SATB2 Induces Malignant Transformation and Cancer Stem Cell Characteristics, and Inhibition of Its Expression Reverses Drug Resistance in Mesothelioma
Highlights
- SATB2 is highly expressed in mesothelioma.
- Inhibition of SATB2 expression reverses drug resistance and enhances the effects of chemotherapy in mesothelioma.
- SATB2 can serve as a diagnostic biomarker for mesothelioma.
- It can be considered a novel target for treating mesothelioma.
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Cell Culture Conditions
2.2. Lentiviral Particle Production and Transduction
2.3. Immunofluorescence
2.4. Motility Assay
2.5. Transwell Migration Assay
2.6. Transwell Invasion Assay
2.7. Immunoblotting
2.8. Immunohistochemical Staining
2.9. Quantitative Real-Time PCR
2.10. Chromatin Immunoprecipitation (ChIP) Assay
2.11. Statistical Analysis
3. Results
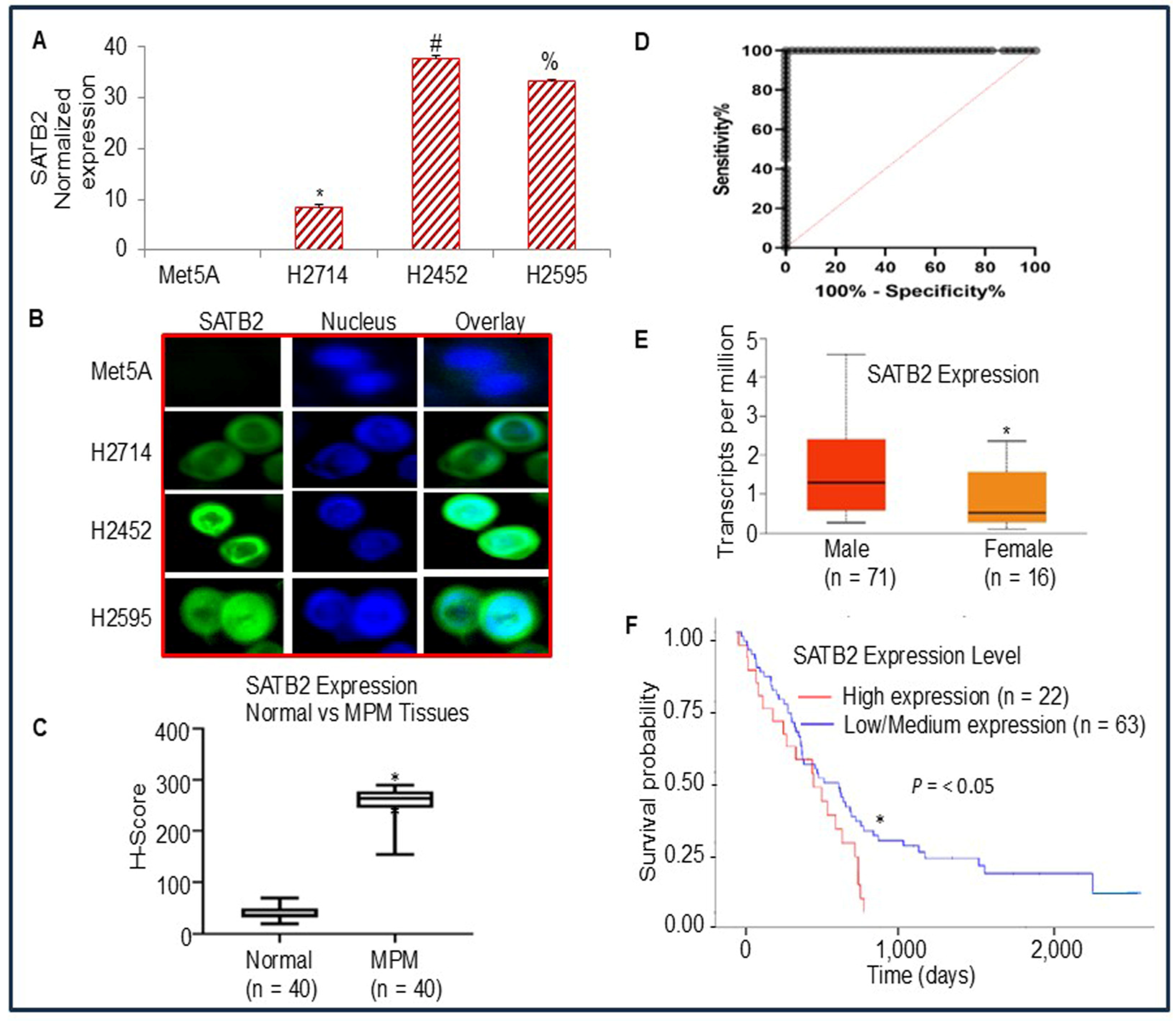
3.1. Differential Expression of SATB2 in Human Normal Mesothelial and Mesothelioma Cells/Tissues, and Survival Probability of Mesothelioma Patients
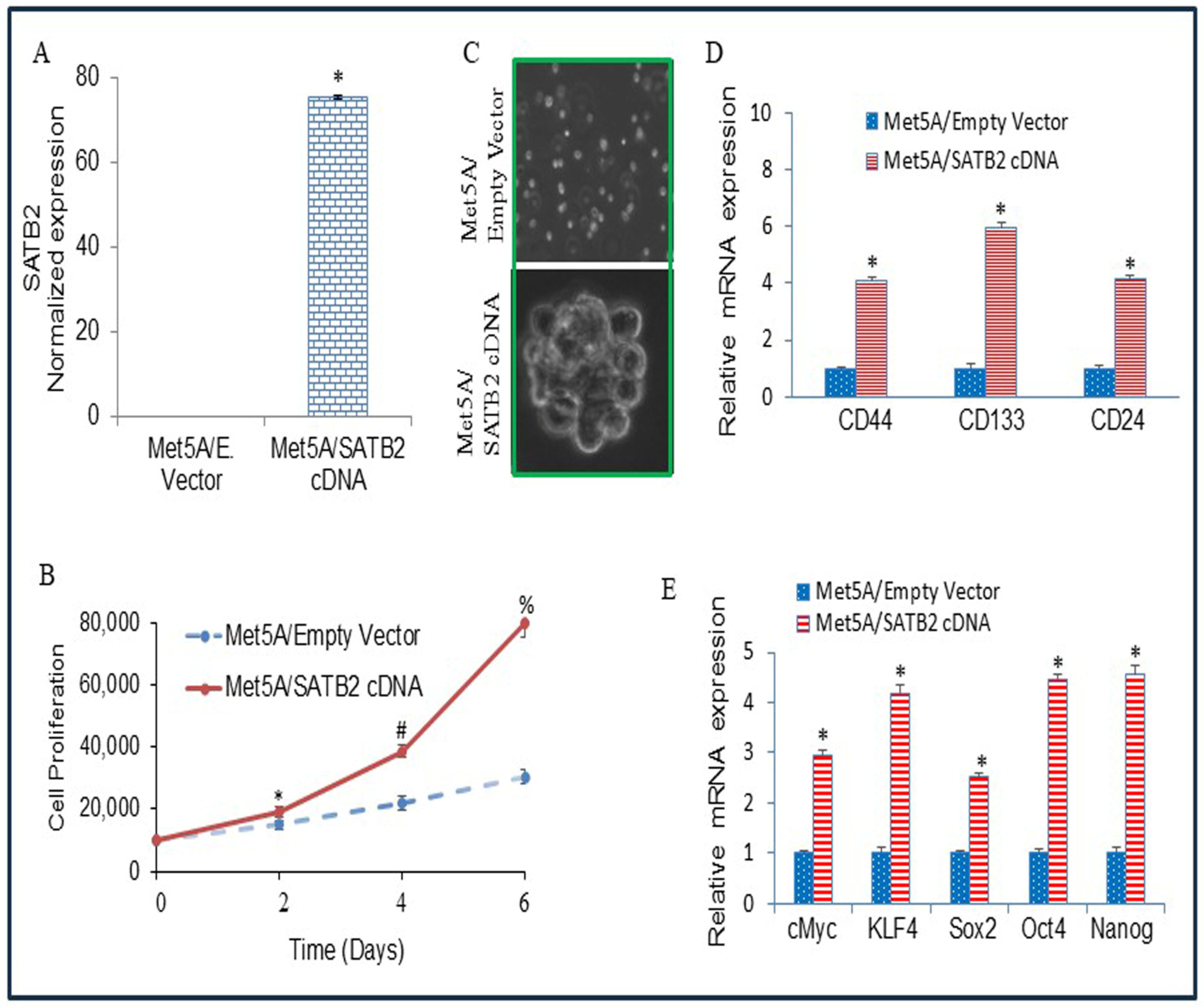
3.2. Overexpression of SATB2 in Met5A Cells Induces Cellular Transformation
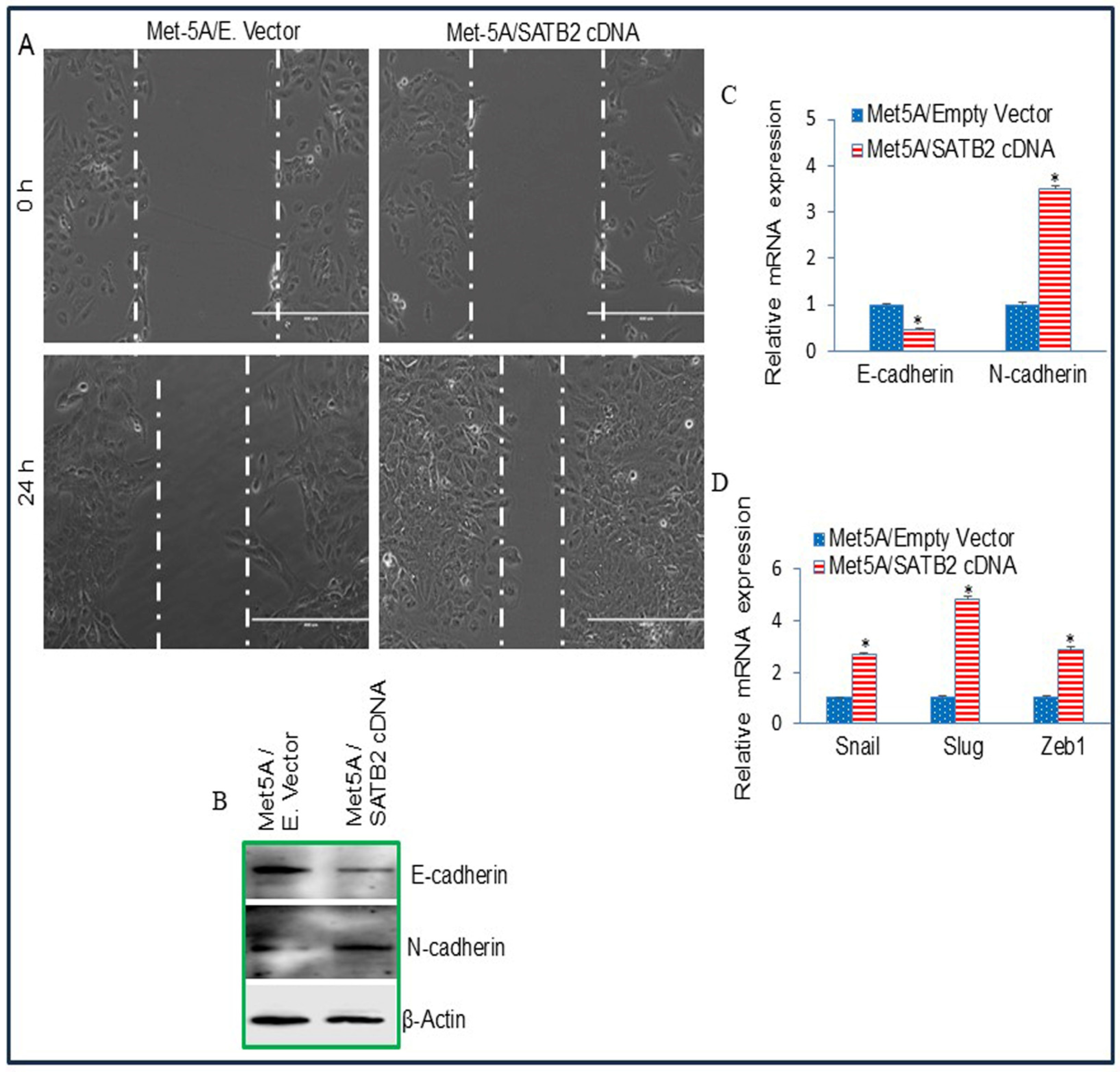
3.3. Overexpression of SATB2 in Met5A Cells Induces Epithelial-to-Mesenchymal Transition
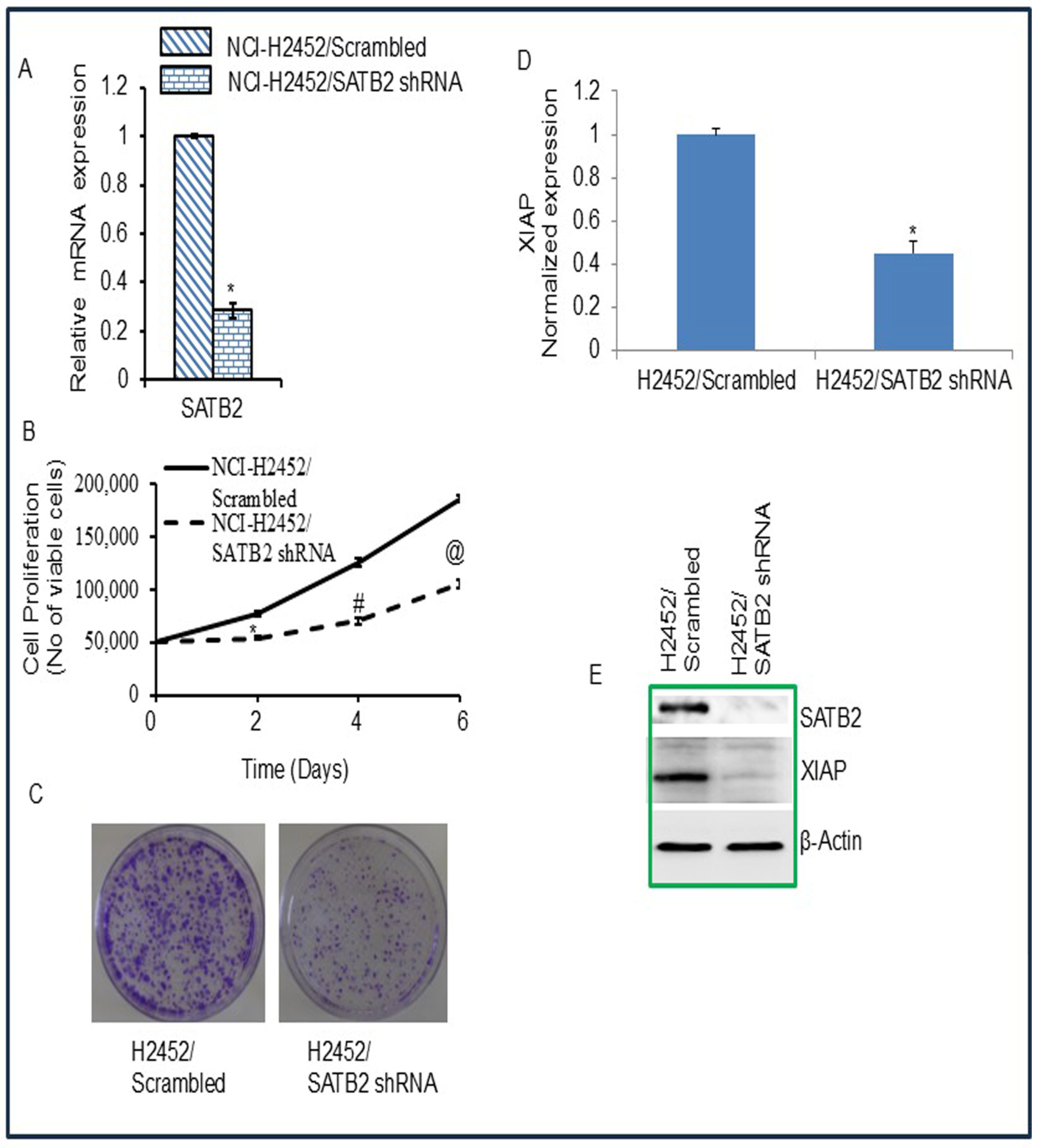
3.4. Inhibition of SATB2 in Mesothelioma Cell Lines Results in Reduced Cell Proliferation, Decreased Colony Formation, and Decreased XIAP Expression
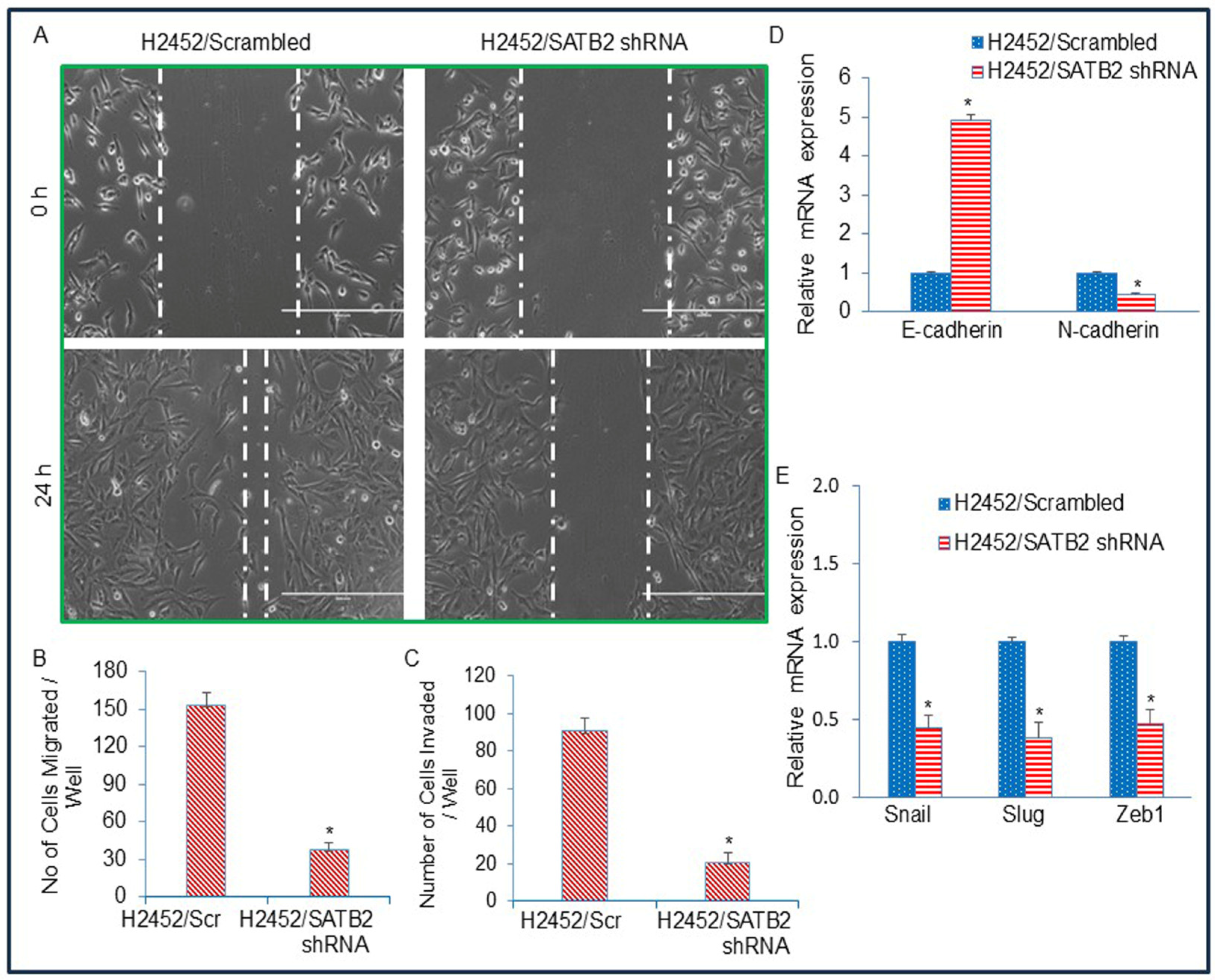
3.5. Inhibition of SATB2 in Mesothelioma Cell Lines Suppresses Epithelial–Mesenchymal Transition
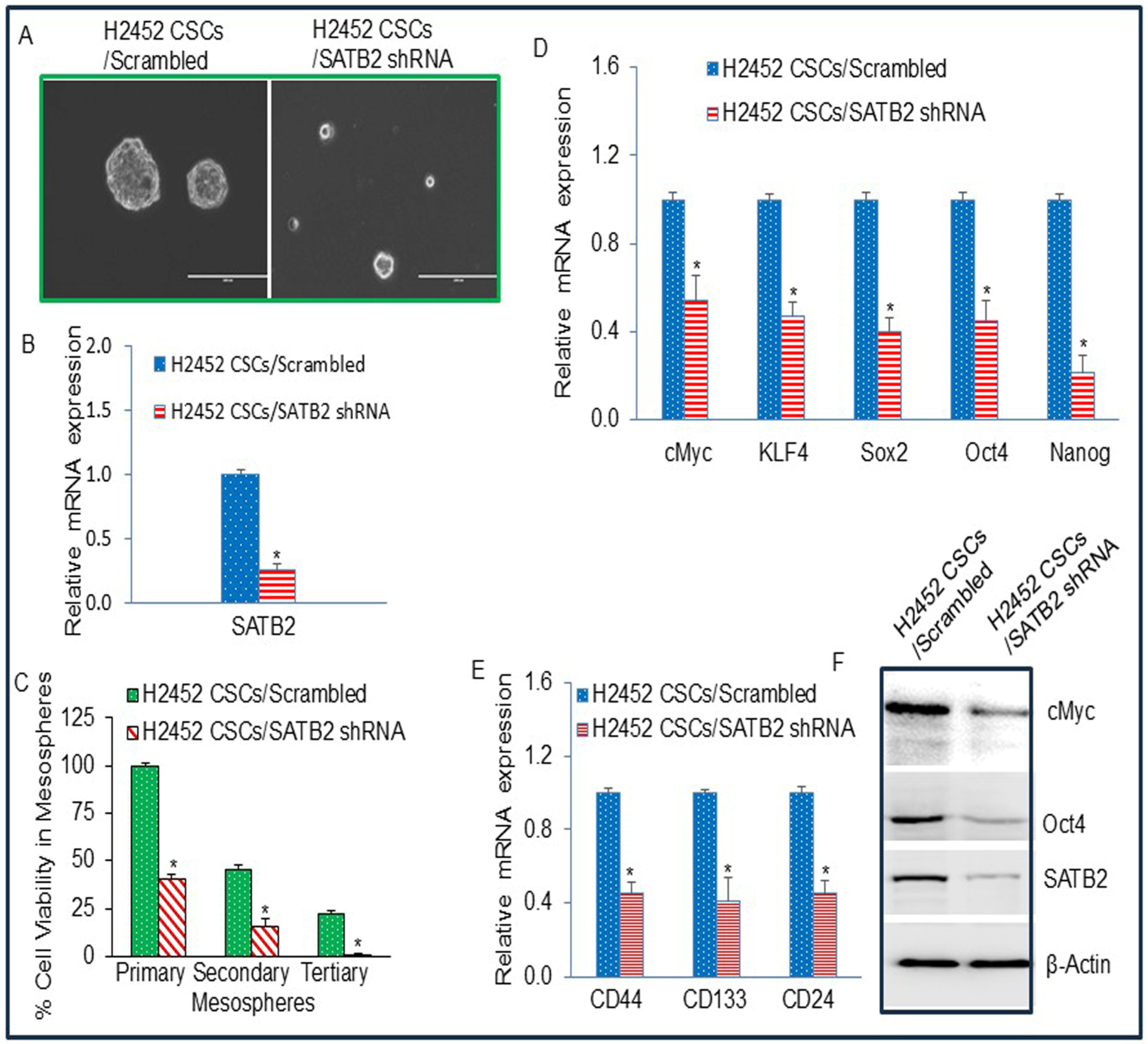
3.6. The Side Population of Malignant Mesothelioma Cells H2452 Contains CSCs, and Inhibition of SATB2 Suppresses Stemness in Mesothelioma
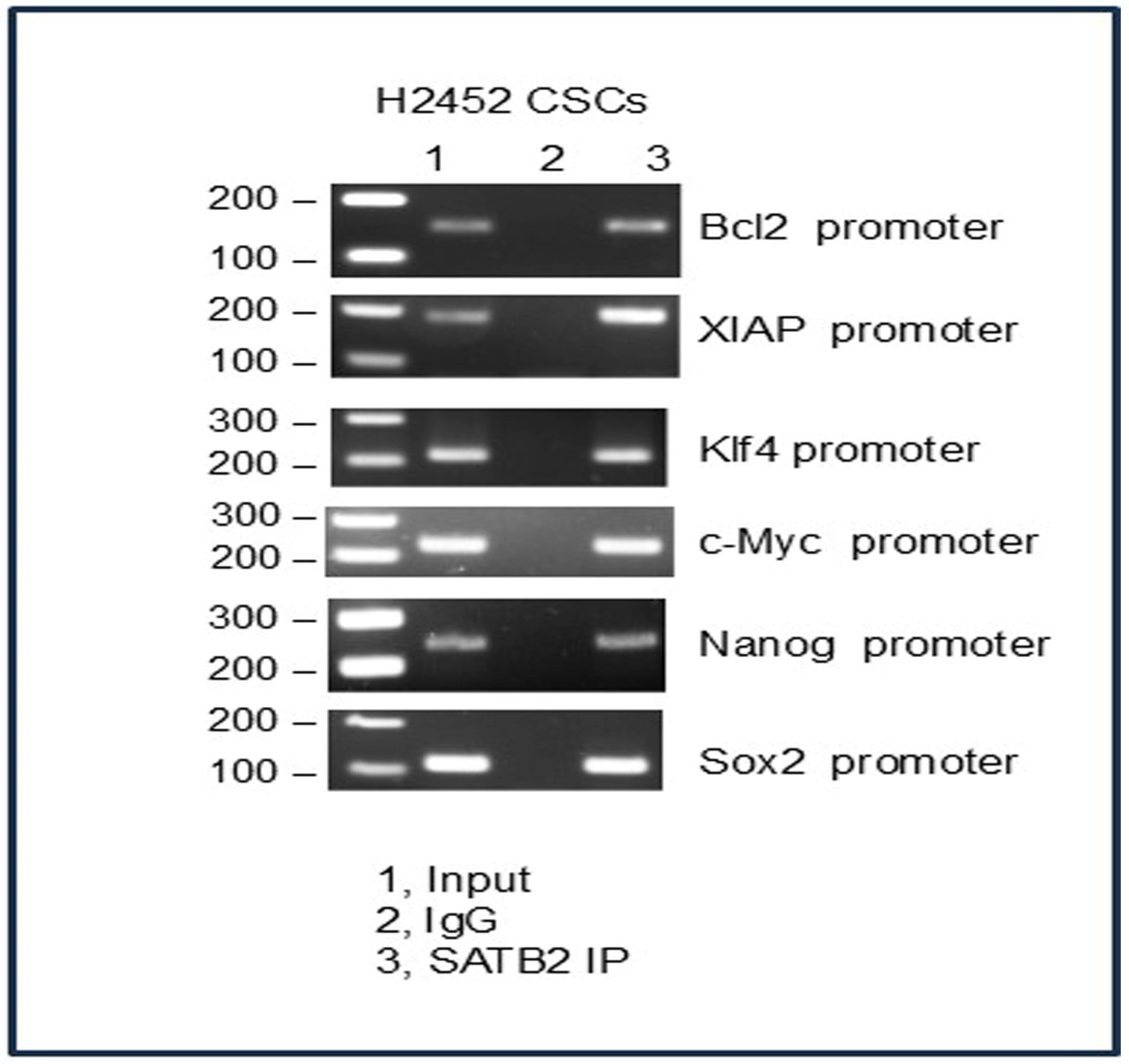
3.7. SATB2 Directly Binds to Bcl2, XIAP, KLF4, cMyc, Nanog, and Sox2 in H2452 CSCs
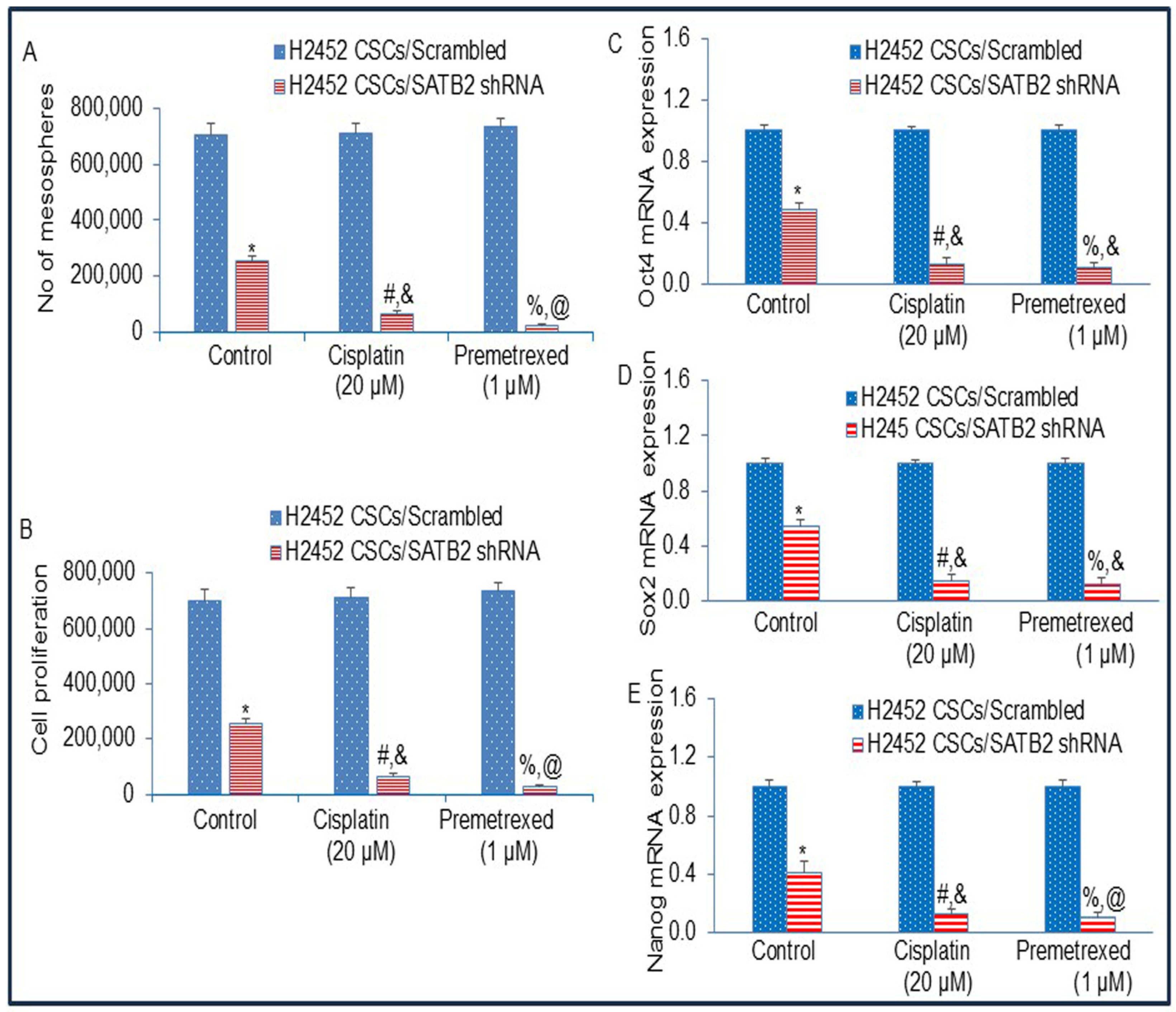
3.8. The Inhibition of SATB2 Expression by shRNA Reverses Chemotherapy Resistance
