Highly Efficient Site-Specific and Cassette Mutagenesis of Plasmids Harboring GC-Rich Sequences
Highlights
- This study reports that P3 and P3a site-directed mutagenesis methods are inefficient with plasmids possessing extremely GC-rich sequences, perhaps due to guanine (G)-quadruplex formation.
- To overcome this problem, we have developed P3b site-specific mutagenesis, which is highly efficient for plasmids with GC-rich regions, as demonstrated for the expression vectors of KAT2B, CDK13 and Cas9.
- Before embarking on site-directed mutagenesis experiments, it is recommended to analyze the GC-content distribution of template plasmids to select P3a or P3b mutagenesis.
- These two methods complement each other and provide a versatile approach for single-site, multisite or cassette mutagenesis, thereby offering two reliable tools for protein, RNA and plasmid engineering.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. GC-Content Calculation
2.3. Heat or Alkali Denaturation of a KAT2B Expression Plasmid
2.4. Isolation of Uracil-Containing Plasmids
2.5. Mutagenic Primers
2.6. P3 Site-Directed Mutagenesis
2.7. P3a Site-Directed Mutagenesis
2.8. P3b Site-Directed Mutagenesis
2.9. Plasmid Sequencing and Analysis
2.10. Statistics
3. Results
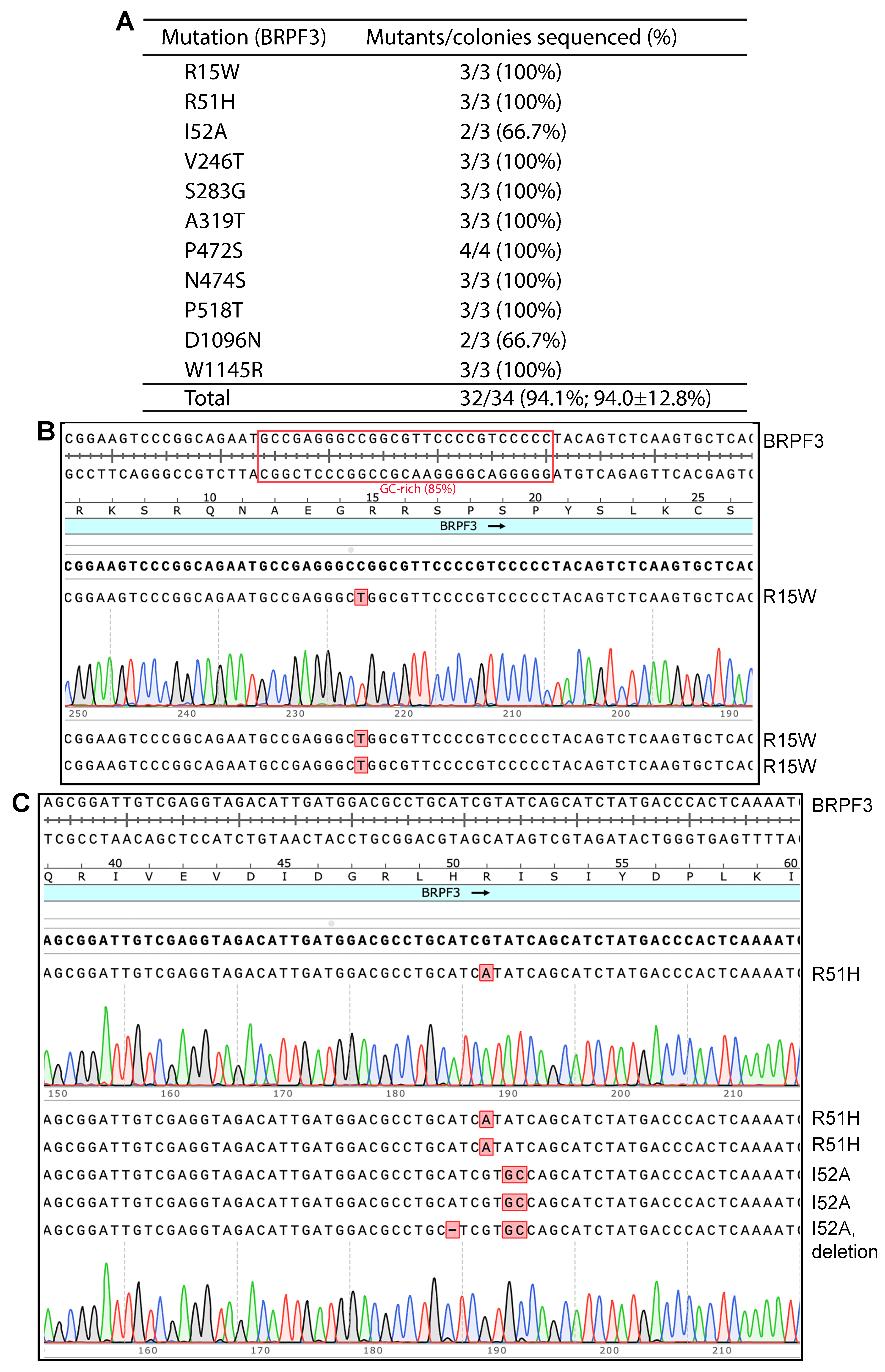
3.1. P3a Mutagenesis for Engineering Point and Deletion Mutants of BRPF3
3.2. Use of Uracil-Containing Templates in P3a Mutagenesis
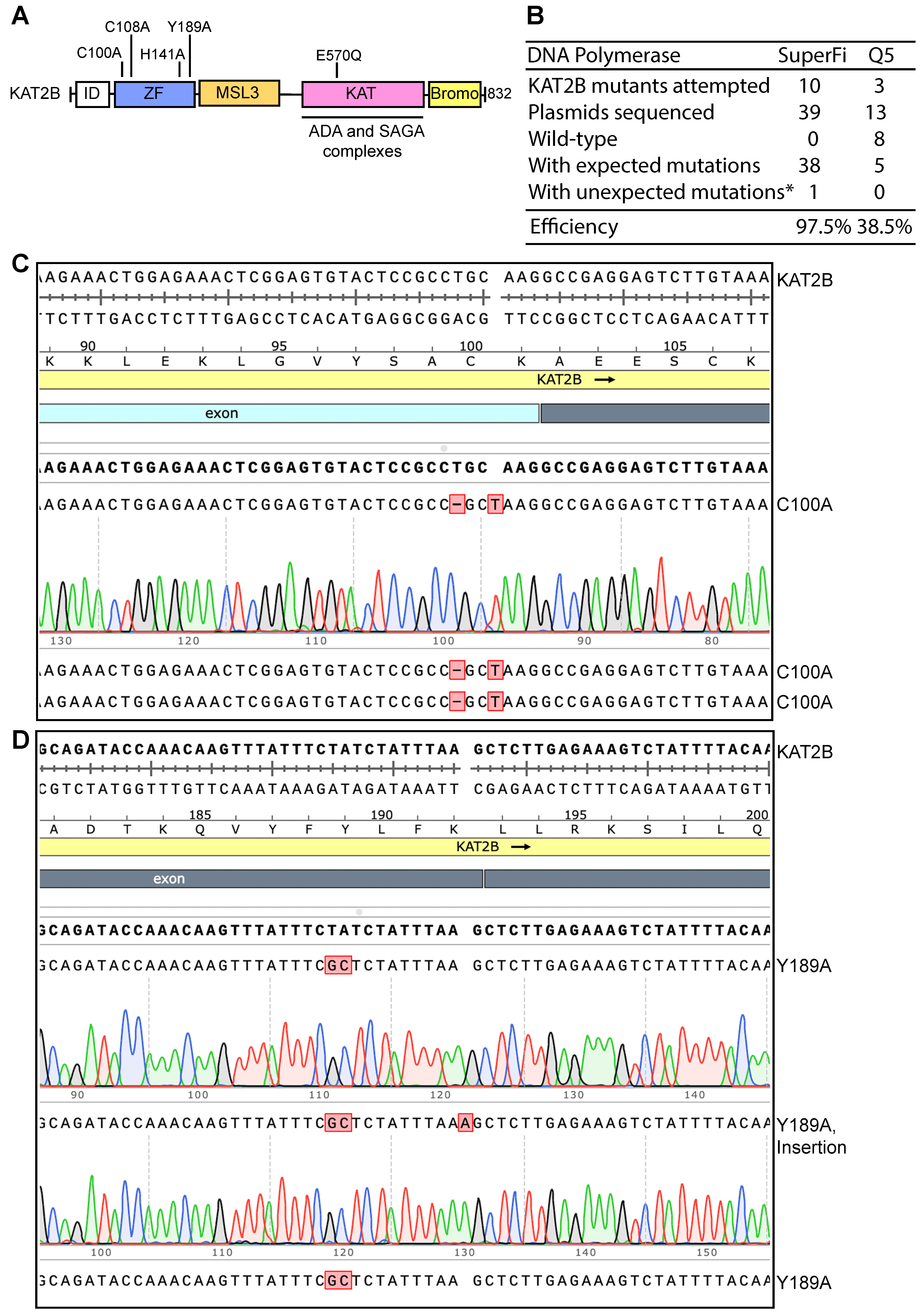
3.3. Developing P3b Mutagenesis for Efficient Generation of KAT2B Mutants
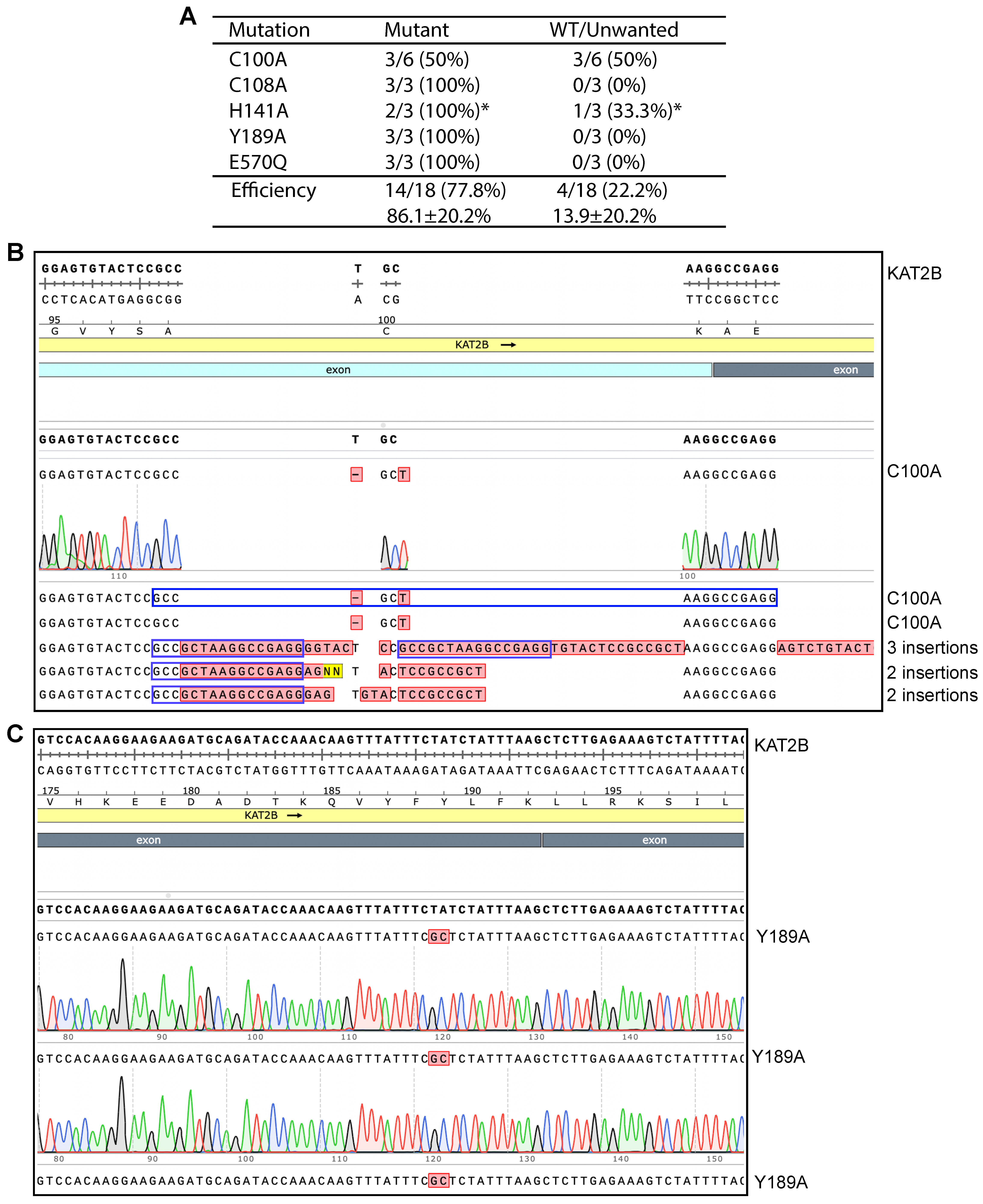
3.4. P3b Mutagenesis for Engineering Different HAT Mutants
3.5. P3a and P3b Mutagenesis Methods to Generate SARS-CoV-2 Spike Mutants at Multiple Sites
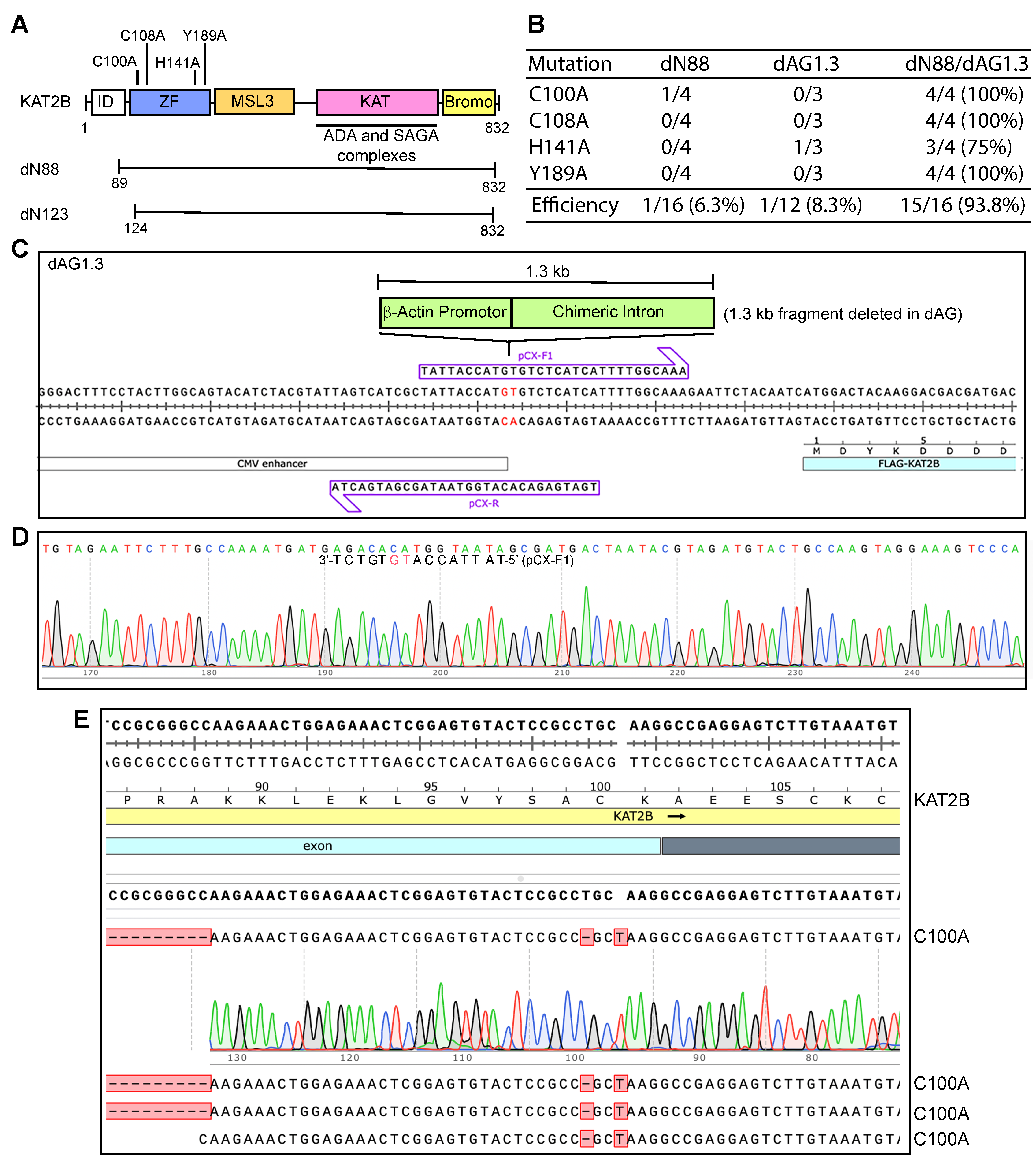
3.6. Two GC-Rich Fragments Render the KAT2B Plasmid Incompatible with P3a Mutagenesis
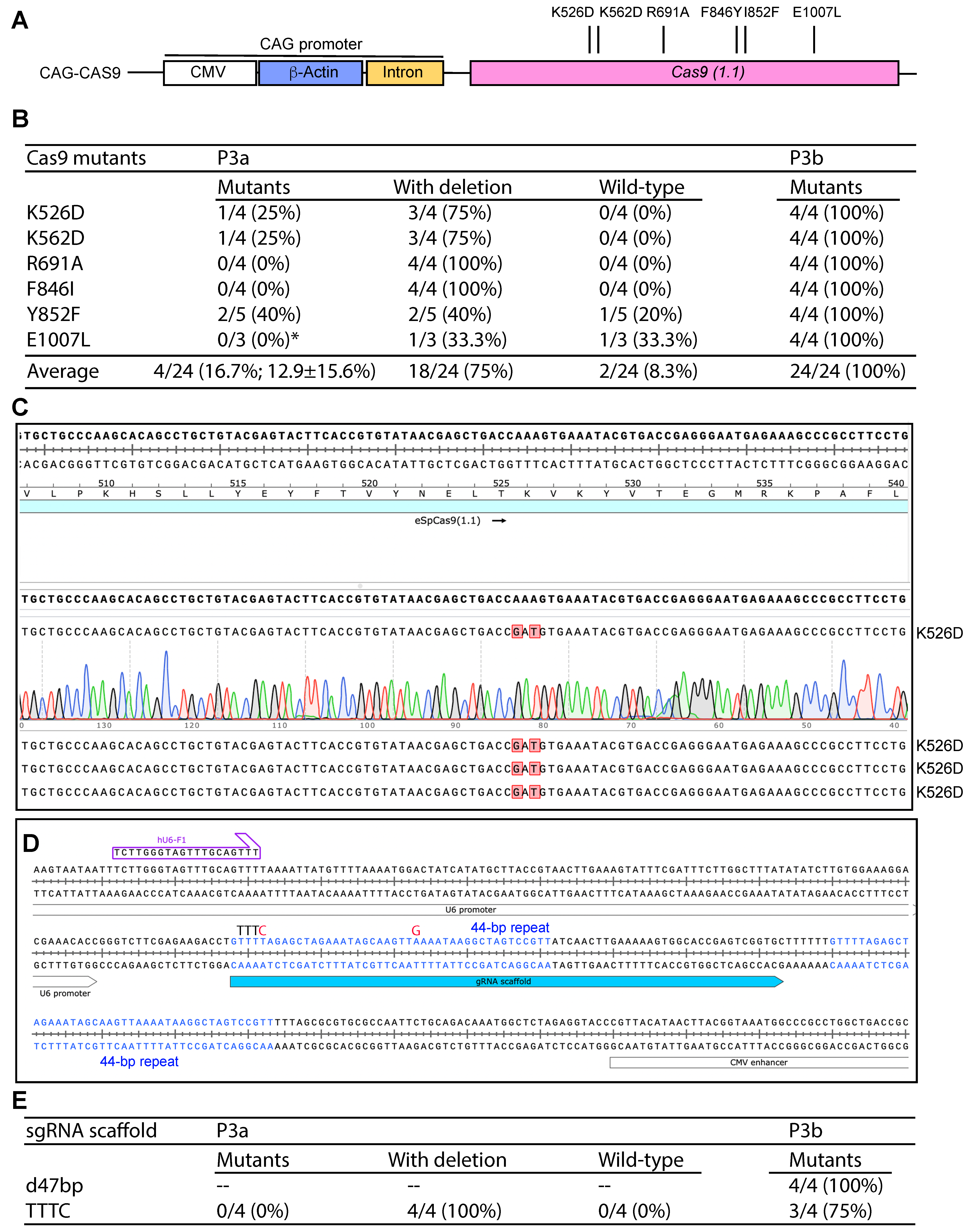
