Mechanisms of Generating Polyubiquitin Chains of Different Topology
Abstract
:1. Introduction
2. Structural Versatility of Protein Ubiquitination
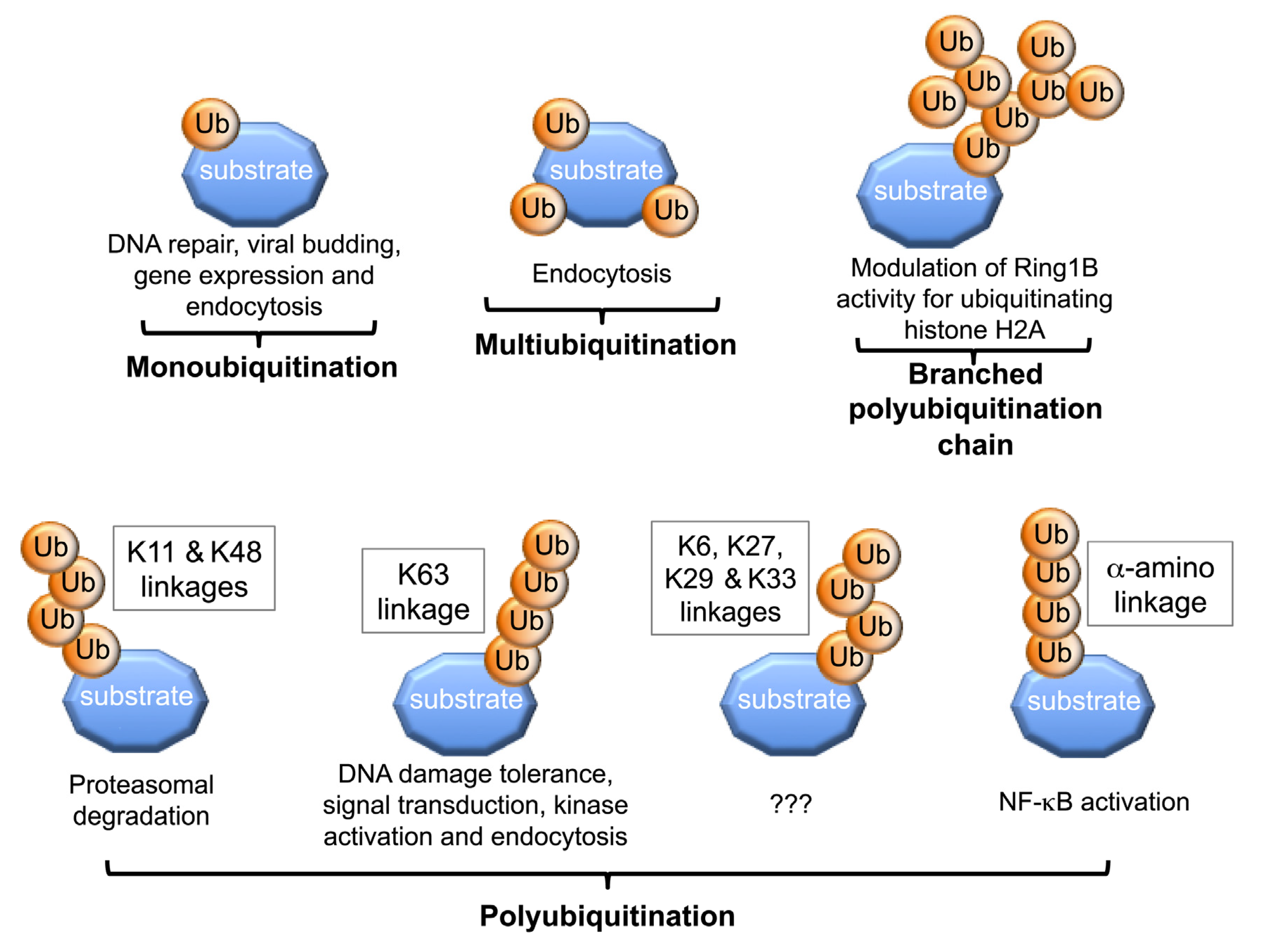
3. Molecular and Structural Insights into Protein Ubiquitination
3.1. Lysine Selection during Protein Mono- and Multi-Ubiquitination
3.2. Mechanisms of Polyubiquitin Chain Formation
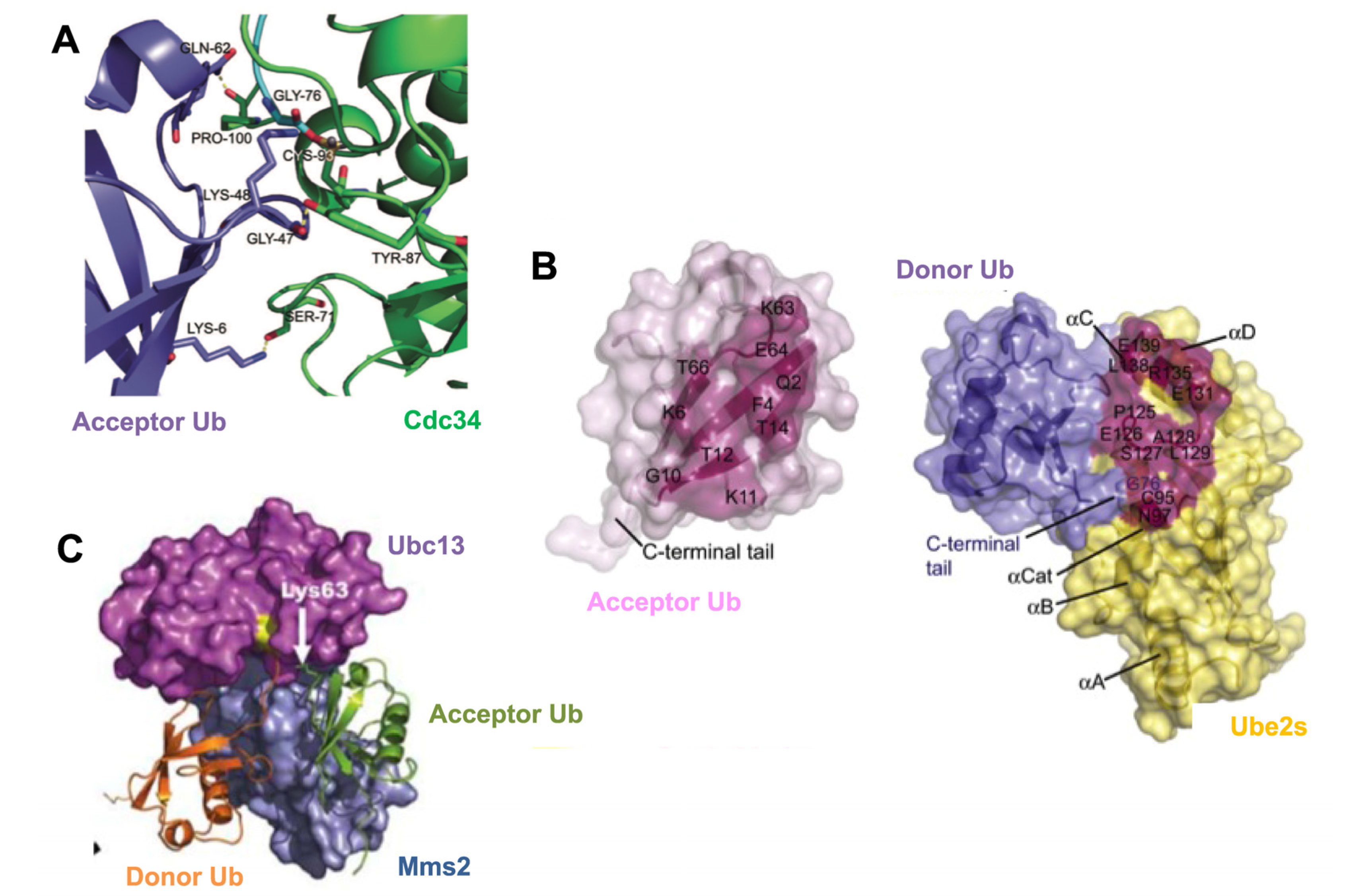
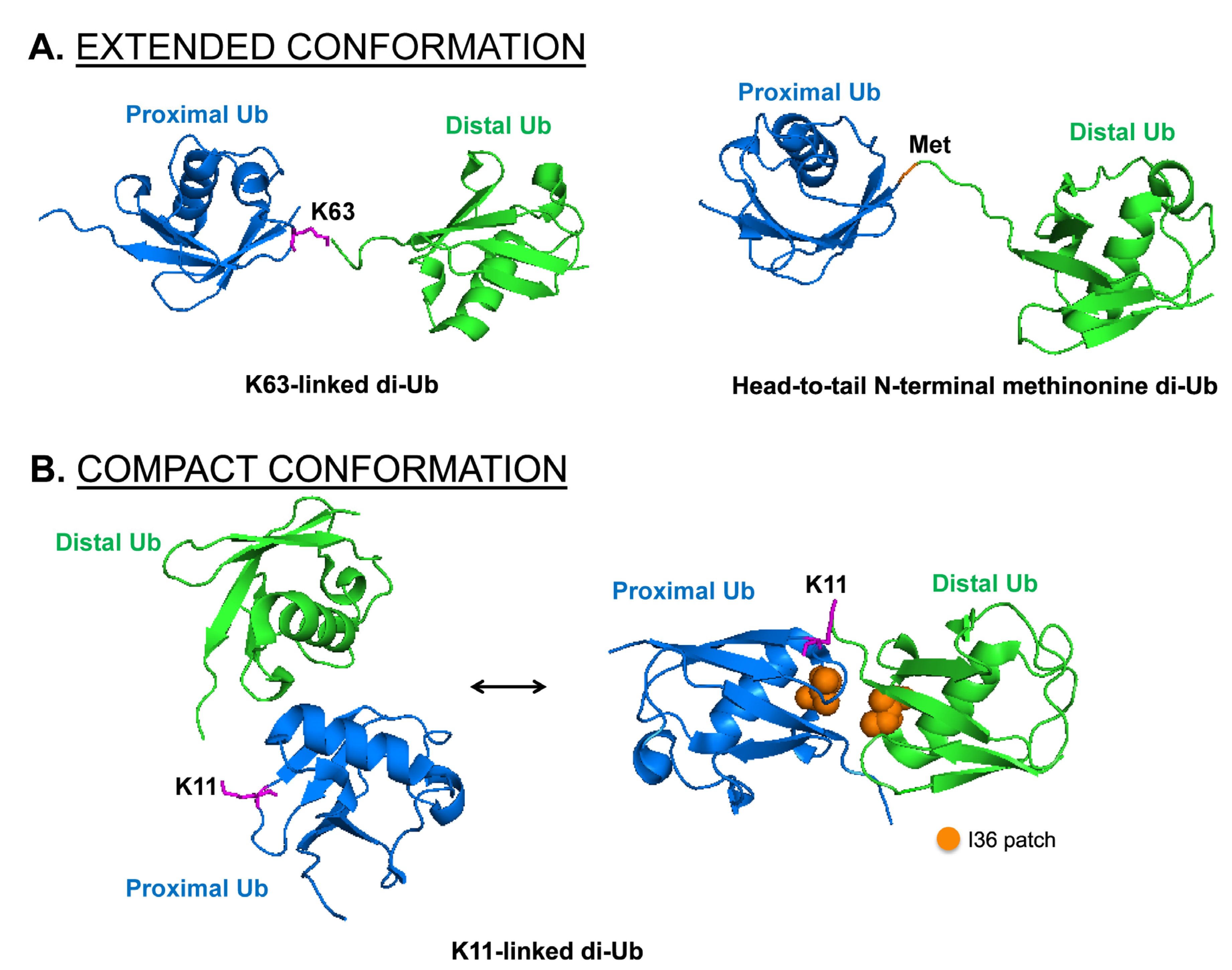
4. The Impact of Different Chain Topology
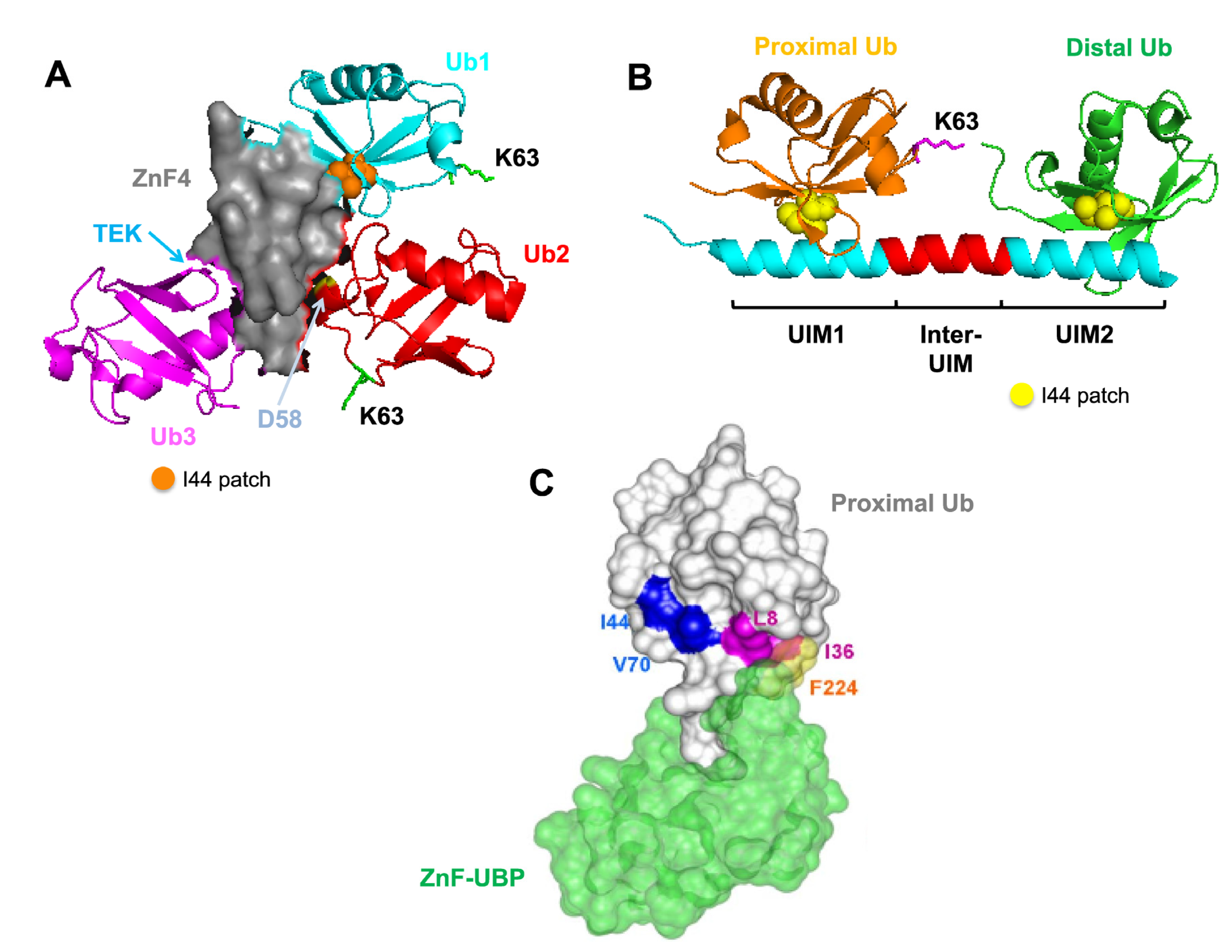
4.1. Recognition Pattern for Different UBD-Containing Proteins
5. Conclusions
Acknowledgements
Author contributions
Conflicts of Interest
References
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Eisenhaber, B.; Chumak, N.; Eisenhaber, F.; Hauser, M.T. The ring between ring fingers (RBR) protein family. Genome. Biol. 2007, 8, 209. [Google Scholar] [CrossRef]
- Wenzel, D.M.; Klevit, R.E. Following Ariadne's thread: A new perspective on RBR ubiquitin ligases. BMC Biol. 2012, 10, 24. [Google Scholar] [CrossRef]
- Sadowski, M.; Suryadinata, R.; Tan, A.R.; Roesley, S.N.; Sarcevic, B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 2012, 64, 136–142. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell 2003, 11, 1435–1444. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Passmore, L.A.; Barford, D. Getting into position: the catalytic mechanisms of protein ubiquitylation. Biochem. J. 2004, 379, 513–525. [Google Scholar] [CrossRef]
- Haglund, K.; Di Fiore, P.P.; Dikic, I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci 2003, 28, 598–603. [Google Scholar] [CrossRef]
- Hicke, L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001, 2, 195–201. [Google Scholar]