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aujayeb, A.; Astoul, P. A Diagnostic Approach to Malignant Pleural Mesothelioma. Pulm. Ther. 2025, 11, 503–517. [Google Scholar] [CrossRef]
- Bertuccio, F.R.; Montini, S.; Fusco, M.A.; Di Gennaro, A.; Sciandrone, G.; Agustoni, F.; Galli, G.; Bortolotto, C.; Saddi, J.; Baietto, G.; et al. Malignant Pleural Mesothelioma: From Pathophysiology to Innovative Actionable Targets. Cancers 2025, 17, 1160. [Google Scholar] [CrossRef]
- Fennell, D.A.; Sekido, Y.; Baas, P.; Husain, A.N.; Curioni-Fontecedro, A.; Lim, E.; Opitz, I.; Simone, C.B., 2nd; Brims, F.; Wong, M.C. Pleural mesothelioma. Nat. Rev. Dis. Primers 2025, 11, 56. [Google Scholar] [CrossRef]
- Jaurand, M.C.; Fleury-Feith, J. Pathogenesis of malignant pleural mesothelioma. Respirology 2005, 10, 2–8. [Google Scholar] [CrossRef]
- Kato, T.; Tanaka, I.; Huang, H.; Okado, S.; Imamura, Y.; Nomata, Y.; Takenaka, H.; Watanabe, H.; Kawasumi, Y.; Nakanishi, K.; et al. Molecular Mechanisms of Tumor Progression and Novel Therapeutic and Diagnostic Strategies in Mesothelioma. Int. J. Mol. Sci. 2025, 26, 4299. [Google Scholar] [CrossRef]
- Cardillo, G.; Waller, D.; Tenconi, S.; Di Noia, V.; Ricciardi, S. Malignant Pleural Mesothelioma: A 2025 Update. J. Clin. Med. 2025, 14, 1004. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Lamb, P.W.; Wiseman, R.W. Multiple mechanisms for the carcinogenic effects of asbestos and other mineral fibers. Environ. Health Perspect. 1989, 81, 81–89. [Google Scholar] [CrossRef]
- Nishimura, S.L.; Broaddus, V.C. Asbestos-induced pleural disease. Clin. Chest Med. 1998, 19, 311–329. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Reguart, N.; Corral, J.; Lianes, P. Malignant pleural mesothelioma: New hope in the horizon with novel therapeutic strategies. Cancer Treat. Rev. 2015, 41, 27–34. [Google Scholar] [CrossRef]
