On the Quest for Biomarkers: A Comprehensive Analysis of Modified Nucleosides in Ovarian Cancer Cell Lines
Abstract
1. Introduction
Aim of This Study
2. Materials and Methods
2.1. Cell Culture
2.2. Preparation of the Cell Culture Medium
2.3. Preparation of the Cells for Targeted Analysis of Modified Nucleosides
2.4. RNA Extraction and Digestion
2.5. Targeted Analysis of the Nucleosides
2.6. Data Processing and Statistical Analysis
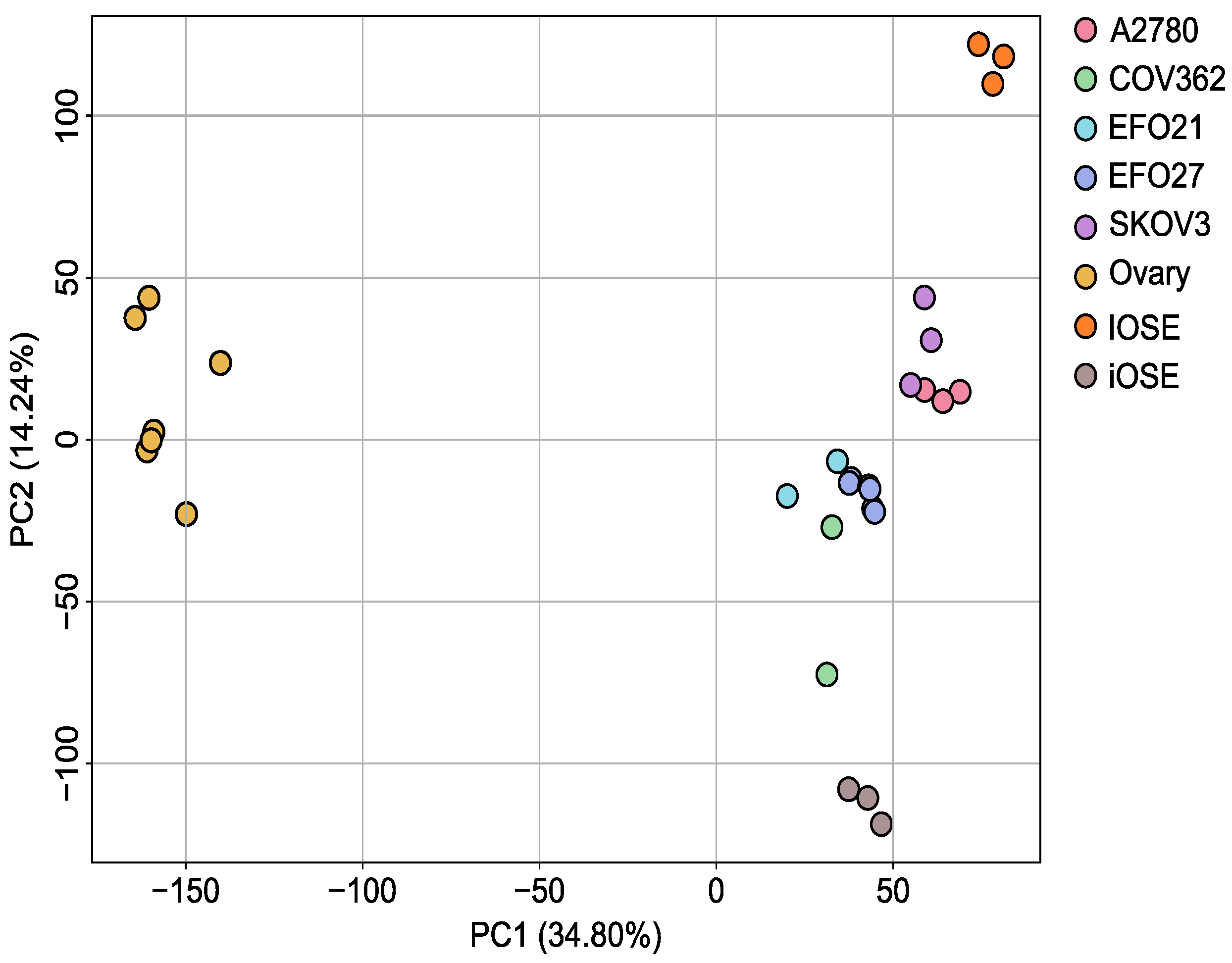
2.7. Transcriptome Analysis of a Set of Ovarian Cancer Cell Lines
- A2780: SRX13261686, SRX13261687, SRX13261688
- SKOV3: SRX25507630, SRX25507631, SRX25507632
- EFO21: SRX7505973, SRX7505974, SRX7505975
- EFO27: SRR7151468, SRR8615287
- COV362: SRR8615949, SRR19537767
- iOSE: iOSE11 (SRR6376807, SRR6376809, SRR6376811); IOSE (SRR27406934, SRR27406935, SRR27406936)
- Ovary: SRX9417156, SRX9417157, SRX9417158, SRX9417159, SRX9417160, SRX9417161, SRX9417162
| Cell Line | Abbrev. | Age | Disease (Cell Type 1) | Tumor Type/Origin | Subtypes */** | Genetic Alteration | Reference |
|---|---|---|---|---|---|---|---|
| A-2780 | A2780 | uk | ovarian endometrioid adenocarcinoma | primary/ovary | ENOC */** | RRAS2; SMARCA4; PIK3CA; MED12 | [44,50,51,52,53] |
| COV362 | COV362 | uk | serous ovarian adenocarcinoma | metastatic/ pleural effusion | HGSOC */** | TP53; BRCA1 | [44,50,52,53,54] |