- Sarcevic, B.; Mawson, A.; Baker, R.T.; Sutherland, R.L. Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation. EMBO J. 2002, 21, 2009–2018. [Google Scholar] [CrossRef]
- Ikeda, F.; Deribe, Y.L.; Skanland, S.S.; Stieglitz, B.; Grabbe, C.; Franz-Wachtel, M.; van Wijk, S.J.; Goswami, P.; Nagy, V.; Terzic, J.; et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 2011, 471, 637–641. [Google Scholar] [CrossRef]
- Iwai, K.; Tokunaga, F. Linear polyubiquitination: A new regulator of NF-kappaB activation. EMBO Rep. 2009, 10, 706–713. [Google Scholar] [CrossRef]
- Kirisako, T.; Kamei, K.; Murata, S.; Kato, M.; Fukumoto, H.; Kanie, M.; Sano, S.; Tokunaga, F.; Tanaka, K.; Iwai, K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006, 25, 4877–4887. [Google Scholar] [CrossRef]
- Tokunaga, F.; Nakagawa, T.; Nakahara, M.; Saeki, Y.; Taniguchi, M.; Sakata, S.; Tanaka, K.; Nakano, H.; Iwai, K. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature 2011, 471, 633–636. [Google Scholar] [CrossRef]
- Stieglitz, B.; Rana, R.R.; Koliopoulos, M.G.; Morris-Davies, A.C.; Schaeffer, V.; Christodoulou, E.; Howell, S.; Brown, N.R.; Dikic, I.; Rittinger, K. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature 2013, 503, 422–426. [Google Scholar] [CrossRef]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000, 19, 94–102. [Google Scholar] [CrossRef]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef]
- Ben-Saadon, R.; Zaaroor, D.; Ziv, T.; Ciechanover, A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol. Cell 2006, 24, 701–711. [Google Scholar] [CrossRef]
- Meyer, H.J.; Rape, M. Enhanced protein degradation by branched ubiquitin chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Dye, B.T.; Schulman, B.A. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys Biomol Struct 2007, 36, 131–150. [Google Scholar] [CrossRef]