- Rivera, Z.; Strianese, O.; Bertino, P.; Yang, H.; Pass, H.; Carbone, M. The relationship between simian virus 40 and mesothelioma. Curr. Opin. Pulm. Med. 2008, 14, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Sekido, Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis 2013, 34, 1413–1419. [Google Scholar] [CrossRef]
- Stenton, S.C. Asbestos, Simian virus 40 and malignant mesothelioma. Thorax 1997, 52, S52–S57. [Google Scholar] [CrossRef]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Didier, A.J.; Li, M.; Gheeya, J.; Alahmadi, A.; Kaufman, J.; Memmott, R.; He, K.; Shields, P.; Carbone, D.P.; Presley, C.; et al. Trends in Mesothelioma Mortality in the United States Between 1999 and 2020. JTO Clin. Res. Rep. 2025, 6, 100804. [Google Scholar] [CrossRef]
- Stevens, M.E.; Paustenbach, D.J.; Korchevskiy, A. Exposure-response analysis of recent epidemiological data: Proposed risk based occupational exposure limits for various mineral types of asbestos. Chem. Biol. Interact. 2025, 419, 111645. [Google Scholar] [CrossRef]
- Yang, H.; Testa, J.R.; Carbone, M. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr. Treat. Options Oncol. 2008, 9, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Krevanko, C.F.; Hernandez, A.M.; Gauthier, A.M.; Vahora, M.S.; Lewis, R.C.; Pierce, J.S. Potential influence of cancer history on mesothelioma incidence: An ecologic analysis in the U.S. population. J. Public Health 2025, 47, e540–e545. [Google Scholar] [CrossRef] [PubMed]
- Bridda, A.; Padoan, I.; Mencarelli, R.; Frego, M. Peritoneal mesothelioma: A review. Medscape Gen. Med. 2007, 9, 32. [Google Scholar]
- Bale, S.S.; Kwon, S.J.; Shah, D.A.; Banerjee, A.; Dordick, J.S.; Kane, R.S. Nanoparticle-mediated cytoplasmic delivery of proteins to target cellular machinery. ACS Nano 2010, 4, 1493–1500. [Google Scholar] [CrossRef]
- Garcia-Carbonero, R.; Paz-Ares, L. Systemic chemotherapy in the management of malignant peritoneal mesothelioma. Eur. J. Surg. Oncol. 2006, 32, 676–681. [Google Scholar] [CrossRef]