| EFO-21 | EFO—21 | 56 | serous ovarian cystadenocarcinoma | metastatic/ ascites | CCOC * / SOC ** | TP53 | [44,50,53,55] |
| EFO-27 | EFO27 | 36 | mucinous ovarian adenocarcinoma | metastatic/ abdomen | ENOC * / MOV ** | TP53; ERBB2 | [44,50,55] |
| OAW-42 | OAW42 | 46 | serous ovarian cystadenocarcinoma | metastatic/ ascites | CCOC * / SOC ** | PIK3CA | [44,50,52,53,56] |
| SK-OV-3 | SKOV3 | 64 | serous ovarian cystadenocarcinoma | metastatic/ ascites | CCOC * / SOC ** | EP300; PIK3CA | [44,50,52,53,57] |
| HOSE 17-1 1 | HOSE | adult | 1 ovarian surface epithelial cells | 1 immortalized control cell line | - | HPV E6, E7 transformed | [58] |
| iOSE11 2 | iOSE | uk | ovarian surface epithelial cells | immortalized control cell line | - | TP49/TP52/TP53; TERT; hph (HygR) | [38,49] |
| IOSE 2 | IOSE | uk | ovarian epithelial cells | immortalized control cell line | - | uk | [38] |
| Ovary 2 | Ovary | uk | Ovary | control cell line | - | - | [38] |
| Nucleoside | Abbreviation | Nucleoside | Abbreviation |
|---|---|---|---|
| Adenosine | A | Guanosine | G |
| N6-Acetyladenosine | ac6A | Isoguanosine (ISTD) | ISOG |
| 2′-O-Methyladenosine | Am | 8-Hydroxy-2′-Desoxyguanosine | ho8dG |
| N6-Isopentenyladenosine | i6A | 8-Hydroxyguanosine | ho8G |
