Multi-Omics Mechanism of Chronic Gout Arthritis and Discovery of the Thyroid Hormone–AMPK–Taurine Metabolic Axis
Highlights
- Multi-omics profiling reveals nine persistently dysregulated proteins and 11 consistently altered metabolites during the transition from acute to chronic gouty arthritis.
- Chronic gout development involves significant perturbations in key pathways—thyroid hormone synthesis, AMPK signaling, and taurine metabolism—and a concomitant shift in the immune response from acute activation to chronic inflammation.
- The study identifies a coordinated disruption of the thyroid hormone–AMPK–taurine metabolic axis and immune microenvironment remodeling as central to chronic gout progression.
- These findings offer potential targets for early diagnosis and targeted interventions to prevent irreversible joint damage in chronic gouty arthritis.
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Blood Sample Collection
2.3. Proteomic Analysis
2.4. Metabolomics Analysis
2.5. Data Processing and Statistical Analysis
3. Results
3.1. Clinical Characteristics of the Selected Subjects
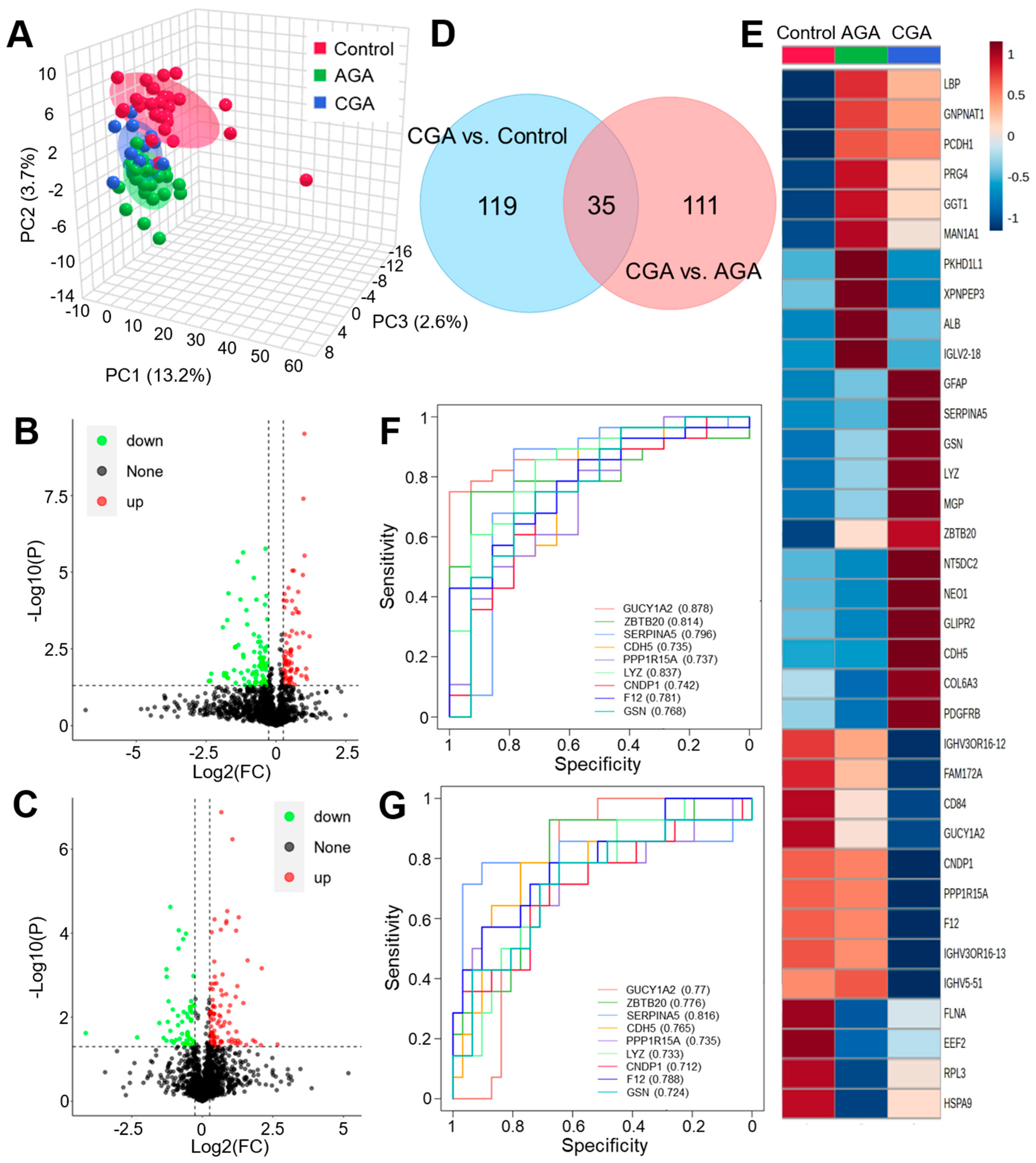
3.2. Proteome Differential Expression Analysis
3.3. Metabolome Difference Analysis
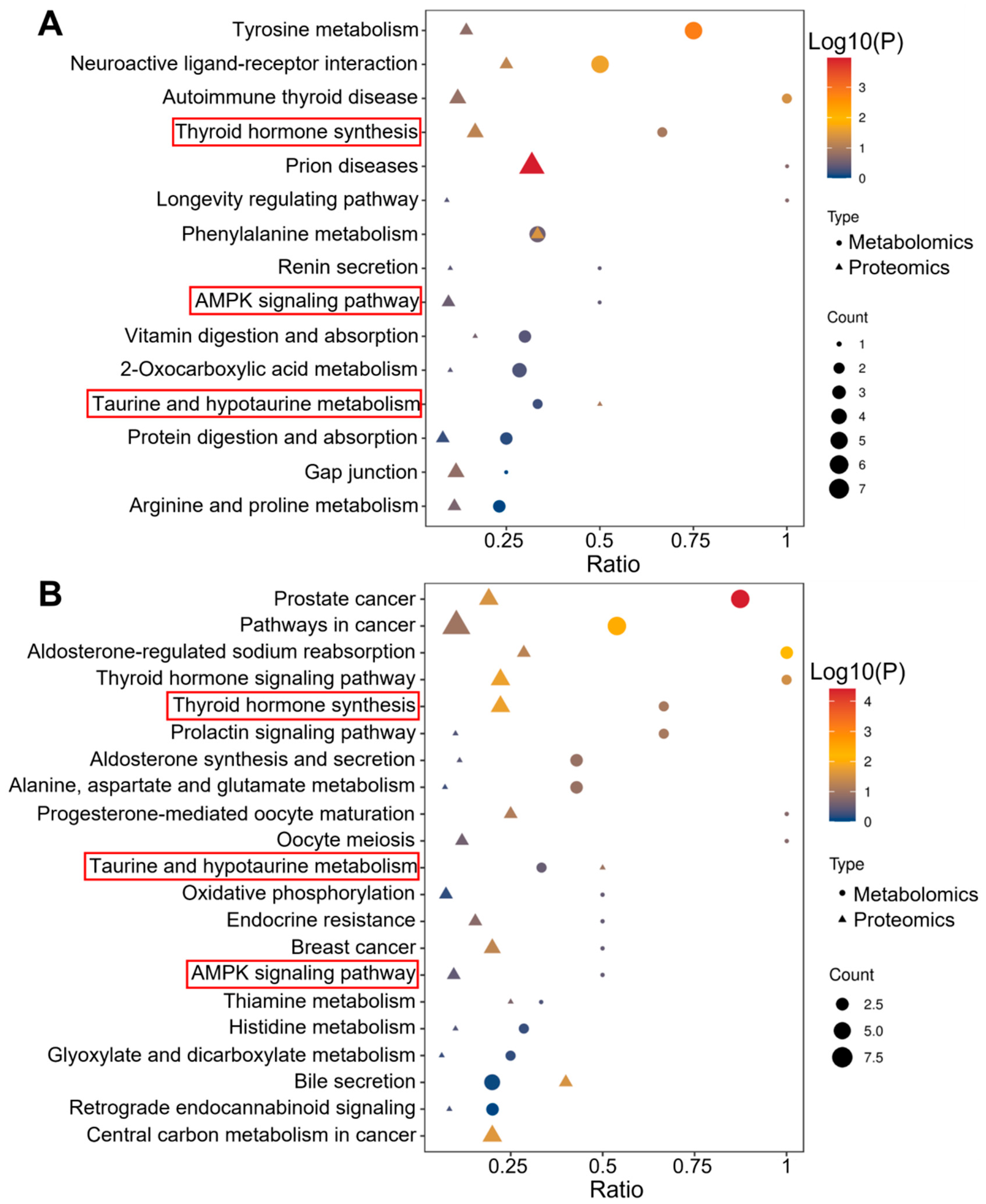
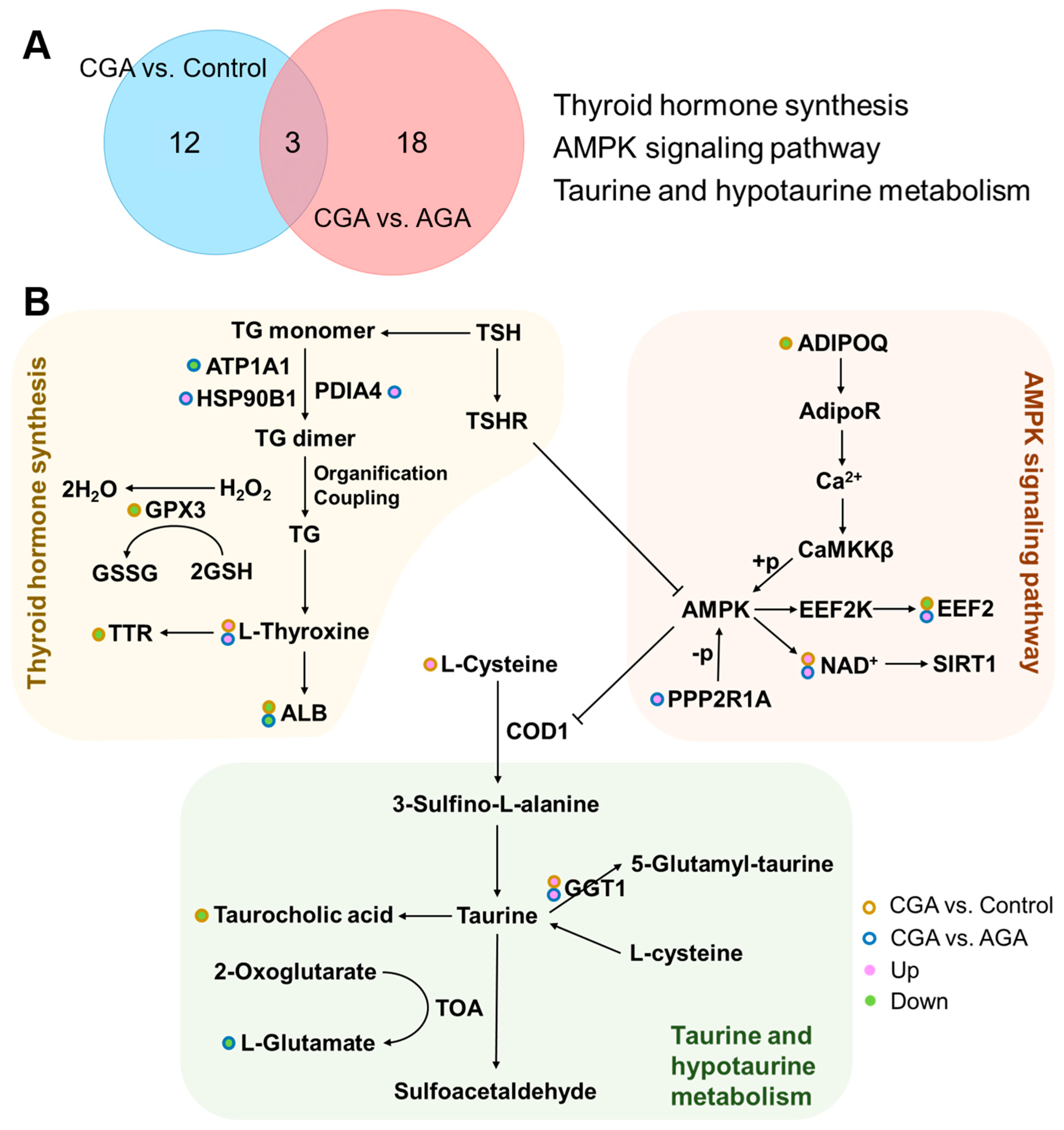
3.4. Pathway Analysis
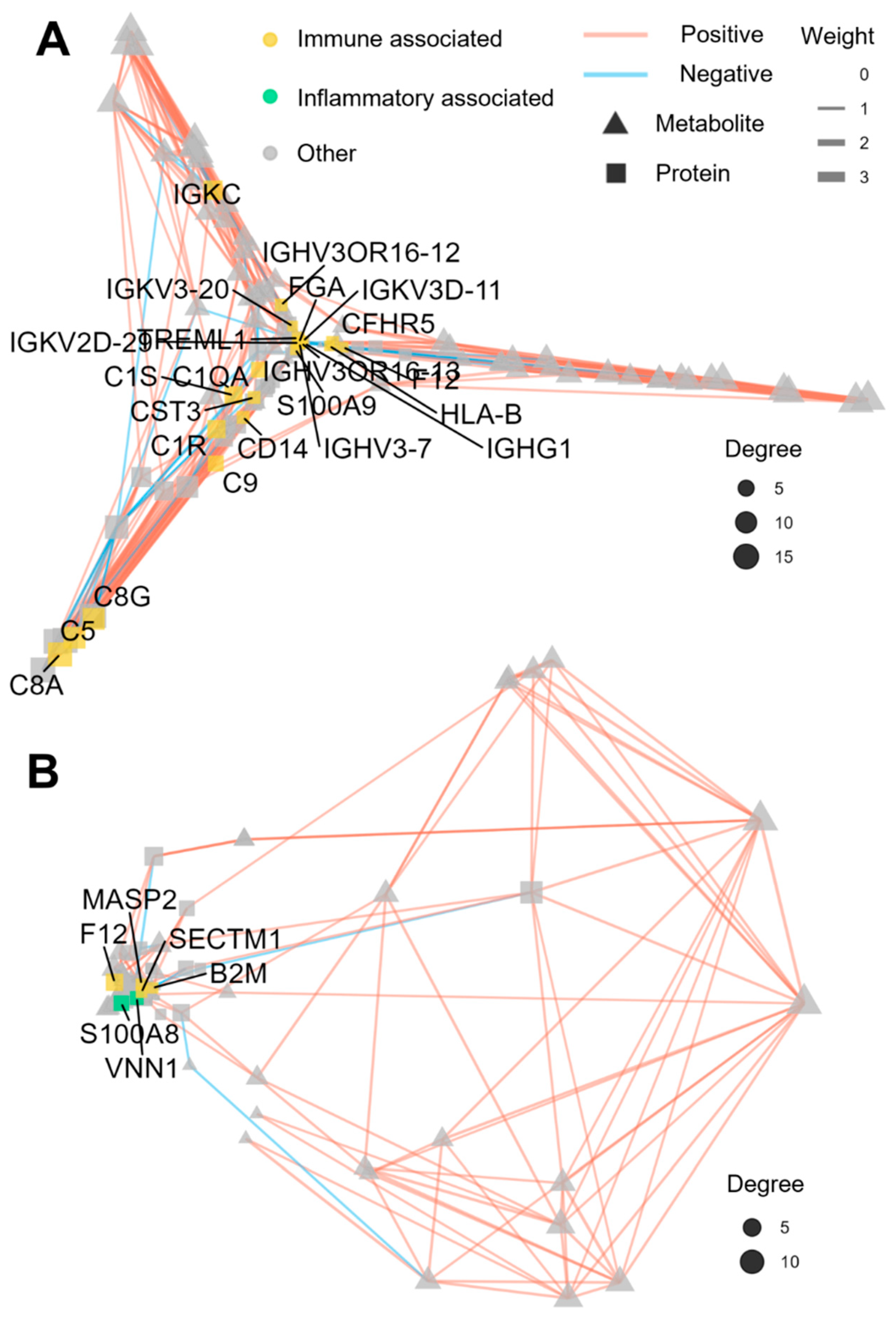
3.5. Regulatory Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGA | acute gouty arthritis |
| CGA | chronic gouty arthritis |
| DIA | data-independent acquisition |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| LC-MS | liquid chromatography–mass spectrometry |
| UHPLC | ultra-high performance liquid chromatography |
| HCD | higher-energy collisional dissociation |
| UHPLC-MS/MS | ultra-high performance liquid chromatography coupled with tandem mass spectrometry |
| QC | quality control |
| PCA | principal component analysis |
| PLS-DA | partial least squares–discriminant analysis |
| VIP | variable importance in the projection |
| FC | fold change |
| IQR | interquartile range |
| BMI | body mass index |