3.7. P3b Mutagenesis of sgRNA/Cas9 Expression Plasmids with the CAG Promoter
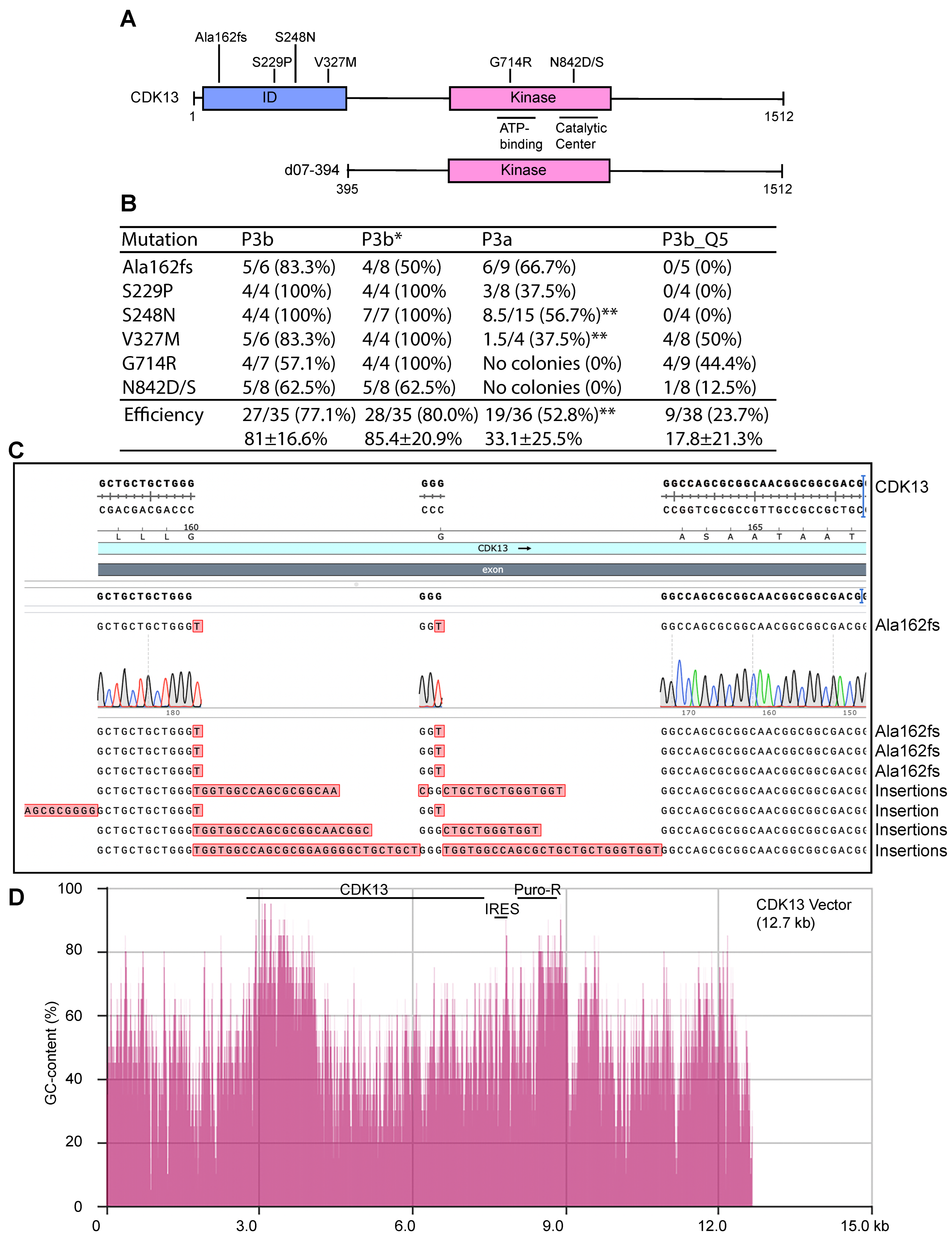
3.8. P3b Mutagenesis of a CDK13 Expression Plasmid with Highly GC-Rich Sequences
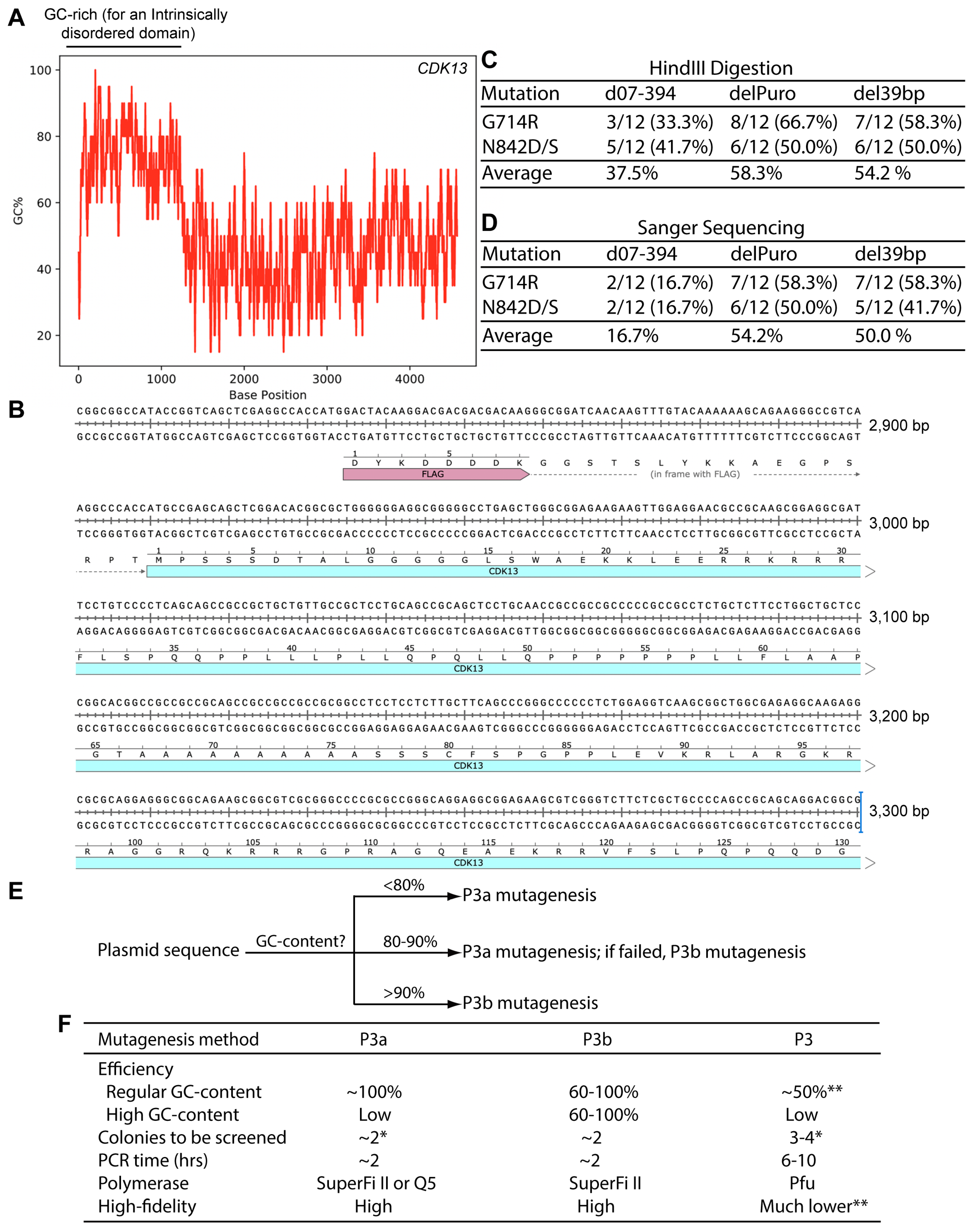
3.9. GC-Rich Sequences Make the CDK13 Plasmid Incompatible with P3a Mutagenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hutchison, C.A., 3rd; Phillips, S.; Edgell, M.H.; Gillam, S.; Jahnke, P.; Smith, M. Mutagenesis at a specific position in a DNA sequence. J. Biol. Chem. 1978, 253, 6551–6560. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.P.; Costa, G.L.; Schoettlin, W.; Cline, J.; Mathur, E.; Bauer, J.C. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 1994, 151, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.L.; Pei, G.K. Modification of a PCR-based site-directed mutagenesis method. Biotechniques 1997, 23, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wilkinson, M.F. Site-directed mutagenesis: A two-step method using PCR and DpnI. Biotechniques 1997, 23, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Baumann, U.; Reymond, J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004, 32, e115. [Google Scholar] [CrossRef]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef]
- Qi, D.; Scholthof, K.B. A one-step PCR-based method for rapid and efficient site-directed fragment deletion, insertion, and substitution mutagenesis. J. Virol. Methods 2008, 149, 85–90. [Google Scholar] [CrossRef]
- Mousavi, N.; Zhou, E.; Razavi, A.; Ebrahimi, E.; Varela-Castillo, P.; Yang, X.J. P3 site-directed mutagenesis: An efficient method based on primer pairs with 3’-overhangs. J. Biol. Chem. 2025, 301, 108219. [Google Scholar] [CrossRef]
- Mousavi, N.; Zhou, E.; Razavi, A.; Ebrahimi, E.; Varela-Castillo, P.; Yang, X.J. Efficient site-directed mutagenesis mediated by primer pairs with 3’-overhangs. Curr. Protoc. Protein Sci. 2025, 5, e70104. [Google Scholar] [CrossRef]