- Britanova, O.; Akopov, S.; Lukyanov, S.; Gruss, P.; Tarabykin, V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci. 2005, 21, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Gyorgy, A.B.; Szemes, M.; de Juan Romero, C.; Tarabykin, V.; Agoston, D.V. SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur. J. Neurosci. 2008, 27, 865–873. [Google Scholar] [CrossRef]
- Huang, X.; Chen, Q.; Luo, W.; Pakvasa, M.; Zhang, Y.; Zheng, L.; Li, S.; Yang, Z.; Zeng, H.; Liang, F.; et al. SATB2: A versatile transcriptional regulator of craniofacial and skeleton development, neurogenesis and tumorigenesis, and its applications in regenerative medicine. Genes. Dis. 2022, 9, 95–107. [Google Scholar] [CrossRef]
- Bell, R.A.V.; Al-Khalaf, M.H.; Brunette, S.; Alsowaida, D.; Chu, A.; Bandukwala, H.; Dechant, G.; Apostolova, G.; Dilworth, F.J.; Megeney, L.A. Chromatin Reorganization during Myoblast Differentiation Involves the Caspase-Dependent Removal of SATB2. Cells 2022, 11, 966. [Google Scholar] [CrossRef]
- Podgornaya, O.I. Nuclear organization by satellite DNA, SAF-A/hnRNPU and matrix attachment regions. Semin. Cell Dev. Biol. 2022, 128, 61–68. [Google Scholar] [CrossRef]
- Dobreva, G.; Chahrour, M.; Dautzenberg, M.; Chirivella, L.; Kanzler, B.; Farinas, I.; Karsenty, G.; Grosschedl, R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 2006, 125, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Britanova, O.; Depew, M.J.; Schwark, M.; Thomas, B.L.; Miletich, I.; Sharpe, P.; Tarabykin, V. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am. J. Hum. Genet. 2006, 79, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Britanova, O.; de Juan Romero, C.; Cheung, A.; Kwan, K.Y.; Schwark, M.; Gyorgy, A.; Vogel, T.; Akopov, S.; Mitkovski, M.; Agoston, D.; et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 2008, 57, 378–392. [Google Scholar] [CrossRef]
- Magnusson, K.; de Wit, M.; Brennan, D.J.; Johnson, L.B.; McGee, S.F.; Lundberg, E.; Naicker, K.; Klinger, R.; Kampf, C.; Asplund, A.; et al. SATB2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am. J. Surg. Pathol. 2011, 35, 937–948. [Google Scholar] [CrossRef]