| ALT | alanine aminotransferase |
| GLU | glucose |
| TG | triglycerides |
| BUN | blood urea nitrogen |
| DEPs | differentially expressed proteins |
| AUC | area under the curve |
References
- Asghari, K.M.; Zahmatyar, M.; Seyedi, F.; Motamedi, A.; Zolfi, M.; Alamdary, S.J.; Fazlollahi, A.; Shamekh, A.; Mousavi, S.E.; Nejadghaderi, S.A.; et al. Gout: Global epidemiology, risk factors, comorbidities and complications: A narrative review. BMC Musculoskelet. Disord. 2024, 25, 1047. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Chen, W.; Qiu, X.; Wang, W. Epidemiology of gout-global burden of disease research from 1990 to 2019 and future trend predictions. Ther. Adv. Endocrinol. Metab. 2024, 15, 20420188241227295. [Google Scholar] [CrossRef] [PubMed]
- Dehlin, M.; Jacobsson, L.; Roddy, E. Global epidemiology of gout: Prevalence, incidence, treatment patterns and risk factors. Nat. Rev. Rheumatol. 2020, 16, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Pétrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241. [Google Scholar] [CrossRef]
- Cha, Y.; Lee, J.; Choy, W.; Lee, J.S.; Lee, H.H.; Chae, D.-S. Pathophysiology and treatment of gout arthritis; including gout arthritis of hip joint: A literature review. Hip Pelvis 2024, 36, 1–11. [Google Scholar] [CrossRef]
- Hansildaar, R.; Vedder, D.; Baniaamam, M.; Tausche, A.-K.; Gerritsen, M.; Nurmohamed, M.T. Cardiovascular risk in inflammatory arthritis: Rheumatoid arthritis and gout. Lancet Rheumatol. 2021, 3, e58–e70. [Google Scholar] [CrossRef]
- Deng, S.-H.; Dang, W.-T.; Liu, J.; Bai, Y.; You, L.-L.; Hu, J.; Luo, H. Differential diagnosis of acute and chronic gouty arthritis by multijoint ultrasound. Ultrasound Med. Biol. 2021, 47, 2853–2859. [Google Scholar] [CrossRef]
- Urano, W.; Yamanaka, H.; Tsutani, H.; Nakajima, H.; Matsuda, Y.; Taniguchi, A.; Hara, M.; Kamatani, N. The inflammatory process in the mechanism of decreased serum uric acid concentrations during acute gouty arthritis. J. Rheumatol. 2002, 29, 1950–1953. [Google Scholar]
