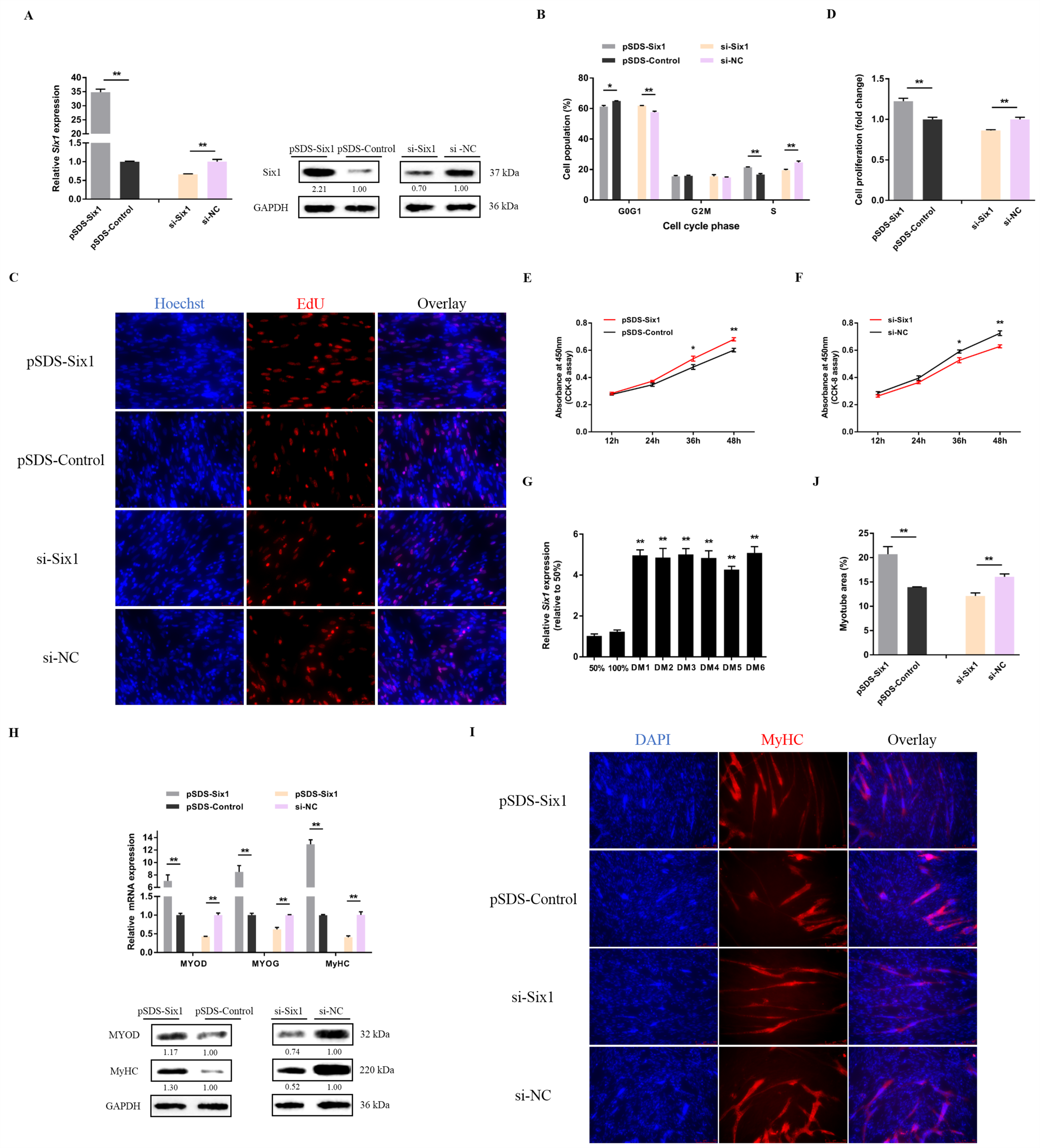
3.3. lncRNA-Six1 Functions to Promote Myoblast Proliferation and Differentiation, and Induces the Fast-Twitch Muscle Phenotype
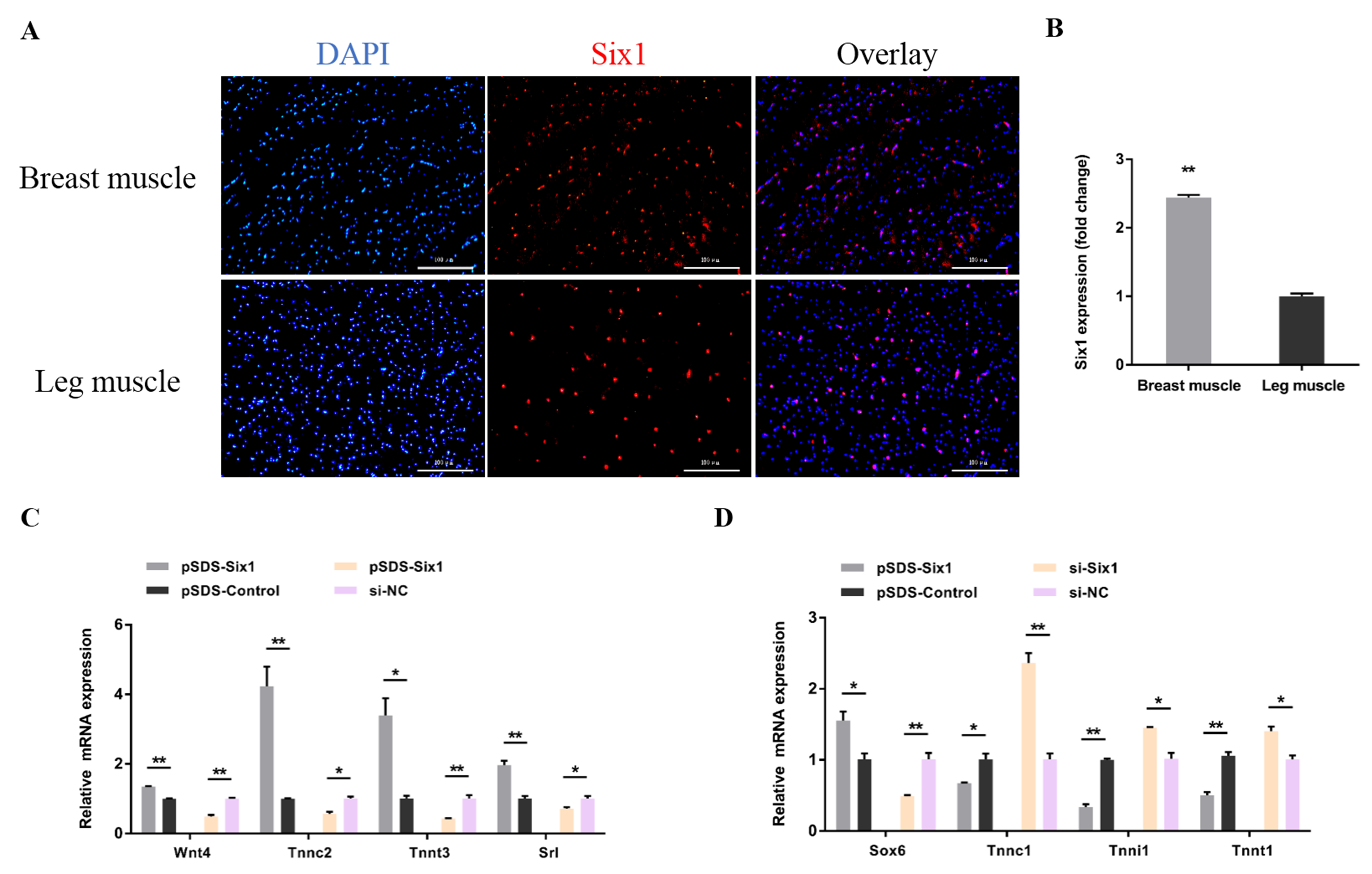
3.4. Six1 Is a Critical Factor in Myogenesis, Participating in the Transformation of Skeletal Muscle Fiber Types
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Maeda, N.; Kasukawa, T.; Oyama, R.; Gough, J.; Frith, M.; Engstrom, P.G.; Lenhard, B.; Aturaliya, R.N.; Batalov, S.; Beisel, K.W.; et al. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet. 2006, 2, e62. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Lian, L.; Li, X.; Zhao, C.; Qu, L.; Liu, C.; Song, J.; Yang, N. Chicken gga-miR-103-3p Targets CCNE1 and TFDP2 and Inhibits MDCC-MSB1 Cell Migration. G3 (Bethesda) 2016, 6, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Abdalla, B.A.; Zheng, M.; He, X.; Cai, B.; Han, P.; Ouyang, H.; Chen, B.; Nie, Q.; Zhang, X. Systematic transcriptome-wide analysis of mRNA-miRNA interactions reveals the involvement of miR-142-5p and its target (FOXO3) in skeletal muscle growth in chickens. Mol. Genet. Genomics 2018, 293, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Luo, W.; Abdalla, B.A.; Ouyang, H.; Yu, J.; Hu, F.; Nie, Q.; Zhang, X. miRNA-223 upregulated by MYOD inhibits myoblast proliferation by repressing IGF2 and facilitates myoblast differentiation by inhibiting ZEB1. Cell Death Dis. 2017, 8, e3094. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, H.; Chen, X.; Wang, Z.; Yu, J.; Jia, X.; Li, Z.; Luo, W.; Abdalla, B.A.; Jebessa, E.; Nie, Q.; et al. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 2017. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Stark, A.; Brennecke, J.; Bushati, N.; Russell, R.B.; Cohen, S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell 2005, 123, 1133–1146. [Google Scholar] [CrossRef]
- Jia, X.; Ouyang, H.; Abdalla, B.A.; Xu, H.; Nie, Q.; Zhang, X. miR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities. Biochim. Biophys. Acta Gene. Regul. Mech. 2017, 1860, 674–684. [Google Scholar] [CrossRef]