- Jiang, G.; Zhou, X.; Chen, S.; Zhong, F.; Huang, G.; Wu, B.; Mou, Q.; Jiang, G.; Lin, T. SATB2 plays a critical role in pancreatic cancer cell proliferation, migration and T cell cytotoxicity. Cancer Genet. 2025, 296–297, 53–64. [Google Scholar] [CrossRef]
- Helal, N.S.; Maher, S.; Samir, S.; Elmeligy, H.A.; Aboul-Ezz, M.A.; Aboushousha, T.; Moussa, M. Assessing the diagnostic potential of SATB2 and beta-catenin as biomarkers and therapeutic targets in pancreatic ductal adenocarcinoma. J. Cancer Res. Clin. Oncol. 2025, 151, 56. [Google Scholar] [CrossRef] [PubMed]
- Ladenheim, A.; Zheng, J.X.; Teklu, A.; Matsukuma, K. PCSK2 can be Useful in a Panel Approach to Distinguish Foregut and Midgut Neuroendocrine Tumors. Int. J. Surg. Pathol. 2025, 33, 76–84. [Google Scholar] [CrossRef]
- Roy, S.K.; Shrivastava, A.; Srivastav, S.; Shankar, S.; Srivastava, R.K. SATB2 is a novel biomarker and therapeutic target for cancer. J. Cell Mol. Med. 2020, 24, 11064–11069. [Google Scholar] [CrossRef]
- Yu, W.; Roy, S.K.; Ma, Y.; LaVeist, T.A.; Shankar, S.; Srivastava, R.K. Higher expression of SATB2 in hepatocellular carcinoma of African Americans determines more aggressive phenotypes than those of Caucasian Americans. J. Cell Mol. Med. 2019, 23, 7999–8009. [Google Scholar] [CrossRef]
- Yu, W.; Srivastava, R.; Srivastava, S.; Ma, Y.; Shankar, S.; Srivastava, R.K. Oncogenic Role of SATB2 In Vitro: Regulator of Pluripotency, Self-Renewal, and Epithelial-Mesenchymal Transition in Prostate Cancer. Cells 2024, 13, 962. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Ochoa, A.C.; Shankar, S.; Srivastava, R.K. Cellular transformation of human mammary epithelial cells by SATB2. Stem Cell Res. 2017, 19, 139–147. [Google Scholar] [CrossRef]