- Liu, S.; Wang, Y.; Liu, H.; Xu, T.; Wang, M.-J.; Lu, J.; Guo, Y.; Chen, W.; Ke, M.; Zhou, G.; et al. Serum lipidomics reveals distinct metabolic profiles for asymptomatic hyperuricemic and gout patients. Rheumatology 2022, 61, 2644–2651. [Google Scholar] [CrossRef]
- Huang, Z.; Zhong, X.; Zhang, Y.; Li, X.; Liu, M.; Huang, Y.; Yue, J.; Yi, G.; Liu, H.; Yuan, B.; et al. A targeted proteomics screen reveals serum and synovial fluid proteomic signature in patients with gout. Front. Immunol. 2024, 15, 1468810. [Google Scholar] [CrossRef]
- Ji, Z.; Zheng, S.; Liang, L.; Huang, L.; Sun, S.; Huang, Z.; He, Y.; Pan, X.; Li, T.; Huang, Y. TMT-based quantitative proteomics analysis of serum-derived exosomes in patients with juvenile gout. Front. Endocrinol. 2025, 16, 1460218. [Google Scholar] [CrossRef] [PubMed]
- Gobena, S.; Admassu, B.; Kinde, M.Z.; Gessese, A.T. Proteomics and its current application in biomedical area: Concise review. Sci. World J. 2024, 2024, 4454744. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Cheng, J.; Yu, H.; Huang, X.; Bao, H.; Qin, L.; Wang, L.; Song, Y.; Liu, X.; Peng, A. Quantitative proteomics by iTRAQ-PRM based reveals the new characterization for gout. Proteome Sci. 2021, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Yang, X.; Wang, X.; Wang, Y.; Zhang, B.; Lin, Z. Exploring exosome contributions to gouty arthritis: A proteomics and experimental study. Int. J. Mol. Sci. 2025, 26, 5320. [Google Scholar] [CrossRef]
- Lv, J.; Pan, C.; Cai, Y.; Han, X.; Wang, C.; Ma, J.; Pang, J.; Xu, F.; Wu, S.; Kou, T.; et al. Plasma metabolomics reveals the shared and distinct metabolic disturbances associated with cardiovascular events in coronary artery disease. Nat. Commun. 2024, 15, 5729. [Google Scholar] [CrossRef]
- Jia, Q.; Dong, Q.; Zhang, J.; Zhao, Q.; Li, Y.; Chao, Z.; Liu, J. Untargeted metabolomics analysis of the urinary metabolic signature of acute and chronic gout. Clin. Chim. Acta 2025, 565, 119968. [Google Scholar] [CrossRef]
- Wang, M.; Li, R.; Qi, H.; Pang, L.; Cui, L.; Liu, Z.; Lu, J.; Wang, R.; Hu, S.; Liang, N.; et al. Metabolomics and machine learning identify metabolic differences and potential biomarkers for frequent versus Infrequent Gout Flares. Arthritis Rheumatol. 2023, 75, 2252–2264. [Google Scholar] [CrossRef]