- Yang, X.J. P3a site-specific and cassette mutagenesis for seamless protein, RNA and plasmid engineering. Genes Cancer 2025, 16, 34–60. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Batra, S.; Allwein, B.; Kumar, C.; Devbhandari, S.; Bruning, J.G.; Bahng, S.; Lee, C.M.; Marians, K.J.; Hite, R.K.; Remus, D. G-quadruplex-stalled eukaryotic replisome structure reveals helical inchworm DNA translocation. Science 2025, 387, eadt1978. [Google Scholar] [CrossRef] [PubMed]
- Insco, M.L.; Abraham, B.J.; Dubbury, S.J.; Kaltheuner, I.H.; Dust, S.; Wu, C.; Chen, K.Y.; Liu, D.; Bellaousov, S.; Cox, A.M.; et al. Oncogenic CDK13 mutations impede nuclear RNA surveillance. Science 2023, 380, eabn7625. [Google Scholar] [CrossRef] [PubMed]
- van den Akker, W.M.R.; Brummelman, I.; Martis, L.M.; Timmermans, R.N.; Pfundt, R.; Kleefstra, T.; Willemsen, M.H.; Gerkes, E.H.; Herkert, J.C.; van Essen, A.J.; et al. De novo variants in CDK13 associated with syndromic ID/DD: Molecular and clinical delineation of 15 individuals and a further review. Clin. Genet. 2018, 93, 1000–1007. [Google Scholar] [CrossRef]
- Thean, D.G.L.; Chu, H.Y.; Fong, J.H.C.; Chan, B.K.C.; Zhou, P.; Kwok, C.C.S.; Chan, Y.M.; Mak, S.Y.L.; Choi, G.C.G.; Ho, J.W.K.; et al. Machine learning-coupled combinatorial mutagenesis enables resource-efficient engineering of CRISPR-Cas9 genome editor activities. Nat. Commun. 2022, 13, 2219. [Google Scholar] [CrossRef]
- Starr, T.N.; Czudnochowski, N.; Liu, Z.; Zatta, F.; Park, Y.J.; Addetia, A.; Pinto, D.; Beltramello, M.; Hernandez, P.; Greaney, A.J.; et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 2021, 597, 97–102. [Google Scholar] [CrossRef]
- Lisanza, S.L.; Gershon, J.M.; Tipps, S.W.K.; Sims, J.N.; Arnoldt, L.; Hendel, S.J.; Simma, M.K.; Liu, G.; Yase, M.; Wu, H.; et al. Multistate and functional protein design using RoseTTAFold sequence space diffusion. Nat. Biotechnol. 2025, 43, 1288–1298. [Google Scholar] [CrossRef]