- Luo, W.; Wu, H.; Ye, Y.; Li, Z.; Hao, S.; Kong, L.; Zheng, X.; Lin, S.; Nie, Q.; Zhang, X. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014, 5, e1347. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, M.; Chen, B.; Li, Z.; Abdalla, B.A.; Nie, Q.; Zhang, X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis. 2018, 9, 367. [Google Scholar] [CrossRef]
- Glazov, E.A.; Cottee, P.A.; Barris, W.C.; Moore, R.J.; Dalrymple, B.P.; Tizard, M.L. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008, 18, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Novikova, I.V.; Hennelly, S.P.; Tung, C.S.; Sanbonmatsu, K.Y. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J. Mol. Biol. 2013, 425, 3731–3746. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Freier, S.M.; Spector, D.L. 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 2008, 135, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Lee, J.T.; Bartolomei, M.S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013, 152, 1308–1323. [Google Scholar] [CrossRef]
- Li, T.; Wang, S.; Wu, R.; Zhou, X.; Zhu, D.; Zhang, Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics 2012, 99, 292–298. [Google Scholar] [CrossRef]
- Li, Z.; Ouyang, H.; Zheng, M.; Cai, B.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. Integrated Analysis of Long Non-coding RNAs (LncRNAs) and mRNA Expression Profiles Reveals the Potential Role of LncRNAs in Skeletal Muscle Development of the Chicken. Front. Physiol. 2016, 7, 687. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Li, Z.; Zhou, Y.; Liu, X.; Han, R.; Wang, Y.; Yan, F.; Sun, G.; Li, H.; Kang, X. Sequencing and characterization of lncRNAs in the breast muscle of Gushi and Arbor Acres chickens. Genome 2018, 61, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef]
- Han, X.; Yang, F.; Cao, H.; Liang, Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015, 29, 3054–3064. [Google Scholar] [CrossRef]
- Liang, L.; Xu, J.; Wang, M.; Xu, G.; Zhang, N.; Wang, G.; Zhao, Y. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018, 9, 372. [Google Scholar] [CrossRef]