| 1-Methyladenosine | m1A | 2′-O-Methylguanosine | Gm |
| 1,2′-O-Dimethyladenosine | m1Am | 1-Methylguanosine | m1G |
| N6,N6-Dimethyladenosine | m6,6A | N2,N2,7-Trimethylguanosine | m2,2,7G |
| N6-Methyladenosine | m6A | N2,N2-Dimethylguanosine | m2,2G |
| N6,2′-O-Dimethyladenosine | m6Am | N2,7-Dimethylguanosine | m2,7G |
| 2-Methylthio-N6-Isopentenyladenosine | ms2i6A | N2-Methylguanosine | m2G |
| N6-Threonylcarbamoyladenosine | t6A | 7-Methylguanosine | m7G |
| S-Adenosylhomocysteine | SAH | Uridine | U |
| S-Adenosylmethionine | SAM | 3-(3-Amino-3-Carboxypropyl)Uridine | acp3U |
| 5′-Methylthioadenosine | MTA | 2′-O-Methyluridine | Um |
| N6-Succinyl Adenosine | N6SAR | 5-Carboxymethyluridine | cm5U |
| Cytidine | C | 5-Methyluridine | m5U |
| N4-Acetylcytidine | ac4C | 5-Methoxycarbonylmethyluridine | mcm5U |
| 2′-O-Methylcytidine | Cm | 5-Methoxyuridine | mo5U |
| 5-Formylcytidine | f5C | 2-Thiouridine | s2U |
| 5-Formyl-2′-O-Methylcytidin | f5Cm | 2-Thio-2′-O-Methyluridine | s2Um |
| 5-Hydroxymethylcytidine | hm5C | 4-Thiouridine | s4U |
| N4,N4-Dimethylcytidine | m4,4C | Pseudouridine | Y |
| 5-Methylcytidine | m5C | 1-Methylpseudouridine | m1Y |
| 5,2′-O-Dimethylcytidine | m5Cm | 3-Methylpseudouridine | m3Y |
| 3-Methylcytidine | m3C | Dihydrouridine | D |
| 2-Thiocytidine | s2C | 5-Hydroxyuridine | ho5U |