- Kortemme, T. De novo protein design—From new structures to programmable functions. Cell 2024, 187, 526–544. [Google Scholar] [CrossRef]
- Wasdin, P.T.; Johnson, N.V.; Janke, A.K.; Held, S.; Marinov, T.M.; Jordaan, G.; Gillespie, R.A.; Vandenabeele, L.; Pantouli, F.; Powers, O.C.; et al. Generation of antigen-specific paired-chain antibodies using large language models. Cell 2025, 188, 1–16. [Google Scholar] [CrossRef]
- Ullah, M.; Pelletier, N.; Xiao, L.; Zhao, S.P.; Wang, K.; Degerny, C.; Tahmasebi, S.; Cayrou, C.; Doyon, Y.; Goh, S.L.; et al. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell. Biol. 2008, 28, 6828–6843. [Google Scholar] [CrossRef]
- Yan, K.; You, L.; Degerny, C.; Ghorbani, M.; Liu, X.; Chen, L.; Li, L.; Miao, D.; Yang, X.J. The chromatin regulator BRPF3 preferentially activates the HBO1 acetyltransferase but is dispensable for mouse development and survival. J. Biol. Chem. 2016, 291, 2647–2663. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Rousseau, J.; Machol, K.; Cross, L.A.; Agre, K.E.; Gibson, C.F.; Goverde, A.; Engleman, K.L.; Verdin, H.; Baere, E.; et al. Deficient histone H3 propionylation by BRPF1-KAT6 complexes in neurodevelopmental disorders and cancer. Sci. Adv. 2020, 6, eaax0021. [Google Scholar] [CrossRef] [PubMed]
- Kitabayashi, I.; Aikawa, Y.; Nguyen, L.A.; Yokoyama, A.; Ohki, M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J. 2001, 20, 7184–7196. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Ogryzko, V.V.; Nishikawa, J.; Howard, B.H.; Nakatani, Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 1996, 382, 319–324. [Google Scholar] [CrossRef]
- Miyazaki, J.; Takaki, S.; Araki, K.; Tashiro, F.; Tominaga, A.; Takatsu, K.; Yamamura, K. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene 1989, 79, 269–277. [Google Scholar]