- Ye, Y.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef]
- Furukawa, M.; Andrews, P.S.; Xiong, Y. Assays for RING family ubiquitin ligases. Methods Mol. Biol. 2005, 301, 37–46. [Google Scholar]
- Jin, J.; Cardozo, T.; Lovering, R.C.; Elledge, S.J.; Pagano, M.; Harper, J.W. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004, 18, 2573–2580. [Google Scholar] [CrossRef]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One 2008, 3, e1487. [Google Scholar] [CrossRef]
- Michelle, C.; Vourc'h, P.; Mignon, L.; Andres, C.R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 2009, 68, 616–628. [Google Scholar] [CrossRef]
- Bergink, S.; Jentsch, S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 2009, 458, 461–467. [Google Scholar] [CrossRef]
- Hoege, C.; Pfander, B.; Moldovan, G.L.; Pyrowolakis, G.; Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002, 419, 135–141. [Google Scholar] [CrossRef]
- Ulrich, H.D.; Jentsch, S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000, 19, 3388–3397. [Google Scholar] [CrossRef]
- Hibbert, R.G.; Huang, A.; Boelens, R.; Sixma, T.K. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. USA 2011, 108, 5590–5595. [Google Scholar] [CrossRef]
- Wu, K.; Kovacev, J.; Pan, Z.Q. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 2011, 37, 784–796. [Google Scholar]
- Rodrigo-Brenni, M.C.; Morgan, D.O. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 2007, 130, 127–139. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 2005, 123, 1107–1120. [Google Scholar] [CrossRef]
- Sadowski, M.; Mawson, A.; Baker, R.; Sarcevic, B. Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression. Biochem. J. 2007, 405, 569–581. [Google Scholar] [CrossRef]