- Yin, C.; Liu, B.; Dong, Z.; Shi, S.; Peng, C.; Pan, Y.; Bi, X.; Nie, H.; Zhang, Y.; Tai, Y.; et al. CXCL5 activates CXCR2 in nociceptive sensory neurons to drive joint pain and inflammation in experimental gouty arthritis. Nat. Commun. 2024, 15, 3263. [Google Scholar] [CrossRef]
- Ahn, E.Y.; So, M.W. The pathogenesis of gout. J. Rheum. Dis. 2025, 32, 8–16. [Google Scholar] [CrossRef]
- Parisa, N.; Kamaluddin, M.T.; Saleh, M.I.; Sinaga, E. The inflammation process of gout arthritis and its treatment. J. Adv. Pharm. Technol. Res. 2023, 14, 166–170. [Google Scholar] [CrossRef]
- Li, C.; Wu, C.; Li, F.; Xu, W.; Zhang, X.; Huang, Y.; Xia, D. Targeting neutrophil extracellular traps in gouty arthritis: Insights into pathogenesis and therapeutic potential. J. Inflamm. Res. 2024, 17, 1735–1763. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Kuriyama, S.; Nakano, T.; Sekine, M.; Toyoda, Y.; Nakayama, A.; Takada, T.; Kawamura, Y.; Nakamura, T.; Matsuo, H.; et al. Urate transporter ABCG2 function and asymptomatic hyperuricemia: A retrospective cohort study of CKD progression. Am. J. Kidney Dis. 2023, 81, 134–144.e1. [Google Scholar] [CrossRef]
- Kanoh, H.; Iwashita, S.; Kuraishi, T.; Goto, A.; Fuse, N.; Ueno, H.; Nimura, M.; Oyama, T.; Tang, C.; Watanabe, R.J.I.; et al. cGMP signaling pathway that modulates NF-κB activation in innate immune responses. iScience 2021, 24, 103473. [Google Scholar] [CrossRef] [PubMed]
- Peters, V.; Yard, B.; Schmitt, C.P. Carnosine and diabetic nephropathy. Curr. Med. Chem. 2020, 27, 1801–1812. [Google Scholar] [CrossRef] [PubMed]
- Ferraboschi, P.; Ciceri, S.; Grisenti, P. Applications of lysozyme, an innate immune defense factor, as an alternative antibiotic. Antibiotics 2021, 10, 1534. [Google Scholar] [CrossRef]
- Su, J.; Li, Z.; Gao, P.; Ahmed, I.; Liu, Q.; Li, R.; Cui, K.; ur Rehman, S. Comparative evolutionary and molecular genetics based study of Buffalo lysozyme gene family to elucidate their antibacterial function. Int. J. Biol. Macromol. 2023, 234, 123646. [Google Scholar] [CrossRef]