- Martsen, E.O.; Zeng, M.; Lapeyre, J.N. Fast plasmid DNA sequencing using a thermal cycler and high temperature alkali denaturation. Biotechniques 1997, 22, 420–422. [Google Scholar] [CrossRef]
- Tao, Y.; Zhong, C.; Zhu, J.; Xu, S.; Ding, J. Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2. Nucleic Acids Res. 2017, 45, 5707–5719. [Google Scholar] [CrossRef]
- Kunkel, T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 1985, 82, 488–492. [Google Scholar] [CrossRef]
- McClary, J.A.; Witney, F.; Geisselsoder, J. Efficient site-directed in vitro mutagenesis using phagemid vectors. Biotechniques 1989, 7, 282–289. [Google Scholar]
- Yang, X.J.; Kaufman, S. High-level expression and deletion mutagenesis of human tryptophan hydroxylase. Proc. Natl. Acad. Sci. USA 1994, 91, 6659–6663. [Google Scholar] [CrossRef]
- Trower, M.K. Site-directed mutagenesis using a uracil-containing phagemid template. Methods Mol. Biol. 1994, 31, 67–77. [Google Scholar]
- Li, F.; Liu, S.L.; Mullins, J.I. Site-directed mutagenesis using uracil-containing double-stranded DNA templates and DpnI digestion. Biotechniques 1999, 27, 734–738. [Google Scholar] [CrossRef]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L.; et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis. 2024, 24, e70–e72. [Google Scholar] [CrossRef]
- Yang, H.; Guo, H.; Wang, A.; Cao, L.; Fan, Q.; Jiang, J.; Wang, M.; Lin, L.; Ge, X.; Wang, H.; et al. Structural basis for the evolution and antibody evasion of SARS-CoV-2 BA.2.86 and JN.1 subvariants. Nat. Commun. 2024, 15, 7715. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef]
- Fischer, V.; Plassard, D.; Ye, T.; Reina-San-Martin, B.; Stierle, M.; Tora, L.; Devys, D. The related coactivator complexes SAGA and ATAC control embryonic stem cell self-renewal through acetyltransferase-independent mechanisms. Cell Rep. 2021, 36, 109598. [Google Scholar] [CrossRef] [PubMed]