| Inosine | I | 5-Methyldihydrouridine | m5D |
| 2′-O-Methylinosine | Im | 5-Methoxycarbonylmethyl-2-Thiouridine | mcm5s2U |
| 1-Methylinosine | m1I | 5-carbamoylmethyluridine | ncm5U |
| Others | |||
| Xanthosine | X | ||
| 5-Aminoimidazole-4-Carboxamide Ribonucleotide | AICAR | ||
| 4-Demethylwyosine | imG-14 |
3. Results and Discussion
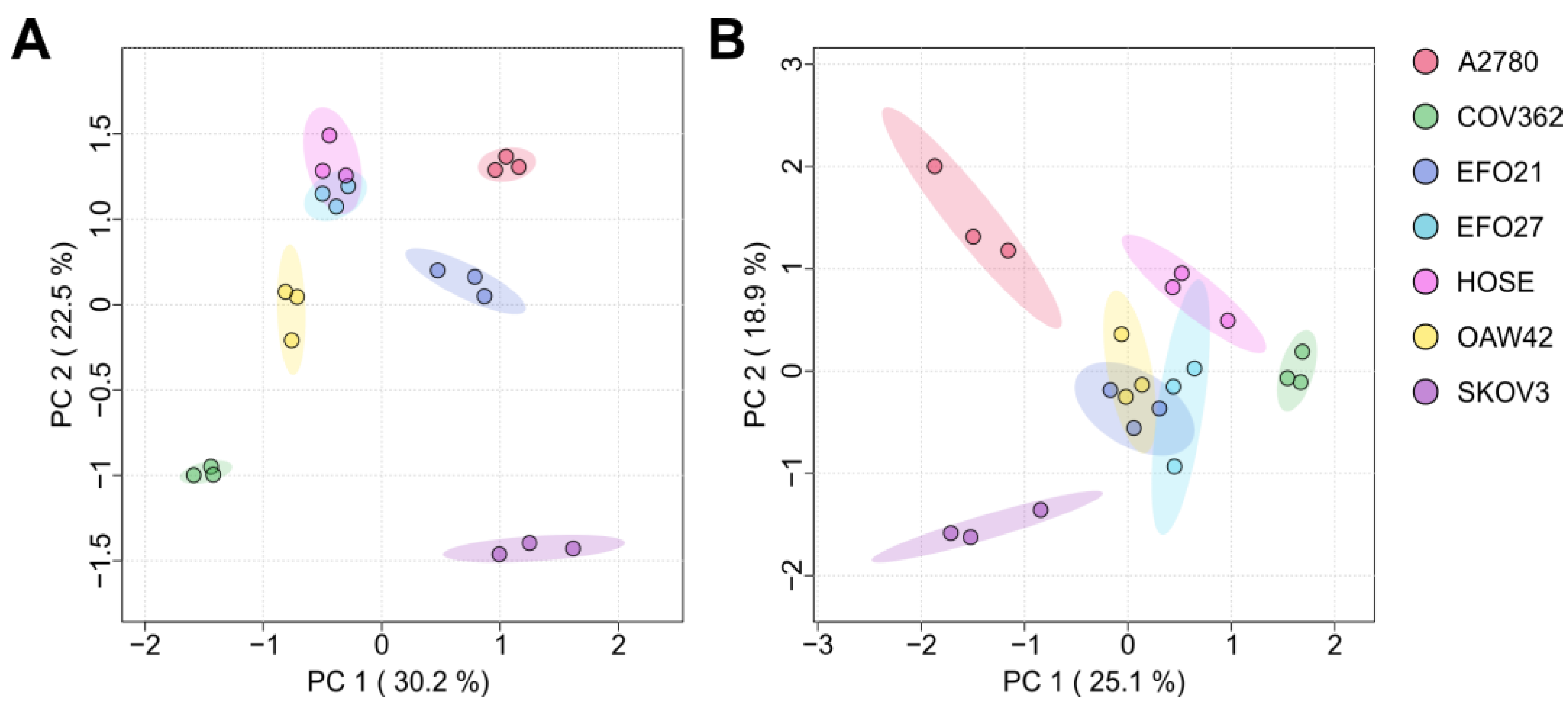
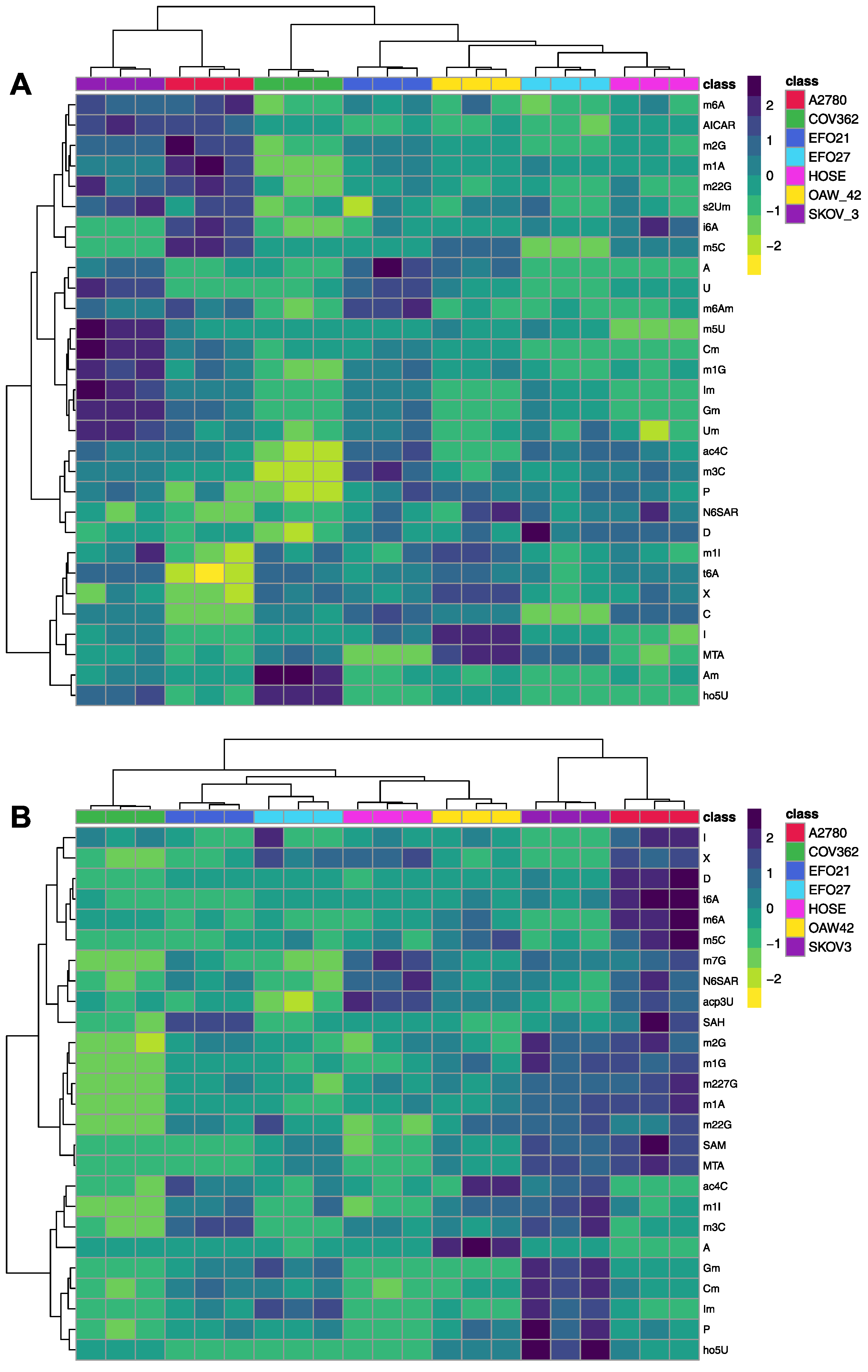
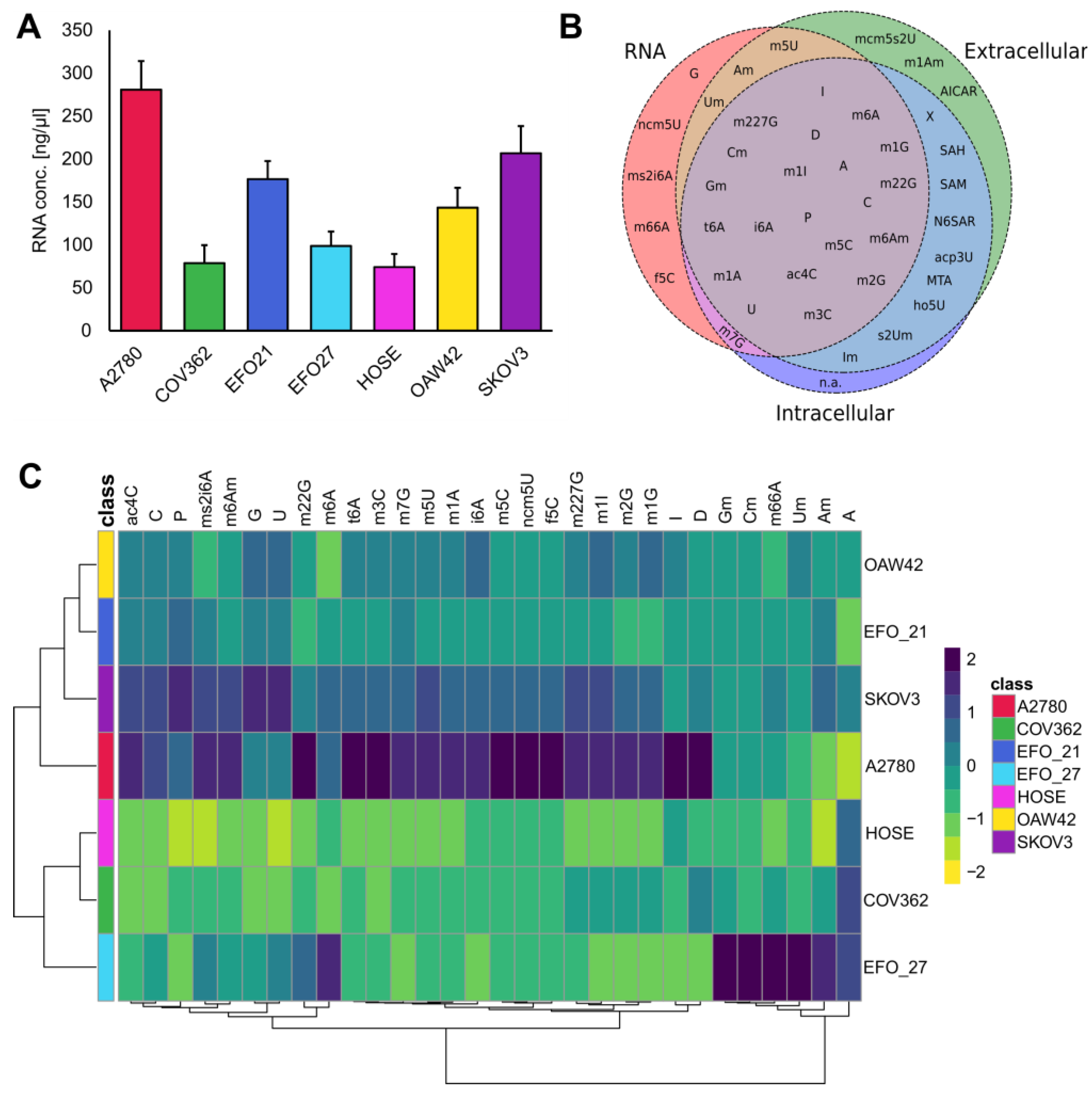
3.1. Exo- and Endo-Metabolomic Analysis of the Cell Culture Medium and Ovarian Cell Lines
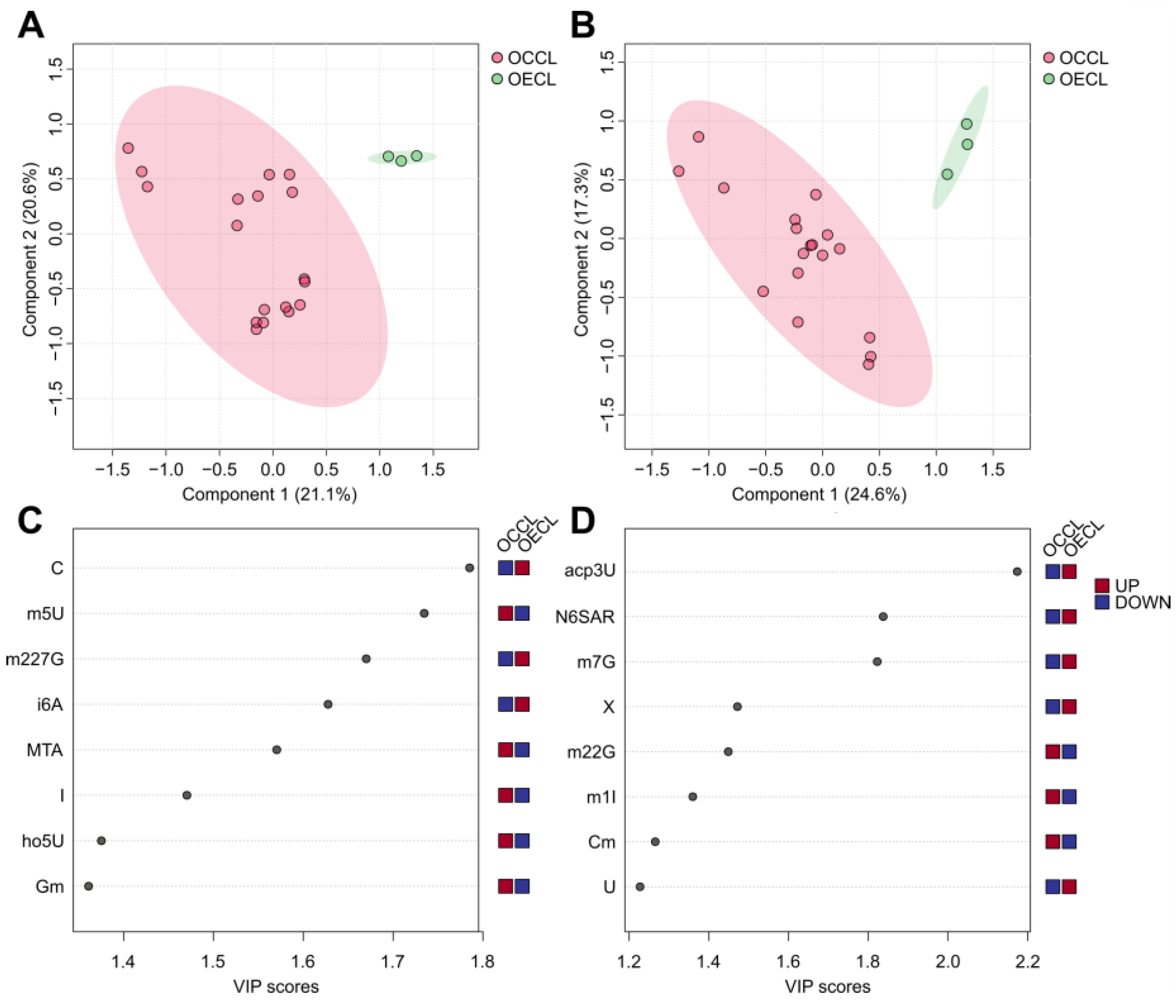
3.2. The Ovarian Cancer Cell Line Versus the Ovarian Epithelial Cell Line: Cancer Versus Control
3.3. Transcriptome Analysis of the Ovarian Cancer Cell Lines Versus Control
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Cancer Facts & Figures. 2024. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2024-cancer-facts-figures.html (accessed on 21 October 2024).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Doubeni, C.A.; Doubeni, A.R.; Myers, A.E. Diagnosis and Management of Ovarian Cancer. Am. Fam. Physician 2016, 93, 937–944. [Google Scholar] [PubMed]
- SEER. U.S. Population Data—SEER Population Data. Available online: https://seer.cancer.gov/popdata/ (accessed on 21 October 2024).
