The Association of XRCC1 Gene Polymorphisms and Chronic Hepatitis C Induced Insulin Resistance in Egyptian Patients
Abstract
1. Introduction
2. Subjects and Methods
2.1. Sampling
2.2. Genotyping of XRCC1 Gene Polymorphisms
3. Statistical Analysis
4. Results
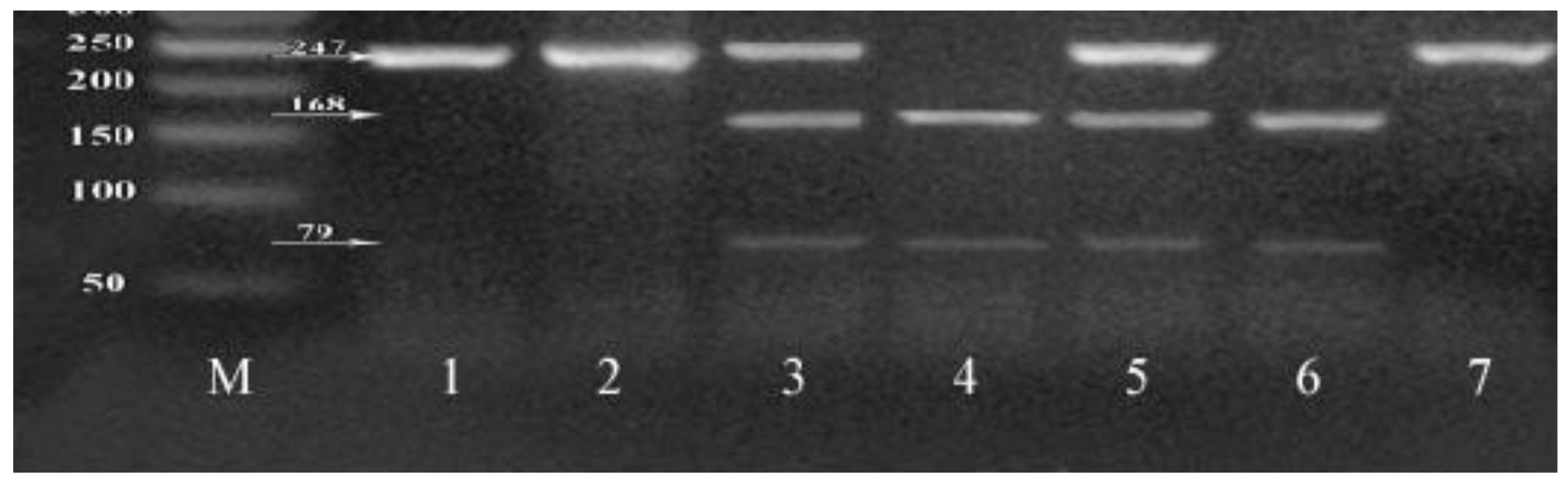
4.1. Genotyping of XRCC1 SNPs
4.2. Association between XRCC1 SNPs and IR Risk in Chronic HCV Patients
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Ethics Committee Approval
Informed Consent
References
- Petta, S. Hepatitis C virus and cardiovascular: A review. J. Adv. Res. 2017, 8, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, A.A.; Helmy, A.M.; El Fayuomy, K.N.; Ahmed, O.M.; El Sayed, E.E. Role of insulin resistance in the development of hepatocellular carcinoma in patients with chronic hepatitis C. AL-Azhar Assiut Med. J. 2010, 8, 294–313. [Google Scholar]
- Singal, A.G.; Volk, M.L.; Jensen, D.; Di Bisceglie, A.M.; Schoenfeld, P.S. A sustained viral response is associated with reduced liver-related morbidity and mortality in patients with hepatitis C virus. Clin. Gastroenterol. Hepatol. 2010, 8, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Negro, F.; Forton, D.; Craxì, A.; Sulkowski, M.S.; Feld, J.J.; Manns, M.P. Extrahepatic morbidity and mortality of chronic hepatitis C. Gastroenterology 2015, 149, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.L. Metabolic alterations and hepatitis C: From bench to bedside. World J. Gastroenterol. 2016, 22, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Stepanova, M.; Estep, M.; Negro, F.; Clark, P.J.; Hunt, S. Dysregulation of distal cholesterol biosynthesis in association with relapse and advanced disease in CHC genotype 2 and 3 treated with sofosbuvir and ribavirin. J. Hepatol. 2016, 64, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Gastaldi, G.; Goossens, N.; Clément, S.; Negro, F. Current level of evidence on causal association between hepatitis C virus and type 2 diabetes: A review. J. Adv. Res. 2017, 8, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Kaddai, V.; Negro, F. Current understanding of insulin resistance in hepatitis C. Expert Rev. Gastroenterol. Hepatol. 2011, 5, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.H.; Brancati, F.L.; Sulkowski, M.S.; Strathdee, S.A.; Szklo, M.; Thomas, D.L. Prevalence of type 2 diabetes mellitus among persons with hepatitis C virus infection in the United States. Ann. Intern. Med. 2000, 133, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Arase, Y.; Suzuki, F.; Suzuki, Y.; Akuta, N.; Kobayashi, M.; Kawamura, Y.; Yatsuji, H.; Sezaki, H.; Hosaka, T.; Hirakawa, M.; et al. Sustained virological response reduces incidence of onset of type 2 diabetes in chronic hepatitis C. Hepatology 2009, 49, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Moucari, R.; Forestier, N.; Larrey, D.; Guyader, D.; Couzigou, P.; Benhamou, Y.; Voitot, H.; Vidaud, M.; Seiwert, S.; Bradford, B.; et al. Danoprevir, an HCV NS3/4A protease inhibitor, improves insulin sensitivity in genotype 1 chronic hepatitis C patients. Gut 2010, 59, 1694–1698. [Google Scholar] [CrossRef] [PubMed]