- Albaugh, B.N.; Denu, J.M. Catalysis by protein acetyltransferase Gcn5. Biochim. Biophys. Acta Gene Regul Mech. 2021, 1864, 194627. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Simonsson, T.; Postberg, J.; Rhodes, D.; Lipps, H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005, 12, 847–854. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Agudelo, D.; Duringer, A.; Bozoyan, L.; Huard, C.C.; Carter, S.; Loehr, J.; Synodinou, D.; Drouin, M.; Salsman, J.; Dellaire, G.; et al. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat. Methods 2017, 14, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Pedrazzoli, E.; Bianchi, A.; Umbach, A.; Amistadi, S.; Brusson, M.; Frati, G.; Ciciani, M.; Badowska, K.A.; Arosio, D.; Miccio, A.; et al. An optimized SpCas9 high-fidelity variant for direct protein delivery. Mol. Ther. 2023, 31, 2257–2265. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.D.; Gandadireja, A.P.; Rossetti, G.; Siira, S.J.; Mantegna, J.L.; Filipovska, A.; Rackham, O. Mutational rescue of the activity of high-fidelity Cas9 enzymes. Cell Rep. Methods 2024, 4, 100756. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.D.; Rossetti, G.; Mantegna, J.L.; Siira, S.J.; Gandadireja, A.P.; Bruce, M.; Raven, S.A.; Khersonsky, O.; Fleishman, S.J.; Filipovska, A.; et al. Computationally designed hyperactive Cas9 enzymes. Nat. Commun. 2022, 13, 3023. [Google Scholar] [CrossRef]
- Kim, Y.H.; Kim, N.; Okafor, I.; Choi, S.; Min, S.; Lee, J.; Bae, S.M.; Choi, K.; Choi, J.; Harihar, V.; et al. Sniper2L is a high-fidelity Cas9 variant with high activity. Nat. Chem. Biol. 2023, 19, 972–980. [Google Scholar] [CrossRef]