- Grover, S.P.; Mackman, N. Anticoagulant SERPINs: Endogenous regulators of hemostasis and thrombosis. Front. Cardiovasc. Med. 2022, 9, 878199. [Google Scholar] [CrossRef]
- Zeng, S.; Liu, Z.; Yin, J.; Li, S.; Jiang, M.; Yang, H.; Long, Y. Improvement in Clinical Features of L-NAME-Induced Preeclampsia-like Rats through Reduced SERPINA5 Expression. Biomolecules 2023, 13, 1792. [Google Scholar] [CrossRef]
- Maas, C.; Renné, T. Coagulation factor XII in thrombosis and inflammation. Blood 2018, 131, 1903–1909. [Google Scholar] [CrossRef]
- Wu, Y.; Zheng, J.; Yan, Y.; Liu, J.; Zhou, Y. Gelsolin can be a prognostic biomarker and correlated with immune infiltrates in gastric cancer. Int. J. Gen. Med. 2022, 15, 927–936. [Google Scholar] [CrossRef]
- Zhen, H.; Gui, F. The role of hyperuricemia on vascular endothelium dysfunction. Biomed. Rep. 2017, 7, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, M.; Huang, S.; Lan, X.; Zheng, J.; Luo, H.; He, Y.; Lei, W. Hyperuricemia: A key contributor to endothelial dysfunction in cardiovascular diseases. FASEB J. 2023, 37, e23012. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zheng, C.; Xu, Y.; Hu, X. Comprehensive molecular and cellular characterization of endoplasmic reticulum stress-related key genes in renal ischemia/reperfusion injury. Front. Immunol. 2024, 15, 1340997. [Google Scholar] [CrossRef] [PubMed]
- Magg, V.; Manetto, A.; Kopp, K.; Wu, C.C.; Naghizadeh, M.; Lindner, D.; Eke, L.; Welsch, J.; Kallenberger, S.M.; Schott, J.; et al. Turnover of PPP1R15A mRNA encoding GADD34 controls responsiveness and adaptation to cellular stress. Cell Rep. 2024, 43, 114069. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, S.; Liu, G.; Pan, A.; Peng, M.; Liao, Y. Association between sex hormones and gout: An analysis of the UK Biobank cohort. Steroids 2024, 207, 109422. [Google Scholar] [CrossRef]
- Singh, A.K.; Durairajan, S.S.K.; Iyaswamy, A.; Williams, L.L. Elucidating the role of gut microbiota dysbiosis in hyperuricemia and gout: Insights and therapeutic strategies. World J. Gastroenterol. 2024, 30, 4404–4410. [Google Scholar] [CrossRef]