- Hagen, T.M.; Huang, S.; Curnutte, J.; Fowler, P.; Martinez, V.; Wehr, C.M.; Ames, B.N.; Chisari, F.V. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 1994, 91, 12808–12812. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. DNA damage and repair. Nature 2003, 421, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 2001, 1, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. XRCCI and DNA strand break repair. DNA Repair 2003, 2, 955–969. [Google Scholar] [CrossRef]
- Kiran, M.; Saxena, R.; Chawla, Y.K.; Kaur, J. Polymorphism of DNA repair gene XRCC1 and hepatitis-related hepatocellular carcinoma risk in Indian population. Mol. Cell. Biochem. 2009, 327, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.Z.; Liang, J.; Yu, Z.; Lun, L.M.; Li, H.; Wang, Q. Polymorphism of DNA repair gene XRCC1 and hepatocellular carcinoma risk in Chinese population. Asian Pac. J. Cancer Prev. 2011, 12, 2947–2950. [Google Scholar] [PubMed]
- Bruce, C.R.; Carey, A.L.; Hawley, J.A.; Febbraio, M.A. Intra-muscular heat shock protein 72 and heme oxygenase-1 mRNA are reduced in patients with type 2 diabetes: Evidence that insulin resistance is associated with a disturbed antioxidant defense mechanism. Diabetes 2003, 52, 2338–2345. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.L.; Goldfine, I.D.; Maddux, B.A.; Grodsky, G.M. Are oxidative stress-activated signaling pathways mediators of insulin resistance and beta-cell dysfunction? Diabetes 2003, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ceriello, A.; Quagliaro, L.; Piconi, L.; Assaloni, R.; Da Ros, R.; Maier, A.; Esposito, K.; Giugliano, D. Effect of postprandial hypertriglyceridemia and hyperglycemia on circulating adhesion molecules and oxidative stress generation and the possible role of simvastatin treatment. Diabetes 2004, 53, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Wongwananuruk, T.; Rattanachaiyanont, M.; Indhavivadhana, S.; Leerasiri, P.; Techatraisak, K.; Tanmahasamut, P.; Angsuwathana, S.; Dangrat, C. Prevalence and clinical predictors of insulin resistance in reproductive-aged thai women with polycystic ovary syndrome. Int. J. Endocrinol. 2012, 2012, 529184. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Li, J.; Li, J.; Gao, X.; Xu, S. SNPs identification and its correlation analysis with milk somatic cell score in bovine MBL1 gene. Mol. Boil. Rep. 2013, 40, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Zhong, C.; Li, K.; Chu, H.; Wang, B. Association study of single nucleotide polymorphisms in XRCC1 gene with risk of hepatocellular carcinoma in Chinese Han population. BioMed Res. Int. 2013, 138785, 1–6. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatitis C virus infection. J. Hepatol. 2011, 55, 245–264. [Google Scholar] [CrossRef] [PubMed]