- Chey, Y.C.J.; Gierus, L.; Lushington, C.; Arudkumar, J.C.; Geiger, A.B.; Staker, L.G.; Robertson, L.J.; Pfitzner, C.; Kennedy, J.G.; Lee, R.H.B.; et al. Optimal SpCas9- and SaCas9-mediated gene editing by enhancing gRNA transcript levels through scaffold poly-T tract reduction. BMC Genom. 2025, 26, 138. [Google Scholar] [CrossRef]
- Sifrim, A.; Hitz, M.P.; Wilsdon, A.; Breckpot, J.; Turki, S.H.; Thienpont, B.; McRae, J.; Fitzgerald, T.W.; Singh, T.; Swaminathan, G.J.; et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat. Genet. 2016, 48, 1060–1065. [Google Scholar] [CrossRef]
- Marsolier-Kergoat, M.C.; Yeramian, E. GC content and recombination: Reassessing the causal effects for the Saccharomyces cerevisiae genome. Genetics 2009, 183, 31–38. [Google Scholar] [CrossRef]
- Kiktev, D.A.; Dominska, M.; Zhang, T.; Dahl, J.; Stepchenkova, E.I.; Mieczkowski, P.; Burgers, P.M.; Lujan, S.; Burkholder, A.; Kunkel, T.A.; et al. The fidelity of DNA replication, particularly on GC-rich templates, is reduced by defects of the Fe-S cluster in DNA polymerase delta. Nucleic Acids Res. 2021, 49, 5623–5636. [Google Scholar] [CrossRef]
- Robinson, J.; Raguseo, F.; Nuccio, S.P.; Liano, D.; Di Antonio, M. DNA G-quadruplex structures: More than simple roadblocks to transcription? Nucleic Acids Res. 2021, 49, 8419–8431. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Kang, Y.M.; Korfmann, C.; Pouyet, F.; Eckford, A.; Palazzo, A.F. The GC-content at the 5’ ends of human protein-coding genes is undergoing mutational decay. Genome Biol. 2024, 25, 219. [Google Scholar] [CrossRef] [PubMed]
- Hanecak, R.; Pattengale, P.K.; Fan, H. Deletion of a GC-rich region flanking the enhancer element within the long terminal repeat sequences alters the disease specificity of Moloney murine leukemia virus. J. Virol. 1991, 65, 5357–5363. [Google Scholar] [CrossRef] [PubMed]
- Schroder, C.; Horsthemke, B.; Depienne, C. GC-rich repeat expansions: Associated disorders and mechanisms. Med. Genet. 2021, 33, 325–335. [Google Scholar] [CrossRef]