- Sadowski, M.; Suryadinata, R.; Lai, X.; Heierhorst, J.; Sarcevic, B. Molecular basis for lysine specificity in the yeast ubiquitin-conjugating enzyme Cdc34. Mol. Cell Biol. 2010, 30, 2316–2329. [Google Scholar] [CrossRef]
- Sadowski, M.; Sarcevic, B. Mechanisms of mono- and poly-ubiquitination: Ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 2010, 5, 19. [Google Scholar] [CrossRef]
- Wu, G.; Xu, G.; Schulman, B.A.; Jeffrey, P.D.; Harper, J.W.; Pavletich, N.P. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell 2003, 11, 1445–1456. [Google Scholar] [CrossRef]
- Tang, X.; Orlicky, S.; Lin, Z.; Willems, A.; Neculai, D.; Ceccarelli, D.; Mercurio, F.; Shilton, B.H.; Sicheri, F.; Tyers, M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 2007, 129, 1165–1176. [Google Scholar] [CrossRef]
- Hao, B.; Oehlmann, S.; Sowa, M.E.; Harper, J.W.; Pavletich, N.P. Structure of a Fbw7-Skp1-cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 2007, 26, 131–143. [Google Scholar] [CrossRef]
- Suryadinata, R.; Holien, J.K.; Yang, G.; Parker, M.W.; Papaleo, E.; Sarcevic, B. Molecular and structural insight into lysine selection on substrate and ubiquitin lysine 48 by the ubiquitin-conjugating enzyme Cdc34. Cell Cycle 2013, 12, 1732–1744. [Google Scholar] [CrossRef]
- Kamadurai, H.B.; Qiu, Y.; Deng, A.; Harrison, J.S.; Macdonald, C.; Actis, M.; Rodrigues, P.; Miller, D.J.; Souphron, J.; Lewis, S.M.; Kurinov, I.; Fujii, N.; Hammel, M.; Piper, R.; Kuhlman, B.; Schulman, B.A. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife 2013, 2, e00828. [Google Scholar] [CrossRef]
- Maspero, E.; Valentini, E.; Mari, S.; Cecatiello, V.; Soffientini, P.; Pasqualato, S.; Polo, S. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat. Struct. Mol. Biol. 2013, 20, 696–701. [Google Scholar] [CrossRef]