- Mullur, R.; Liu, Y.-Y.; Brent, G.A. Thyroid hormone regulation of metabolism. Physiol. Rev. 2014, 94, 355–382. [Google Scholar] [CrossRef]
- Arioli, F.; Gamberini, M.C.; Pavlovic, R.; Di Cesare, F.; Draghi, S.; Bussei, G.; Mungiguerra, F.; Casati, A.; Fidani, M. Quantification of cortisol and its metabolites in human urine by LC-MSn: Applications in clinical diagnosis and anti-doping control. Anal. Bioanal. Chem. 2022, 414, 6841–6853. [Google Scholar] [CrossRef]
- Johannsen, D.L.; Galgani, J.E.; Johannsen, N.M.; Zhang, Z.; Covington, J.D.; Ravussin, E. Effect of short-term thyroxine administration on energy metabolism and mitochondrial efficiency in humans. PLoS ONE 2012, 7, e40837. [Google Scholar] [CrossRef]
- Fan, Y.-Y.; Wang, Y.-J.; Guo, J.; Wu, M.-N.; Zhang, M.-S.; Niu, B.-L.; Li, Y.; Zhao, J.; Yang, C.-H.; Li, Y.; et al. Delayed metformin treatment improves functional recovery following traumatic brain injury via central AMPK-dependent brain tissue repair. Brain Res. Bull. 2020, 164, 146–156. [Google Scholar] [CrossRef]
- Baliou, S.; Adamaki, M.; Ioannou, P.; Pappa, A.; Panayiotidis, M.I.; Spandidos, D.A.; Christodoulou, I.; Kyriakopoulos, A.M.; Zoumpourlis, V. Protective role of taurine against oxidative stress. Brain Res. Bull. 2021, 24, 605. [Google Scholar] [CrossRef]


| Normal Controls (n = 28) | Patients with AGA (n = 31) | Participants with CGA (n = 14) | |
|---|---|---|---|
| Age, years, median (IQR) | 26 (24–28) | 33 (22–51) † | 28 (21–54) † |
| BMI, kg/m2, median (IQR) | 23.2 (19.6–28.4) | 26.5 (17.0–32.0) † | 24.8 (19.6–34.3) |
| Smoking, n(%) a | 2 (10.5) | 20 (66.7) † | 9 (64.3) † |
| Drinking, n(%) b | 0 (0) | 11 (36.7) † | 2 (14.3) |
| Beverage, n(%) c | 8 (42.1) | 13 (43.3) | 1 (7.1) †,‡ |
| Sleep time, hours/day, median (IQR) | 7 (6–8) | 7 (4–8) † | 7 (6–9) |
| ALT, units/liter, median (IQR) | 25.1 (7.7–70.8) | 42.4 (16.9–91.6) † | 30.7 (11.1–92.5) |
| AST, units/liter, median (IQR) | 20.6 (8.9–67.6) | 25.5 (10.4–54) | 21.9 (15.4–41.5) |