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Breevoort, S.; Gibson, S.; Figueroa, K.; Bromberg, M.; Pulst, S. Expanding Clinical Spectrum of C9ORF72-Related Disorders and Promising Therapeutic Strategies: A Review. Neurol. Genet. 2022, 8, e670. [Google Scholar] [CrossRef]
- Trollet, C.; Anvar, S.Y.; Venema, A.; Hargreaves, I.P.; Foster, K.; Vignaud, A.; Ferry, A.; Negroni, E.; Hourde, C.; Baraibar, M.A.; et al. Molecular and phenotypic characterization of a mouse model of oculopharyngeal muscular dystrophy reveals severe muscular atrophy restricted to fast glycolytic fibres. Hum. Mol. Genet. 2010, 19, 2191–2207. [Google Scholar] [CrossRef]
- Verkerk, A.J.; Pieretti, M.; Sutcliffe, J.S.; Fu, Y.H.; Kuhl, D.P.; Pizzuti, A.; Reiner, O.; Richards, S.; Victoria, M.F.; Zhang, F.P.; et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 1991, 65, 905–914. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varela-Castillo, P.; Zhou, E.; Razavi, A.; Ebrahimi, E.; Yang, X.-J. Highly Efficient Site-Specific and Cassette Mutagenesis of Plasmids Harboring GC-Rich Sequences. Cells 2025, 14, 2016. https://doi.org/10.3390/cells14242016
Varela-Castillo P, Zhou E, Razavi A, Ebrahimi E, Yang X-J. Highly Efficient Site-Specific and Cassette Mutagenesis of Plasmids Harboring GC-Rich Sequences. Cells. 2025; 14(24):2016. https://doi.org/10.3390/cells14242016
Chicago/Turabian StyleVarela-Castillo, Paulina, Ethan Zhou, Arezousadat Razavi, Elham Ebrahimi, and Xiang-Jiao Yang. 2025. "Highly Efficient Site-Specific and Cassette Mutagenesis of Plasmids Harboring GC-Rich Sequences" Cells 14, no. 24: 2016. https://doi.org/10.3390/cells14242016
APA StyleVarela-Castillo, P., Zhou, E., Razavi, A., Ebrahimi, E., & Yang, X.-J. (2025). Highly Efficient Site-Specific and Cassette Mutagenesis of Plasmids Harboring GC-Rich Sequences. Cells, 14(24), 2016. https://doi.org/10.3390/cells14242016