- Patani, N.; Jiang, W.; Mansel, R.; Newbold, R.; Mokbel, K. The mRNA expression of SATB1 and SATB2 in human breast cancer. Cancer Cell Int. 2009, 9, 18. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Shankar, S.; Srivastava, R.K. Role of SATB2 in human pancreatic cancer: Implications in transformation and a promising biomarker. Oncotarget 2016, 7, 57783–57797. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Ma, Y.; Shankar, S.; Srivastava, R.K. SATB2/beta-catenin/TCF-LEF pathway induces cellular transformation by generating cancer stem cells in colorectal cancer. Sci. Rep. 2017, 7, 10939. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Ma, Y.; Shrivastava, S.K.; Srivastava, R.K.; Shankar, S. Chronic alcohol exposure induces hepatocyte damage by inducing oxidative stress, SATB2 and stem cell-like characteristics, and activating lipogenesis. J. Cell. Mol. Med. 2022, 26, 2119–2131. [Google Scholar] [CrossRef]
- Chew, S.H.; Toyokuni, S. Malignant mesothelioma as an oxidative stress-induced cancer: An update. Free Radic. Biol. Med. 2015, 86, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Dericks, L.; Galetta, D. An Overview of Cellular and Molecular Determinants Regulating Chemoresistance in Pleural Mesothelioma. Cancers 2025, 17, 979. [Google Scholar] [CrossRef] [PubMed]
- Ghani, F.I.; Yamazaki, H.; Iwata, S.; Okamoto, T.; Aoe, K.; Okabe, K.; Mimura, Y.; Fujimoto, N.; Kishimoto, T.; Yamada, T.; et al. Identification of cancer stem cell markers in human malignant mesothelioma cells. Biochem. Biophys. Res. Commun. 2011, 404, 735–742. [Google Scholar] [CrossRef]
- Kai, K.; D’Costa, S.; Yoon, B.I.; Brody, A.R.; Sills, R.C.; Kim, Y. Characterization of side population cells in human malignant mesothelioma cell lines. Lung Cancer 2010, 70, 146–151. [Google Scholar] [CrossRef]
- Thellung, S.; Favoni, R.E.; Wurth, R.; Nizzari, M.; Pattarozzi, A.; Daga, A.; Florio, T.; Barbieri, F. Molecular Pharmacology of Malignant Pleural Mesothelioma: Challenges and Perspectives from Preclinical and Clinical Studies. Curr. Drug Targets 2016, 17, 824–849. [Google Scholar] [CrossRef]
- Varghese, S.; Whipple, R.; Martin, S.S.; Alexander, H.R. Multipotent cancer stem cells derived from human malignant peritoneal mesothelioma promote tumorigenesis. PLoS ONE 2012, 7, e52825. [Google Scholar] [CrossRef]