- Kamadurai, H.B.; Souphron, J.; Scott, D.C.; Duda, D.M.; Miller, D.J.; Stringer, D.; Piper, R.C.; Schulman, B.A. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell 2009, 36, 1095–1102. [Google Scholar] [CrossRef]
- Bosanac, I.; Phu, L.; Pan, B.; Zilberleyb, I.; Maurer, B.; Dixit, V.M.; Hymowitz, S.G.; Kirkpatrick, D.S. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J. Mol. Biol. 2011, 408, 420–431. [Google Scholar] [CrossRef]
- Rodrigo-Brenni, M.C.; Foster, S.A.; Morgan, D.O. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 2010, 39, 548–559. [Google Scholar] [CrossRef]
- Wickliffe, K.E.; Lorenz, S.; Wemmer, D.E.; Kuriyan, J.; Rape, M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 2011, 144, 769–781. [Google Scholar] [CrossRef]
- VanDemark, A.P.; Hofmann, R.M.; Tsui, C.; Pickart, C.M.; Wolberger, C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 2001, 105, 711–720. [Google Scholar] [CrossRef]
- Eddins, M.J.; Carlile, C.M.; Gomez, K.M.; Pickart, C.M.; Wolberger, C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006, 13, 915–920. [Google Scholar] [CrossRef]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009, 10, 466–473. [Google Scholar]
- Tenno, T.; Fujiwara, K.; Tochio, H.; Iwai, K.; Morita, E.H.; Hayashi, H.; Murata, S.; Hiroaki, H.; Sato, M.; Tanaka, K.; et al. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells 2004, 9, 865–875. [Google Scholar] [CrossRef]
- Varadan, R.; Assfalg, M.; Haririnia, A.; Raasi, S.; Pickart, C.; Fushman, D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J. Biol. Chem. 2004, 279, 7055–7063. [Google Scholar]
- Sloper-Mould, K.E.; Jemc, J.C.; Pickart, C.M.; Hicke, L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 2001, 276, 30483–30489. [Google Scholar] [CrossRef]