- Alberti, K.G.; Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z.; Cleeman, J.I.; Donato, K.A.; Fruchart, J.C.; James, W.P.; Loria, C.M.; Smith, S.C., Jr.; et al. Harmonizing the metabolic syndrome: A joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009, 120, 1640–1645. [Google Scholar] [PubMed]
- Negro, F. HCV infection and metabolic syndrome: Which is the chicken and which is the egg? Gastroenterology 2012, 142, 1288–1292. [Google Scholar] [CrossRef] [PubMed]
- Hui, J.M.; Sud, A.; Farrell, G.C.; Bandara, P.; Byth, K.; Kench, J.G.; McCaughan, G.W.; George, J. Insulin resistance is associated with chronic hepatitis C and virus infection fibrosis progression. Gastroenterology 2003, 125, 1695–1704. [Google Scholar] [CrossRef] [PubMed]
- Abdelsatar, H.; Mahfouz, M.; Rashed, L.; Ahmad, D. Insulin Resistance in Patients with Chronic Hepatitis C Infection. Med. J. Cairo Univ. 2010, 78, 163–167. [Google Scholar]
- Imazeki, F.; Yokosuka, O.; Fukai, K.; Kanda, T.; Kojima, H.; Saisho, H. Prevalence of diabetes mellitus and insulin resistance in patients with chronic hepatitis C: Comparison with hepatitis B virus-infected and hepatitis C virus-cleared patients. Liver Int. 2008, 28, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.A.; Loutfy, S.A.; el Sayed, N.R.; Siam, I.; Hashem, A.E. The Role of Insulin Resistance (HOMA-IR) in the Development of Hepatocellular Carcinoma associated with Chronic Hepatitis C Genotype-4 Infection. Kasr El Aini Med. J. 2011, 17, 47–59. [Google Scholar]
- Xu, P.; Xu, C.F.; Wan, X.Y.; Yu, C.H.; Shen, C.; Chen, P.; Xu, G.Y.; Li, Y.M. Association between serum alpha-fetoprotein levels and fatty liver disease: A cross-sectional study. World J. Gastroenterol. 2014, 20, 11865–11870. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Feng, L.; Zhang, J.; Feng, G. Association between alpha-fetoprotein and metabolic syndrome in a Chinese asymptomatic population: A cross-sectional study. Lipids Health Dis. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Mizuta, T.; Eguchi, Y.; Sakurai, E.; Motomura, Y.; Isoda, H.; Kuwashiro, T.; Oeda, S.; Iwane, S.; Takahashi, H.; et al. Whole-body insulin resistance is associated with elevated serum α-fetoprotein levels in patients with chronic hepatitis C. Intern. Med. 2013, 52, 2393–2400. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.H.; Brookman, K.W.; Jones, N.J.; Allen, S.A.; Carrano, A.V. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 1990, 10, 6160–6171. [Google Scholar] [CrossRef] [PubMed]