- Menon, U.; Gentry-Maharaj, A.; Hallett, R.; Ryan, A.; Burnell, M.; Sharma, A.; Lewis, S.; Davies, S.; Philpott, S.; Lopes, A.; et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: Results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Lancet Oncol. 2009, 10, 327–340. [Google Scholar] [CrossRef]
- Karam, A.K.; Karlan, B.Y. Ovarian cancer: The duplicity of CA125 measurement. Nat. Rev. Clin. Oncol. 2010, 7, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Bonifácio, V.D. Ovarian Cancer Biomarkers: Moving Forward in Early Detection. In Tumor Microenvironment: The Main Driver of Metabolic Adaptation; Springer: Cham, Switzerland, 2020; Volume 1219, pp. 355–363. [Google Scholar] [CrossRef]
- Zhang, R.; Siu, M.K.Y.; Ngan, H.Y.S.; Chan, K.K.L. Molecular Biomarkers for the Early Detection of Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 12041. [Google Scholar] [CrossRef]
- Tiersma, J.F.; Evers, B.; Bakker, B.M.; Reijngoud, D.J.; de Bruyn, M.; de Jong, S.; Jalving, M. Targeting tumour metabolism in melanoma to enhance response to immune checkpoint inhibition: A balancing act. Cancer Treat. Rev. 2024, 129, 102802. [Google Scholar] [CrossRef]
- Liu, X.; Ren, B.; Ren, J.; Gu, M.; You, L.; Zhao, Y. The significant role of amino acid metabolic reprogramming in cancer. Cell Commun. Signal. 2024, 22, 380. [Google Scholar] [CrossRef]
- Woo, H.M.; Kim, K.M.; Choi, M.H.; Jung, B.H.; Lee, J.; Kong, G.; Nam, S.J.; Kim, S.; Bai, S.W.; Chung, B.C. Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clin. Chim. Acta 2009, 400, 63–69. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, L.; Zhang, X.; Lu, X.; Cao, R.; Xu, C.; Xu, G. Urinary hydrophilic and hydrophobic metabolic profiling based on liquid chromatography-mass spectrometry methods: Differential metabolite discovery specific to ovarian cancer. Electrophoresis 2012, 33, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wu, X.; Ke, C.; Yin, M.; Li, Z.; Fan, L.; Zhang, W.; Zhang, H.; Zhao, F.; Zhou, X.; et al. Identification of Potential Biomarkers for Ovarian Cancer by Urinary Metabolomic Profiling. J. Proteome Res. 2012, 12, 505–512. [Google Scholar] [CrossRef]
- Jiang, T.; Lin, Y.; Yin, H.; Wang, S.; Sun, Q.; Zhang, P.; Bi, W. Correlation analysis of urine metabolites and clinical staging in patients with ovarian cancer. Int. J. Clin. Exp. Med. 2015, 8, 18165–18171. [Google Scholar] [PubMed]
- Monoe, Y.; Miyamoto, S.; Jingushi, K.; Tanimoto, M.; Tanaka, T.; Taniguchi, K.; Komura, K.; Ohmichi, M.; Tsujikawa, K. Hypoxia regulates tumour characteristic RNA modifications in ovarian cancers. FEBS J. 2023, 290, 2085–2096. [Google Scholar] [CrossRef]
- Mohl, D.A.; Lagies, S.; Zodel, K.; Zumkeller, M.; Peighambari, A.; Ganner, A.; Plattner, D.A.; Neumann-Haefelin, E.; Adlesic, M.; Frew, I.J.; et al. Integrated Metabolomic and Transcriptomic Analysis of Modified Nucleosides for Biomarker Discovery in Clear Cell Renal Cell Carcinoma. Cells 2023, 12, 1102. [Google Scholar] [CrossRef]
- Hsu, W.-Y.; Lin, W.-D.; Tsai, Y.; Lin, C.-T.; Wang, H.-C.; Jeng, L.-B.; Lee, C.-C.; Lin, Y.-C.; Lai, C.-C.; Tsai, F.-J. Analysis of urinary nucleosides as potential tumor markers in human breast cancer by high performance liquid chromatography/electrospray ionization tandem mass spectrometry. Clin. Chim. Acta 2011, 412, 1861–1866. [Google Scholar] [CrossRef] [PubMed]
- Willmann, L.; Erbes, T.; Halbach, S.; Brummer, T.; Jäger, M.; Hirschfeld, M.; Fehm, T.; Neubauer, H.; Stickeler, E.; Kammerer, B. Exometabolom analysis of breast cancer cell lines: Metabolic signature. Sci. Rep. 2015, 5, 13374. [Google Scholar] [CrossRef]
- Braun, L.M.; Lagies, S.; Klar, R.F.U.; Hussung, S.; Fritsch, R.M.; Kammerer, B.; Wittel, U.A. Metabolic Profiling of Early and Late Recurrent Pancreatic Ductal Adenocarcinoma Using Patient-Derived Organoid Cultures. Cancers 2020, 12, 1440. [Google Scholar] [CrossRef]
- Cappannini, A.; Ray, A.; Purta, E.; Mukherjee, S.; Boccaletto, P.; Moafinejad, S.N.; Lechner, A.; Barchet, C.; Klaholz, B.P.; Stefaniak, F.; et al. MODOMICS: A database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 2023, 52, D239–D244. [Google Scholar] [CrossRef]
- McCown, P.J.; Ruszkowska, A.; Kunkler, C.N.; Breger, K.; Hulewicz, J.P.; Wang, M.C.; Springer, N.A.; Brown, J.A. Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA 2020, 11, e1595. [Google Scholar] [CrossRef]