- Fu, J.; Rodova, M.; Nanta, R.; Meeker, D.; Van Veldhuizen, P.J.; Srivastava, R.K.; Shankar, S. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128, and miR-200. Neuro Oncol. 2013, 15, 691–706. [Google Scholar] [CrossRef]
- Iwatsuki, M.; Mimori, K.; Yokobori, T.; Ishi, H.; Beppu, T.; Nakamori, S.; Baba, H.; Mori, M. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010, 101, 293–299. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, J.A.; Ballif, B.C.; Lucas, A.; Spence, E.J.; Powell, C.; Aylsworth, A.S.; Torchia, B.A.; Shaffer, L.G. Small deletions of SATB2 cause some of the clinical features of the 2q33.1 microdeletion syndrome. PLoS ONE 2009, 4, e6568. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Shankar, S.; Srivastava, R.K. Chronic ethanol exposure of human pancreatic normal ductal epithelial cells induces cancer stem cell phenotype through SATB2. J. Cell Mol. Med. 2018, 22, 3920–3928. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Nall, D.; Tang, S.N.; Meeker, D.; Passarini, J.; Sharma, J.; Srivastava, R.K. Resveratrol inhibits pancreatic cancer stem cell characteristics in human and KrasG12D transgenic mice by inhibiting pluripotency maintaining factors and epithelial-mesenchymal transition. PLoS ONE 2011, 6, e16530. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.Q.; Wu, J.; Wang, W.T.; Yu, W.; Zhao, G.N.; Zhang, P.; Xiong, J.; Li, M.; Xue, Z.; Wang, X.; et al. The AT-rich DNA-binding protein SATB2 promotes expression and physical association of human (G)gamma- and (A)gamma-globin genes. J. Biol. Chem. 2012, 287, 30641–30652. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Tian, C.; Wang, W.; Zhou, Y.; Wang, X.; Zhang, L. Advances in research on SATB2 and its role in tumor development. Cell Biosci. 2025, 15, 111. [Google Scholar] [CrossRef]
- Favoni, R.E.; Daga, A.; Malatesta, P.; Florio, T. Preclinical studies identify novel targeted pharmacological strategies for treatment of human malignant pleural mesothelioma. Br. J. Pharmacol. 2012, 166, 532–553. [Google Scholar] [CrossRef][Green Version]