- Basso, D.; Navaglia, F.; Fogar, P.; Zambon, C.F.; Greco, E.; Schiavon, S.; Fasolo, M.; Stranges, A.; Falda, A.; Padoan, A.; et al. DNA repair pathways and mitochondrial DNA mutations in gastrointestinal carcinogenesis. Clin. Chim. Acta 2007, 381, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Tudek, B. Base excision repair modulation as a risk factor for human cancers. Mol. Asp. Med. 2007, 28, 258–275. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.P.; Zhang, X.Y.; Wang, X.L.; Shi, L.Y.; Li, Y.Y.; Li, F.; Su, Y.H.; Wang, Y.J.; Lu, B.; Sun, X.; et al. DNA repair gene XRCC1 polymorphisms, smoking, and esophageal cancer risk. Cancer Detect. Prev. 2004, 28, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Xing, Q.; Li, Y.; Sun, J.; Ji, H.; Huazheng, P.; Jun, L. Study on the DNA repair gene XRCC1 and XRCC3 polymorphism in prediction and prognosis of hepatocellular carcinoma risk. Hepato-Gastroenterology 2012, 59, 2285–2289. [Google Scholar] [CrossRef] [PubMed]
- Long, X.D.; Ma, Y.; Huang, H.D.; Yao, J.G.; Qu, D.Y.; Lu, Y.L. Polymorphism of XRCC1 and the frequency of mutation in codon 249 of the p53 gene in hepatocellular carcinoma among Guangxi population, China. Mol. Carcinog. 2008, 47, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.W.; Lu, C.R.; Ye, M.; Xiao, W.H.; Liang, J. Evaluation of DNA repair gene XRCC1 polymorphism in prediction and prognosis of hepatocellular carcinoma risk. Asian Pac. J. Cancer Prev. 2012, 13, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Talior, I.; Yarkoni, M.; Bashan, N.; Eldar-Finkelman, H. Increased glucose uptake promotes oxidative stress and PKC-delta activation in adipocytes of obese insulin-resistant mice. Am. J. Physiol. Endocrinol. Metab. 2003, 285, E295–E302. [Google Scholar] [CrossRef] [PubMed]
- Pekow, J.R.; Bhan, A.K.; Zheng, H.; Chung, R.T. Hepatic steatosis is associated with increased frequency of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis. Cancer 2007, 109, 2490–2496. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.F.; Pace, F.H.; Chebli, J.M.F.; Ferreira, L.E. Insulin resistance in non-diabetic patients with chronic hepatitis C: What does it mean? Arq. Bras. Endocrinol. Metabol. 2011, 55, 412–418. [Google Scholar] [CrossRef] [PubMed]

| Control | Non-IR | IR | P | P1 | P2 | P3 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age | 56.6 | 7.2 | 58.7 | 6.7 | 56.9 | 5.6 | 0.2 | |||
| Female | 22 | 31.4% | 16 | 30.2% | 26 | 29.9% | 0.97 | |||
| Male | 48 | 68.6% | 37 | 69.8% | 61 | 70.1% | ||||
| BMI | 25.11 | 2.77 | 26.3 | 4.64 | 26.63 | 5.1 | 0.09 | |||
| AFP | 6.20 | 2.60–11.60 | 19.20 | 2.70–4233.60 | 33.50 | 6.00–5000.00 | <0.001 | 0.06 | <0.001 | 0.015 |
| HOMA-IR | 1.5 | 2.0–0.3 | 0.9 | 2.0–0.2 | 3.4 | 13.8–2.1 | <0.001 | 0.09 | <0.001 | <0.001 |
| Genotype and Allele Analysis of c.1254 C > T Polymorphism of XRCC1 Gene | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All Patients | Control Group (70) No (%) | P1 | OR1 (CI 95%) | P2 | OR2 (CI 95%) | P3 | OR3 (CI 95%) | |||