| GLU, mmoles/liter, median (IQR) | 4.7 (3.5–5.6) | 5.0 (3.8–6.8) † | 4.7 (4.1–5.6) ‡ |
| TG, mmoles/liter, median (IQR) | 0.9 (0.5–2.2) | 1.4 (0.7–4.2) † | 1.3 (0.7–1.5) |
| TCH, mmoles/liter, median (IQR) | 4.7 (3.4–6.1) | 4.6 (1.8–6.9) | 3.9 (3.1–4.9) † |
| BUN, mmoles/liter, median (IQR) | 5.5 (3.7–8.4) | 5.1 (3.9–7.7) | 4.3 (3.0–7.0) †,‡ |
| CR, mmoles/liter, median (IQR) | 78.5 (62.0–93.0) | 82.1 (65.0–106.0) | 93.0 (60.0–112.0) |
| SUA, µmoles/liter, median (IQR) | 342.0 (231.6–427.3) | 481.0 (336.0–696.0) † | 538.0 (388.2–718.0) † |
| No. | Protein | CGA vs. Control | CGA vs. AGA | Up. Down | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p | FC | log2FC | AUC | p | FC | log2FC | AUC | |||
| 1 | GUCY1A2 | 4.52 × 10−6 | 0.39 | −1.37 | 0.878 | 2.37 × 10−5 | 0.46 | −1.13 | 0.77 | down |
| 2 | ZBTB20 | 2.10 × 10−4 | 1.77 | 0.82 | 0.814 | 1.55 × 10−3 | 1.41 | 0.50 | 0.776 | up |
| 3 | SERPINA5 | 1.99 × 10−4 | 1.69 | 0.76 | 0.796 | 2.96 × 10−3 | 1.63 | 0.70 | 0.816 | up |
| 4 | CDH5 | 1.06 × 10−2 | 1.28 | 0.35 | 0.735 | 5.65 × 10−3 | 1.29 | 0.37 | 0.765 | up |
| 5 | PPP1R15A | 7.01 × 10−3 | 0.72 | −0.48 | 0.737 | 6.88 × 10−3 | 0.74 | −0.43 | 0.735 | down |
| 6 | LYZ | 8.69 × 10−5 | 1.30 | 0.38 | 0.837 | 7.33 × 10−3 | 1.20 | 0.27 | 0.733 | up |
| 7 | CNDP1 | 8.64 × 10−3 | 0.73 | −0.45 | 0.742 | 1.14 × 10−2 | 0.76 | −0.40 | 0.712 | down |
| 8 | F12 | 9.37 × 10−3 | 0.73 | −0.46 | 0.781 | 1.21 × 10−2 | 0.76 | −0.39 | 0.788 | down |
| 9 | GSN | 1.45 × 10−2 | 1.48 | 0.56 | 0.768 | 4.07 × 10−2 | 1.35 | 0.44 | 0.724 | up |
| No. | Metabolite | CGA vs. Control | CGA vs. AGA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FC | log2FC | p | VIP | AUC | FC | log2FC | p | VIP | AUC | Up. Down | ||
| 1 | 5α-Dihydrotestosterone glucuronide | 0.33 | −1.60 | 8.24 × 10−13 | 2.31 | 0.982 | 0.69 | −0.54 | 8.13 × 10−5 | 1.09 | 0.866 | down |
| 2 | Normorphine | 0.39 | −1.37 | 2.97 × 10−4 | 1.16 | 0.911 | 0.55 | −0.86 | 8.60 × 10−5 | 1.13 | 0.892 | down |
| 3 | Indole-3-lactic acid | 5.46 | 2.45 | 1.28 × 10−8 | 2.51 | 1 | 1.90 | 0.93 | 5.79 × 10−4 | 1.60 | 0.823 | up |