- Ogawa, A.; Nagiri, C.; Shihoya, W.; Inoue, A.; Kawakami, K.; Hiratsuka, S.; Aoki, J.; Ito, Y.; Suzuki, T.; Suzuki, T.; et al. N6-methyladenosine (m6A) is an endogenous A3 adenosine receptor ligand. Mol. Cell 2021, 81, 659–674.e7. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.-L.; Fukuda, H.; Chujo, T.; Kouwaki, T.; Oshiumi, H.; Tomizawa, K.; Wei, F.-Y. Export of RNA-derived modified nucleosides by equilibrative nucleoside transporters defines the magnitude of autophagy response and Zika virus replication. RNA Biol. 2021, 18, 478–495. [Google Scholar] [CrossRef] [PubMed]
- Artymowicz, M.; Struck-Lewicka, W.; Wiczling, P.; Markuszewski, M.; Markuszewski, M.J.; Siluk, D. Targeted quantitative metabolomics with a linear mixed-effect model for analysis of urinary nucleosides and deoxynucleosides from bladder cancer patients before and after tumor resection. Anal. Bioanal. Chem. 2023, 415, 5511–5528. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Woo, H.-H.; Chambers, S.K. Human ALKBH3-induced m1A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim. Et Biophys. Acta (BBA)—Gene Regul. Mech. 2019, 1862, 35–46. [Google Scholar] [CrossRef]
- Lorenz, C.; Lünse, C.E.; Mörl, M. tRNA Modifications: Impact on Structure and Thermal Adaptation. Biomolecules 2017, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef]
- Deng, X.; Qing, Y.; Horne, D.; Huang, H.; Chen, J. The roles and implications of RNA m6A modification in cancer. Nat. Rev. Clin. Oncol. 2023, 20, 507–526. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Lin, S.; Kuang, M. RNA modification-mediated mRNA translation regulation in liver cancer: Mechanisms and clinical perspectives. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 267–281. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, Q.; Qin, Y.; Zhong, Q.; Lv, D.; Wu, X.; Fu, P.; Lin, H. Potential Misidentification of Natural Isomers and Mass-Analogs of Modified Nucleosides by Liquid Chromatography–Triple Quadrupole Mass Spectrometry. Genes 2022, 13, 878. [Google Scholar] [CrossRef] [PubMed]
- Kellner, S.; Ochel, A.; Thüring, K.; Spenkuch, F.; Neumann, J.; Sharma, S.; Entian, K.-D.; Schneider, D.; Helm, M. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 2014, 42, e142. [Google Scholar] [CrossRef]
- Haug, S.; Schnerch, D.; Halbach, S.; Mastroianni, J.; Dumit, V.I.; Follo, M.; Hasenburg, A.; Köhler, M.; Dierbach, H.; Herzog, S.; et al. Metadherin exon 11 skipping variant enhances metastatic spread of ovarian cancer. Int. J. Cancer 2015, 136, 2328–2340. [Google Scholar] [CrossRef]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics—FastQC A Quality Control tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 February 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Putri, G.H.; Anders, S.; Pyl, P.T.; Pimanda, J.E.; Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 2022, 38, 2943–2945. [Google Scholar] [CrossRef]
- Muzellec, B.; Teleńczuk, M.; Cabeli, V.; Andreux, M. PyDESeq2: A python package for bulk RNA-seq differential expression analysis. Bioinformatics 2023, 39, btad547. [Google Scholar] [CrossRef]
- McCabe, A.; Zaheed, O.; McDade, S.S.; Dean, K. Investigating the suitability of in vitro cell lines as models for the major subtypes of epithelial ovarian cancer. Front. Cell Dev. Biol. 2023, 11, 1104514. [Google Scholar] [CrossRef]
- DepMap: The Cancer Dependency Map Project at Broad Institute. Available online: https://depmap.org/portal/ (accessed on 16 October 2024).
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576. [Google Scholar] [CrossRef]
- Suehnholz, S.P.; Nissan, M.H.; Zhang, H.; Kundra, R.; Nandakumar, S.; Lu, C.; Carrero, S.; Dhaneshwar, A.; Fernandez, N.; Xu, B.W.; et al. Quantifying the Expanding Landscape of Clinical Actionability for Patients with Cancer. Cancer Discov. 2024, 14, 49–65. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, 63, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef]
- Barnes, B.M.; Nelson, L.; Tighe, A.; Morgan, R.D.; McGrail, J.; Taylor, S.S. Classification of Ovarian Cancer Cell Lines Using Transcriptional Profiles Defines the Five Major Pathological Subtypes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Eva, A.; Robbins, K.C.; Andersen, P.R.; Srinivasan, A.; Tronick, S.R.; Reddy, E.P.; Ellmore, N.W.; Galen, A.T.; Lautenberger, J.A.; Papas, T.S.; et al. Cellular genes analogous to retroviral onc genes are transcribed in human tumour cells. Nature 1982, 295, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Beaufort, C.M.; Helmijr, J.C.A.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van IJcken, W.F.J.; Heine, A.A.J.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef]