| IR (87) No (%) | Non-IR (53) No (%) | |||||||||
| Genotypes | CC | 31 (35.6%) | 24 (45.3%) | 38 (54.3%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| CT | 44 (50.6%) | 20 (37.7%) | 26 (37.1%) | 0.16 | 1.7 (0.8–3.6) | 0.03 | 2.07 (1.05–4.09) | 0.6 | 1.2 (0.56–2.6) | |
| TT | 12 (13.8%) | 9 (17.0%) | 6 (8.6%) | 0.95 | 1.03 (0.37–2.8) | 0.1 | 2.45 (0.8–7.3) | 0.13 | 2.37 (0.75–7.5) | |
| CT + TT | 56 (64.4%) | 29 (54.7%) | 32 (45.7%) | 0.25 | 1.5 (0.7–3.00) | 0.019 | 2.14 (1.13–4.08) | 0.3 | 1.4 (0.7–2.9) | |
| Alleles | C | 106 (61.0%) | 68 (64.0%) | 102 (73.0%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| T | 68 (39.0%) | 38 (36.0%) | 38 (27.0%) | 0.58 | 1.14 (0.7–1.9) | 0.026 | 1.7 (1.06–2.78) | 0.14 | 1.5 (0.87–2.58) | |
| Genotype and Allele Analysis of c.1517 G > C Polymorphism of XRCC1 Gene | ||||||||||
| Genotypes | GG | 36 (41.4%) | 16 (30.2%) | 42 (60.0%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| GC | 44 (50.6%) | 28 (52.8%) | 24 (34.3%) | 0.35 | 0.69 (0.1–1.09) | 0.025 | 2.1 (1.097–4.16) | 0.005 | 3.06 (1.4–6.76) | |
| CC | 7 (8.0%) | 9 (17.0%) | 4 (5.7%) | 0.065 | 0.34 (0.37–2.8) | 0.3 | 2.04 (0.55–7.5) | 0.008 | 5.9 (1.6–21.9) | |
| GC + CC | 51 (58.6%) | 37 (69.8%) | 28 (40.0%) | 0.18 | 0.6 (0.3–1.26) | 0.02 | 2.14 (1.13–4.08) | 0.001 | 3.5 (1.6–7.4) | |
| Alleles | G | 116 (67.0%) | 60 (57.0%) | 108 (77.0%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| C | 58 (33.0%) | 46 (43.0%) | 32 (23.0%) | 0.09 | 0.65 (0.4–1.07) | 0.04 | 1.68 (1.02–2.8) | 0.001 | 2.55 (1.5–4.48) | |
| Genotype and Allele Analysis of c.1254 C > T Polymorphism of XRCC1 Gene | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IR | Control Group (70) No (%) | P1 | OR1 (CI 95%) | P2 | OR2 (CI 95%) | P3 | OR3 (CI 95%) | |||
| IR–HCC (43) No (%) | IR-Cirrhotic (44) No (%) | |||||||||
| Genotypes | CC | 11 (25.6%) | 20 (63.6%) | 38 (54.3%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| CT | 22 (51.2%) | 22 (33.3%) | 26 (37.1%) | 0.2 | 1.8 (0.7–4.67) | 0.015 | 2.9 (1.2–7.04) | 0.2 | 1.6 (0.7–3.5) | |
| TT | 10 (23.3%) | 2 (3.0%) | 6 (8.6%) | 0.007 | 9.09 (1.68–49.12) | 0.003 | 5.7 (1.7–19.4) | 0.7 | 0.6 (0.11–3.4) | |
| CT + TT | 32 (74.5%) | 24 (36.3%) | 32 (45.7%) | 0.053 | 2.4 (0.97–6.00) | 0.003 | 3.45 (1.5–7.9) | 0.35 | 1.4 (0.66–3.04) | |
| Alleles | C | 44 (51.0%) | 62 (70.0%) | 102 (73.0%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| T | 42 (49.0%) | 26 (30.0%) | 38 (27.0%) | 0.009 | 2.27 (1.2–4.2) | 0.001 | 2.56 (1.45–4.5) | 0.7 | 1.12 (0.6–2.03) | |
| Genotype and Allele Analysis of c.1517 G > C Polymorphism of XRCC1 Gene | ||||||||||
| Genotypes | GG | 14 (32.6%) | 22 (50.0%) | 42 (60.0%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| GC | 26 (60.5%) | 18 (40.9%) | 24 (34.3%) | 0.07 | 2.27 (0.9–5.58) | 0.004 | 3.25 (1.4–7.4) | 0.37 | 1.4 (0.6–3.2) | |