| 4 | 3-hydroxy-3-methylpentanedioic acid | 20.06 | 4.33 | 8.46 × 10−13 | 2.32 | 1 | 2.38 | 1.25 | 1.61 × 10−3 | 1.04 | 0.77 | up |
| 5 | β-Cortolone | 4.67 | 2.22 | 9.77 × 10−6 | 2.05 | 0.982 | 2.46 | 1.30 | 3.11 × 10−3 | 1.74 | 0.765 | up |
| 6 | Tangeritin | 0.56 | −0.84 | 1.56 × 10−8 | 1.94 | 0.995 | 0.65 | −0.61 | 3.34 × 10−3 | 1.48 | 0.747 | down |
| 7 | Citrinin | 1.51 | 0.60 | 4.61 × 10−5 | 1.47 | 0.906 | 1.32 | 0.40 | 3.92 × 10−3 | 1.48 | 0.747 | up |
| 8 | Syringic acid | 3.44 | 1.78 | 1.38 × 10−4 | 2.16 | 0.913 | 1.96 | 0.97 | 4.53 × 10−3 | 2.09 | 0.765 | up |
| 9 | Dehydroepiandrosterone | 2.35 | 1.23 | 3.89 × 10−4 | 1.73 | 0.872 | 1.64 | 0.71 | 1.90 × 10−2 | 1.53 | 0.735 | up |
| 10 | L-Thyroxine | 1.39 | 0.47 | 7.26 × 10−3 | 1.21 | 0.827 | 1.28 | 0.35 | 3.34 × 10−2 | 1.38 | 0.749 | up |
| 11 | Glycodeoxycholic acid | 0.21 | −2.28 | 3.47 × 10−6 | 2.01 | 0.934 | 0.48 | −1.06 | 3.51 × 10−2 | 1.22 | 0.7 | down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, G.; Luo, Y.; Zheng, X.; Mei, Z.; Ye, Q.; Peng, J.; Duan, F.; Cui, Y.; An, P.; Song, Y.; et al. Multi-Omics Mechanism of Chronic Gout Arthritis and Discovery of the Thyroid Hormone–AMPK–Taurine Metabolic Axis. Cells 2026, 15, 41. https://doi.org/10.3390/cells15010041
Zhu G, Luo Y, Zheng X, Mei Z, Ye Q, Peng J, Duan F, Cui Y, An P, Song Y, et al. Multi-Omics Mechanism of Chronic Gout Arthritis and Discovery of the Thyroid Hormone–AMPK–Taurine Metabolic Axis. Cells. 2026; 15(1):41. https://doi.org/10.3390/cells15010041
Chicago/Turabian StyleZhu, Guizhen, Yuan Luo, Xiangyi Zheng, Zhusong Mei, Qiao Ye, Jie Peng, Fengsen Duan, Yueying Cui, Peiyu An, Yangqian Song, and et al. 2026. "Multi-Omics Mechanism of Chronic Gout Arthritis and Discovery of the Thyroid Hormone–AMPK–Taurine Metabolic Axis" Cells 15, no. 1: 41. https://doi.org/10.3390/cells15010041
APA StyleZhu, G., Luo, Y., Zheng, X., Mei, Z., Ye, Q., Peng, J., Duan, F., Cui, Y., An, P., Song, Y., Li, H., Zhang, H., & Wang, G. (2026). Multi-Omics Mechanism of Chronic Gout Arthritis and Discovery of the Thyroid Hormone–AMPK–Taurine Metabolic Axis. Cells, 15(1), 41. https://doi.org/10.3390/cells15010041