- van den Berg-Bakker, C.A.; Hagemeijer, A.; Franken-Postma, E.M.; Smit, V.T.; Kuppen, P.J.; Claasen, H.H.V.R.; Cornelisse, C.J.; Schrier, P.I. Establishment and characterization of 7 ovarian carcinoma cell lines and one granulosa tumor cell line: Growth features and cytogenetics. Int. J. Cancer 1993, 53, 613–620. [Google Scholar] [CrossRef]
- Simon, W.E.; Albrecht, M.; Hänsel, M.; Dietel, M.; Hölzel, F. Cell lines derived from human ovarian carcinomas: Growth stimulation by gonadotropic and steroid hormones. J. Natl. Cancer Inst. 1983, 70, 839–845. [Google Scholar]
- Wilson, A.P.; Dent, M.; Pejovic, T.; Hubbold, L.; Radford, H. Characterisation of seven human ovarian tumour cell lines. Br. J. Cancer 1996, 74, 722–727. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fogh, J. (Ed.) Human Tumor Cells In Vitro; Springer Science+Business Media: New York, NY, USA, 1975; ISBN 978-1-4757-1649-8. [Google Scholar]
- Tsao, S.-W.; Mok, S.C.; Fey, E.G.; Fletcher, J.A.; Wan, T.S.; Chew, E.-C.; Muto, M.G.; Knapp, R.C.; Berkowitz, R.S. Characterization of Human Ovarian Surface Epithelial Cells Immortalized by Human Papilloma Viral Oncogenes (HPV-E6E7 ORFs). Exp. Cell Res. 1995, 218, 499–507. [Google Scholar] [CrossRef]
- Abou Assi, H.; Rangadurai, A.K.; Shi, H.; Liu, B.; Clay, M.C.; Erharter, K.; Kreutz, C.; Holley, C.L.; Al-Hashimi, H.M. 2′-O-Methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucleic Acids Res. 2020, 48, 12365–12379. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y.; Kimura, S.; Suzuki, T. Dual pathways of tRNA hydroxylation ensure efficient translation by expanding decoding capability. Nat. Commun. 2019, 10, 2858. [Google Scholar] [CrossRef]
- Smith, D.A.; Visser, D.W. Studies on 5-Hydroxyuridine. J. Biol. Chem. 1965, 240, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Mazziotta, C.; Rotondo, J.C.; Lanzillotti, C.; Campione, G.; Martini, F.; Tognon, M. Cancer biology and molecular genetics of A3 adenosine receptor. Oncogene 2022, 41, 301–308. [Google Scholar] [CrossRef]
- Bullinger, D.; Neubauer, H.; Fehm, T.; Laufer, S.; Gleiter, C.H.; Kammerer, B. Metabolic signature of breast cancer cell line MCF-7: Profiling of modified nucleosides via LC-IT MS coupling. BMC Biochem. 2007, 8, 25. [Google Scholar] [CrossRef]
- Akyol, S.; Ashrafi, N.; Yilmaz, A.; Turkoglu, O.; Graham, S.F. Metabolomics: An Emerging “Omics” Platform for Systems Biology and Its Implications for Huntington Disease Research. Metabolites 2023, 13, 1203. [Google Scholar] [CrossRef]
- Lakshminarasimhan, R.; Liang, G. The Role of DNA Methylation in Cancer. In DNA Methyltransferases-Role and Function; Springer: Cham, Switzerland, 2016; Volume 945, pp. 151–172. [Google Scholar] [CrossRef]
- Globisch, D.; Pearson, D.; Hienzsch, A.; Brückl, T.; Wagner, M.; Thoma, I.; Thumbs, P.; Reiter, V.; Kneuttinger, A.C.; Müller, M.; et al. Systems-based analysis of modified tRNA bases. Angew. Chem. Int. Ed. 2011, 50, 9739–9742. [Google Scholar] [CrossRef]
- Reiter, V.; Matschkal, D.M.S.; Wagner, M.; Globisch, D.; Kneuttinger, A.C.; Müller, M.; Carell, T. The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic Acids Res. 2012, 40, 6235–6240. [Google Scholar] [CrossRef]
- Jenner, L.B.; Demeshkina, N.; Yusupova, G.; Yusupov, M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 2010, 17, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.-N.; Gai, B.-X.; Liu, L.; Cheng, L. Advances in the investigation of N6-isopentenyl adenosine i6A RNA modification. Bioorg. Med. Chem. 2024, 110, 117838. [Google Scholar] [CrossRef]
- Freund, I.; Buhl, D.K.; Boutin, S.; Kotter, A.; Pichot, F.; Marchand, V.; Vierbuchen, T.; Heine, H.; Motorin, Y.; Helm, M.; et al. 2′-O-methylation within prokaryotic and eukaryotic tRNA inhibits innate immune activation by endosomal Toll-like receptors but does not affect recognition of whole organisms. RNA 2019, 25, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Höfler, S.; Carlomagno, T. Structural and functional roles of 2′-O-ribose methylations and their enzymatic machinery across multiple classes of RNAs. Curr. Opin. Struct. Biol. 2020, 65, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Daignan-Fornier, B.; Pinson, B. 5-Aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5′-Monophosphate (AICAR), a Highly Conserved Purine Intermediate with Multiple Effects. Metabolites 2012, 2, 292–302. [Google Scholar] [CrossRef]