- Blum, W.; Pecze, L.; Felley-Bosco, E.; Wu, L.; de Perrot, M.; Schwaller, B. Stem Cell Factor-Based Identification and Functional Properties of In Vitro-Selected Subpopulations of Malignant Mesothelioma Cells. Stem Cell Rep. 2017, 8, 1005–1017. [Google Scholar] [CrossRef]
- Cortes-Dericks, L.; Froment, L.; Boesch, R.; Schmid, R.A.; Karoubi, G. Cisplatin-resistant cells in malignant pleural mesothelioma cell lines show ALDH(high)CD44(+) phenotype and sphere-forming capacity. BMC Cancer 2014, 14, 304. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Brown, C.; Srivastava, S.; Srivastava, R.; Srivastava, R.; Morvant, J.; Shrivastava, A.; Srivastava, R.K. SATB2 Induces Malignant Transformation and Cancer Stem Cell Characteristics, and Inhibition of Its Expression Reverses Drug Resistance in Mesothelioma. Cells 2026, 15, 283. https://doi.org/10.3390/cells15030283
Brown C, Srivastava S, Srivastava R, Srivastava R, Morvant J, Shrivastava A, Srivastava RK. SATB2 Induces Malignant Transformation and Cancer Stem Cell Characteristics, and Inhibition of Its Expression Reverses Drug Resistance in Mesothelioma. Cells. 2026; 15(3):283. https://doi.org/10.3390/cells15030283
Chicago/Turabian StyleBrown, Cynthia, Shivam Srivastava, Rohit Srivastava, Rashmi Srivastava, Jason Morvant, Anju Shrivastava, and Rakesh K. Srivastava. 2026. "SATB2 Induces Malignant Transformation and Cancer Stem Cell Characteristics, and Inhibition of Its Expression Reverses Drug Resistance in Mesothelioma" Cells 15, no. 3: 283. https://doi.org/10.3390/cells15030283
APA StyleBrown, C., Srivastava, S., Srivastava, R., Srivastava, R., Morvant, J., Shrivastava, A., & Srivastava, R. K. (2026). SATB2 Induces Malignant Transformation and Cancer Stem Cell Characteristics, and Inhibition of Its Expression Reverses Drug Resistance in Mesothelioma. Cells, 15(3), 283. https://doi.org/10.3390/cells15030283