| CC | 3 (7.0%) | 4 (9.1%) | 4 (5.7%) | 1.00 | 1.17 (0.22–6.07) | 0.37 | 2.25 (0.44–11.3) | 0.44 | 1.9 (0.4–8.4) | |
| GC + CC | 32 (74.5%) | 24 (36.3%) | 28 (40.0%) | 0.087 | 2.095 (0.9–4.9) | 0.001 | 4.00 (1.8–8.9) | 0.19 | 1.6 (0.77–3.5) | |
| Alleles | G | 54 (63.0%) | 62 (70.0%) | 108 (77.0%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| C | 32 (37.0%) | 26 (30.0%) | 32 (23.0%) | 0.28 | 1.4 (0.75–2.66) | 0.02 | 2.00 (1.11–3.6) | 0.27 | 1.4 (0.77–2.6) | |
| IR (87) No (%) | Non-IR (53) No (%) | Control Group (70) No (%) | P1 | OR1 (CI 95%) | P2 | OR2 (CI 95%) | P3 | OR3 (CI 95%) | |
|---|---|---|---|---|---|---|---|---|---|
| CCGG | 10 (11.5%) | 10 (18.9%) | 20 (28.6%) | - | 1 (Ref) | - | 1 (Ref) | - | 1 (Ref) |
| CCGC | 16 (18.4%) | 8 (15.1%) | 14 (20.0%) | 0.26 | 2.0 (0.6–6.77) | 0.11 | 2.28 (0.8–6.5) | 0.8 | 1.14 (0.36–3.6) |
| CCCC | 5 (5.7%) | 6 (11.3%) | 4 (5.7%) | 0.8 | 0.83 (0.19–3.6) | 0.22 | 2.5 (0.5–11.4) | 0.16 | 3.00 (0.69–13.11) |
| CTGG | 24 (27.6%) | 3 (5.7%) | 18 (25.7%) | 0.003 | 8.00 (1.8–35.36) | 0.046 | 2.66 (1.006–7.07) | 0.19 | 0.33 (0.08–1.4) |
| CTGC | 18 (20.7%) | 15 (28.3%) | 8 (11.4%) | 0.75 | 1.2 (0.39–3.65) | 0.007 | 4.5 (1.45–13.88) | 0.02 | 3.75 (1.19–11.8) |
| CTCC | 2 (2.3%) | 2 (3.8%) | 0 (0%) | 1.00 | 1.00 (0.11–8.55) | 0.13 | - | 0.13 | - |
| TTGG | 2 (2.3%) | 3 (5.7%) | 4 (5.7%) | 0.69 | 0.66 (0.09–4.88) | 1.00 | 1.00 (0.16–6.4) | 0.68 | 1.5 (0.28–8.04) |
| TTGC | 10 (11.5%) | 5 (9.4%) | 2 (2.9%) | 0.3 | 2.00 (0.5–7.99) | 0.006 | 10.00 (1.8–54.6) | 0.095 | 5.00 (0.8–30.46) |
| TTCC | 0 (0%) | 1 (1.9%) | 0 (0%) | 0.3 | - | - | - | 0.35 | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abo El-khair, S.M.; Arafa, M.; Besheer, T.; El-Eraky, A.M.; Elsamanoudy, A.Z. The Association of XRCC1 Gene Polymorphisms and Chronic Hepatitis C Induced Insulin Resistance in Egyptian Patients. Cells 2018, 7, 185. https://doi.org/10.3390/cells7110185
Abo El-khair SM, Arafa M, Besheer T, El-Eraky AM, Elsamanoudy AZ. The Association of XRCC1 Gene Polymorphisms and Chronic Hepatitis C Induced Insulin Resistance in Egyptian Patients. Cells. 2018; 7(11):185. https://doi.org/10.3390/cells7110185
Chicago/Turabian StyleAbo El-khair, Salwa M., Mona Arafa, Tarek Besheer, Ahmed M. El-Eraky, and Ayman Z. Elsamanoudy. 2018. "The Association of XRCC1 Gene Polymorphisms and Chronic Hepatitis C Induced Insulin Resistance in Egyptian Patients" Cells 7, no. 11: 185. https://doi.org/10.3390/cells7110185
APA StyleAbo El-khair, S. M., Arafa, M., Besheer, T., El-Eraky, A. M., & Elsamanoudy, A. Z. (2018). The Association of XRCC1 Gene Polymorphisms and Chronic Hepatitis C Induced Insulin Resistance in Egyptian Patients. Cells, 7(11), 185. https://doi.org/10.3390/cells7110185