- Višnjić, D.; Lalić, H.; Dembitz, V.; Tomić, B.; Smoljo, T. AICAr, a Widely Used AMPK Activator with Important AMPK-Independent Effects: A Systematic Review. Cells 2021, 10, 1095. [Google Scholar] [CrossRef]
- Noma, A.; Sakaguchi, Y.; Suzuki, T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009, 37, 1335–1352. [Google Scholar] [CrossRef]
- Tsao, S.W.; Wong, N.; Wang, X.; Liu, Y.; Wan, T.S.K.; Fung, L.F.; Lancaster, W.D.; Gregoire, L.; Wong, Y.C. Nonrandom chromosomal imbalances in human ovarian surface epithelial cells immortalized by HPV16-E6E7 viral oncogenes. Cancer Genet. Cytogenet. 2001, 130, 141–149. [Google Scholar] [CrossRef]
- Lapins, M.; Wikberg, J.E. Kinome-wide interaction modelling using alignment-based and alignment-independent approaches for kinase description and linear and non-linear data analysis techniques. BMC Bioinform. 2010, 11, 339. [Google Scholar] [CrossRef]
- Vigano, S.; Alatzoglou, D.; Irving, M.; Ménétrier-Caux, C.; Caux, C.; Romero, P.; Coukos, G. Targeting Adenosine in Cancer Immunotherapy to Enhance T-Cell Function. Front. Immunol. 2019, 10, 925. [Google Scholar] [CrossRef]
- Siddiqui, A.; Ceppi, P. A non-proliferative role of pyrimidine metabolism in cancer. Mol. Metab. 2020, 35, 100962. [Google Scholar] [CrossRef] [PubMed]
- Aria, H.; Rezaei, M.; Nazem, S.; Daraei, A.; Nikfar, G.; Mansoori, B.; Bahmanyar, M.; Tavassoli, A.; Vakil, M.K.; Mansoori, Y. Purinergic receptors are a key bottleneck in tumor metabolic reprogramming: The prime suspect in cancer therapeutic resistance. Front. Immunol. 2022, 13, 947885. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.-M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 2010, 34, 433–443. [Google Scholar] [CrossRef] [PubMed]
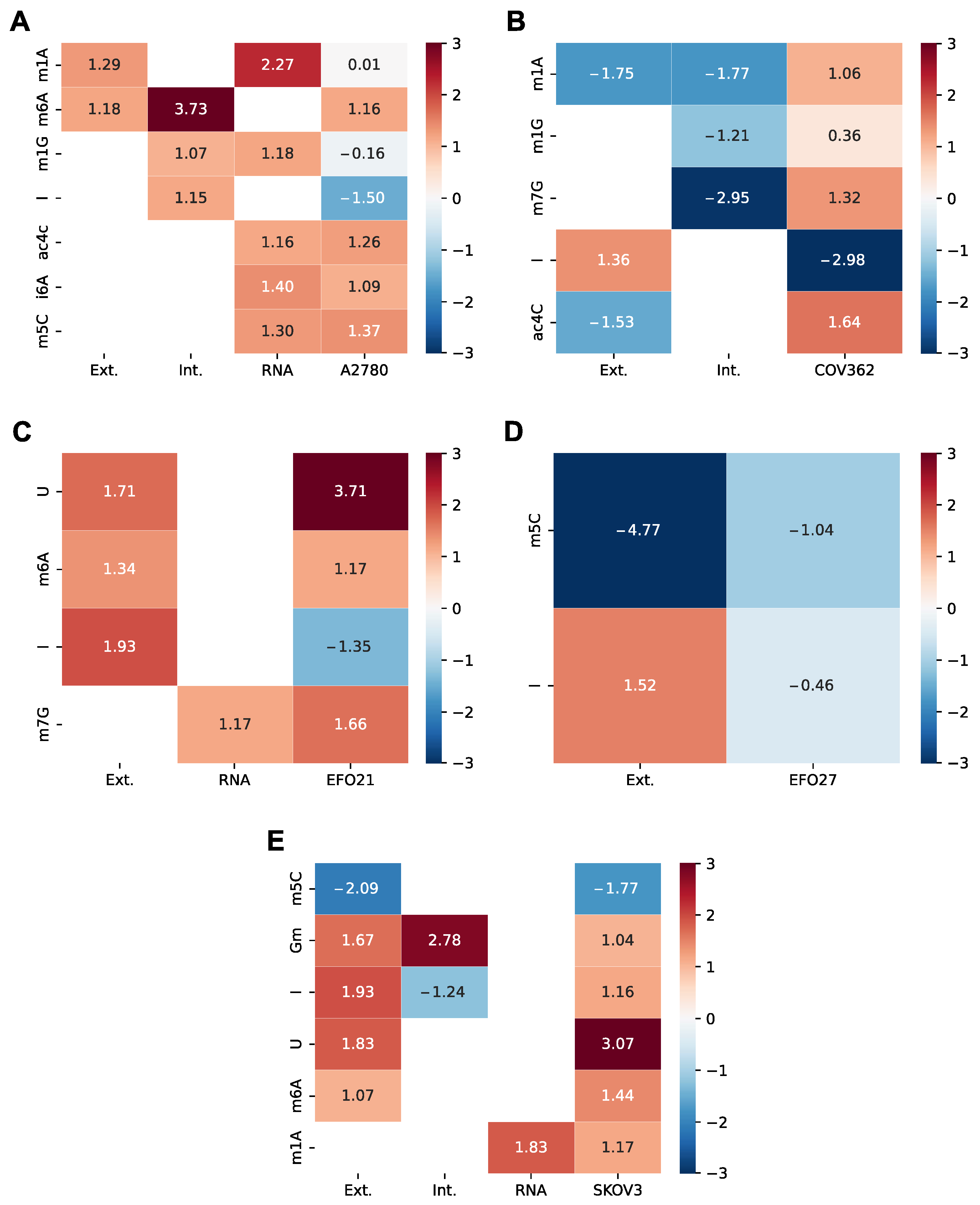
| Enzyme | mNS | Log2FC A2780 | Log2FC COV362 | Log2FC EFO21 | Log2FC EFO27 | Log2FC SKOV3 |
|---|---|---|---|---|---|---|
| ADARB1 | I | −1.50 | −2.91 | −1.27 | −1.32 | |
| ADAT2 | I | 1.04 | ||||
| ADAT3 | I | −3.04 | −1.43 | −1.10 | 1.16 | |
| AICDA | U | 3.71 | 3.07 | |||
| ALKBH1 | hm5C; f5C; f5Cm | 1.29 | ||||
| ALKBH8 | nchm5U; mcm5U | 1.22 | ||||
| APOBEC1 | U | 2.01 | ||||
| CTU1 | mcm5s2U | −2.54 | −2.31 | −1.01 | −2.16 | |
| DIMT1 | m6A | 1.16 | 1.04 | |||
| FTO | hm6A; f6A | −1.11 | −2.64 | −1.07 | ||
| METTL1 | m7G | 1.32 | 1.66 | |||
| MRM1 | Gm | −1.66 | 1.04 | |||
| NAT10 | ac4C | 1.26 | 1.64 | |||
| NSUN2 | m5C | 1.37 | ||||
| PUS1 | Y | 1.62 | 1.31 | |||
| TRDMT1 | m5C | −1.61 | −2.91 | −1.04 | −1.77 | |
| TRMO | m6t6A | 1.01 | 1.02 | |||
| TRMT10A | m1G | 1.08 | 2.17 | |||
| TRMT10B | m1G | −1.39 | −2.17 | −1.31 | −1.50 | |
| TRMT10C | m1G; m1A | 1.22 | 1.06 | 1.03 | ||
| TRMT112 | m6A; m2G; mcm5U | 1.52 | 1.17 | 1.44 | ||
| TRMT61A | m1A | −1.21 | 1.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohl, D.A.; Lagies, S.; Lonzer, A.; Pfäffle, S.P.; Groß, P.; Benka, M.; Jäger, M.; Huber, M.C.; Günther, S.; Plattner, D.A.; et al. On the Quest for Biomarkers: A Comprehensive Analysis of Modified Nucleosides in Ovarian Cancer Cell Lines. Cells 2025, 14, 626. https://doi.org/10.3390/cells14090626
Mohl DA, Lagies S, Lonzer A, Pfäffle SP, Groß P, Benka M, Jäger M, Huber MC, Günther S, Plattner DA, et al. On the Quest for Biomarkers: A Comprehensive Analysis of Modified Nucleosides in Ovarian Cancer Cell Lines. Cells. 2025; 14(9):626. https://doi.org/10.3390/cells14090626
Chicago/Turabian StyleMohl, Daniel A., Simon Lagies, Alexander Lonzer, Simon P. Pfäffle, Philipp Groß, Moritz Benka, Markus Jäger, Matthias C. Huber, Stefan Günther, Dietmar A. Plattner, and et al. 2025. "On the Quest for Biomarkers: A Comprehensive Analysis of Modified Nucleosides in Ovarian Cancer Cell Lines" Cells 14, no. 9: 626. https://doi.org/10.3390/cells14090626
APA StyleMohl, D. A., Lagies, S., Lonzer, A., Pfäffle, S. P., Groß, P., Benka, M., Jäger, M., Huber, M. C., Günther, S., Plattner, D. A., Juhasz-Böss, I., Backhaus, C., & Kammerer, B. (2025). On the Quest for Biomarkers: A Comprehensive Analysis of Modified Nucleosides in Ovarian Cancer Cell Lines. Cells, 14(9), 626. https://doi.org/10.3390/cells14090626