- Cai, B.; Li, Z.; Ma, M.; Wang, Z.; Han, P.; Abdalla, B.A.; Nie, Q.; Zhang, X. LncRNA-Six1 Encodes a Micropeptide to Activate Six1 in Cis and Is Involved in Cell Proliferation and Muscle Growth. Front. Physiol. 2017, 8, 230. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, L.; Liu, H.H.; Xu, F.; Si, J.M.; Jia, J.; Wang, J.W. MyoD expression profile and developmental differences of leg and breast muscle in Peking duck (Anas platyrhynchos Domestica) during embryonic to neonatal stages. Micron 2010, 41, 847–852. [Google Scholar] [CrossRef]
- Turner, D.L.; Butler, P.J. The aerobic capacity of locomotory muscles in the tufted duck, Aythya fuligula. J. Exp. Biol. 1988, 135, 445–460. [Google Scholar] [PubMed]
- Ying, F.; Zhang, L.; Bu, G.; Xiong, Y.; Zuo, B. Muscle fiber-type conversion in the transgenic pigs with overexpression of PGC1alpha gene in muscle. Biochem. Biophys. Res. Commun. 2016, 480, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef]
- Fan, R.F.; Cao, C.Y.; Chen, M.H.; Shi, Q.X.; Xu, S.W. Gga-let-7f-3p promotes apoptosis in selenium deficiency-induced skeletal muscle by targeting selenoprotein K. Metallomics 2018, 10, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017, 8, 14718. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.T. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 2018, 9, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Simionescu-Bankston, A.; Kumar, A. Noncoding RNAs in the regulation of skeletal muscle biology in health and disease. J. Mol. Med. (Berl) 2016, 94, 853–866. [Google Scholar] [CrossRef]
- Neguembor, M.V.; Jothi, M.; Gabellini, D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet. Muscle 2014, 4, 8. [Google Scholar] [CrossRef]
- Caretti, G.; Schiltz, R.L.; Dilworth, F.J.; Di Padova, M.; Zhao, P.; Ogryzko, V.; Fuller-Pace, F.V.; Hoffman, E.P.; Tapscott, S.J.; Sartorelli, V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell 2006, 11, 547–560. [Google Scholar] [CrossRef]
- Hube, F.; Velasco, G.; Rollin, J.; Furling, D.; Francastel, C. Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 2011, 39, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Sun, K.; Chen, X.; Zhao, Y.; Wang, L.; Zhou, L.; Sun, H.; Wang, H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 2013, 32, 2575–2588. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Ripoche, M.A.; Le Digarcher, A.; Watrin, F.; Ziyyat, A.; Forne, T.; Jammes, H.; Ainscough, J.F.; Surani, M.A.; Journot, L.; et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 2009, 136, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Huang, R.; Wu, Q.; Li, P.; Chen, J.; Li, B.; Liu, H. The role of Six1 in the genesis of muscle cell and skeletal muscle development. Int. J. Biol. Sci. 2014, 10, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Le Grand, F.; Grifone, R.; Mourikis, P.; Houbron, C.; Gigaud, C.; Pujol, J.; Maillet, M.; Pages, G.; Rudnicki, M.; Tajbakhsh, S.; et al. Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. J. Cell Biol. 2012, 198, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, X.; Liu, H.; Sun, L.; Zhang, R.; Li, L.; Wangding, M.; Wang, J. Six1 induces protein synthesis signaling expression in duck myoblasts mainly via up-regulation of mTOR. Genet. Mol. Biol. 2016, 39, 151–161. [Google Scholar] [CrossRef]
- Sakakibara, I.; Wurmser, M.; Dos, S.M.; Santolini, M.; Ducommun, S.; Davaze, R.; Guernec, A.; Sakamoto, K.; Maire, P. Six1 homeoprotein drives myofiber type IIA specialization in soleus muscle. Skelet. Muscle 2016, 6, 30. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, M.; Cai, B.; Jiang, L.; Abdalla, B.A.; Li, Z.; Nie, Q.; Zhang, X. lncRNA-Six1 Is a Target of miR-1611 That Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis. Cells 2018, 7, 243. https://doi.org/10.3390/cells7120243
Ma M, Cai B, Jiang L, Abdalla BA, Li Z, Nie Q, Zhang X. lncRNA-Six1 Is a Target of miR-1611 That Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis. Cells. 2018; 7(12):243. https://doi.org/10.3390/cells7120243
Chicago/Turabian StyleMa, Manting, Bolin Cai, Liang Jiang, Bahareldin Ali Abdalla, Zhenhui Li, Qinghua Nie, and Xiquan Zhang. 2018. "lncRNA-Six1 Is a Target of miR-1611 That Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis" Cells 7, no. 12: 243. https://doi.org/10.3390/cells7120243
APA StyleMa, M., Cai, B., Jiang, L., Abdalla, B. A., Li, Z., Nie, Q., & Zhang, X. (2018). lncRNA-Six1 Is a Target of miR-1611 That Functions as a ceRNA to Regulate Six1 Protein Expression and Fiber Type Switching in Chicken Myogenesis. Cells, 7(12), 243. https://doi.org/10.3390/cells7120243