- Hu, M.; Li, P.; Li, M.; Li, W.; Yao, T.; Wu, J.W.; Gu, W.; Cohen, R.E.; Shi, Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 2002, 111, 1041–1054. [Google Scholar] [CrossRef]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—from structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- Bremm, A.; Freund, S.M.; Komander, D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Struct. Mol. Biol. 2010, 17, 939–947. [Google Scholar] [CrossRef]
- Matsumoto, M.L.; Wickliffe, K.E.; Dong, K.C.; Yu, C.; Bosanac, I.; Bustos, D.; Phu, L.; Kirkpatrick, D.S.; Hymowitz, S.G.; Rape, M.; Kelley, R.F.; et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 2010, 39, 477–484. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Cook, W.J.; Jeffrey, L.C.; Carson, M.; Chen, Z.; Pickart, C.M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J. Biol. Chem. 1992, 267, 16467–16471. [Google Scholar]
- Varadan, R.; Walker, O.; Pickart, C.; Fushman, D. Structural properties of polyubiquitin chains in solution. J. Mol. Biol. 2002, 324, 637–647. [Google Scholar] [CrossRef]
- Ryabov, Y.; Fushman, D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins 2006, 63, 787–796. [Google Scholar]
- Ryabov, Y.E.; Fushman, D. A model of interdomain mobility in a multidomain protein. J. Am. Chem. Soc. 2007, 129, 3315–3327. [Google Scholar] [CrossRef]
- Ye, Y.; Blaser, G.; Horrocks, M.H.; Ruedas-Rama, M.J.; Ibrahim, S.; Zhukov, A.A.; Orte, A.; Klenerman, D.; Jackson, S.E.; Komander, D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 2012, 492, 266–270. [Google Scholar] [CrossRef]
- Ikeda, F.; Crosetto, N.; Dikic, I. What determines the specificity and outcomes of ubiquitin signaling? Cell 2010, 143, 677–681. [Google Scholar]
- Bosanac, I.; Wertz, I.E.; Pan, B.; Yu, C.; Kusam, S.; Lam, C.; Phu, L.; Phung, Q.; Maurer, B.; Arnott, D.; et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol. Cell 40, 548–557.
- Trempe, J.F.; Brown, N.R.; Lowe, E.D.; Gordon, C.; Campbell, I.D.; Noble, M.E.; Endicott, J.A. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005, 24, 3178–3189. [Google Scholar] [CrossRef]
- Varadan, R.; Assfalg, M.; Raasi, S.; Pickart, C.; Fushman, D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell 2005, 18, 687–698. [Google Scholar]
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; Randow, F.; Wakatsuki, S.; Dikic, I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 2009, 136, 1098–1109. [Google Scholar] [CrossRef]
- Yoshikawa, A.; Sato, Y.; Yamashita, M.; Mimura, H.; Yamagata, A.; Fukai, S. Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett. 2009, 583, 3317–3322. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshikawa, A.; Mimura, H.; Yamashita, M.; Yamagata, A.; Fukai, S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009, 28, 2461–2468. [Google Scholar] [CrossRef]
- Sims, J.J.; Cohen, R.E. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 2009, 33, 775–783. [Google Scholar] [CrossRef]
- Swanson, K.A.; Kang, R.S.; Stamenova, S.D.; Hicke, L.; Radhakrishnan, I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003, 22, 4597–4606. [Google Scholar] [CrossRef]
- Kanayama, A.; Seth, R.B.; Sun, L.; Ea, C.K.; Hong, M.; Shaito, A.; Chiu, Y.H.; Deng, L.; Chen, Z.J. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol. Cell 2004, 15, 535–548. [Google Scholar] [CrossRef]
- Kulathu, Y.; Akutsu, M.; Bremm, A.; Hofmann, K.; Komander, D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct Mol. Biol. 2009, 16, 1328–1330. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshikawa, A.; Yamashita, M.; Yamagata, A.; Fukai, S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009, 28, 3903–3909. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Horton, J.R.; Mullally, J.E.; Heroux, A.; Cheng, X.; Wilkinson, K.D. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 2006, 124, 1197–1208. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Shanks, J.R.; Komander, D.; Wilkinson, K.D. Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J. Biol. Chem. 2008, 283, 19581–19592. [Google Scholar]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef]
- Ocio, E.M.; Mateos, M.V.; Maiso, P.; Pandiella, A.; San-Miguel, J.F. New drugs in multiple myeloma: mechanisms of action and phase I/II clinical findings. Lancet Oncol. 2008, 9, 1157–1165. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Suryadinata, R.; Roesley, S.N.A.; Yang, G.; Šarčević, B. Mechanisms of Generating Polyubiquitin Chains of Different Topology. Cells 2014, 3, 674-689. https://doi.org/10.3390/cells3030674
Suryadinata R, Roesley SNA, Yang G, Šarčević B. Mechanisms of Generating Polyubiquitin Chains of Different Topology. Cells. 2014; 3(3):674-689. https://doi.org/10.3390/cells3030674
Chicago/Turabian StyleSuryadinata, Randy, Siti Nur Ain Roesley, George Yang, and Boris Šarčević. 2014. "Mechanisms of Generating Polyubiquitin Chains of Different Topology" Cells 3, no. 3: 674-689. https://doi.org/10.3390/cells3030674
APA StyleSuryadinata, R., Roesley, S. N. A., Yang, G., & Šarčević, B. (2014). Mechanisms of Generating Polyubiquitin Chains of Different Topology. Cells, 3(3), 674-689. https://doi.org/10.3390/cells3030674



