Sibling-Derived Cell Lines of Whole Larval Siberian Sturgeon as an In Vitro Model System for Studying Inter-Individual Differences Within the Same Genomic Heritage
Highlights
- Five sibling-derived cell lines were successfully generated and maintained for long-term culture, cryopreservation, and transfer.
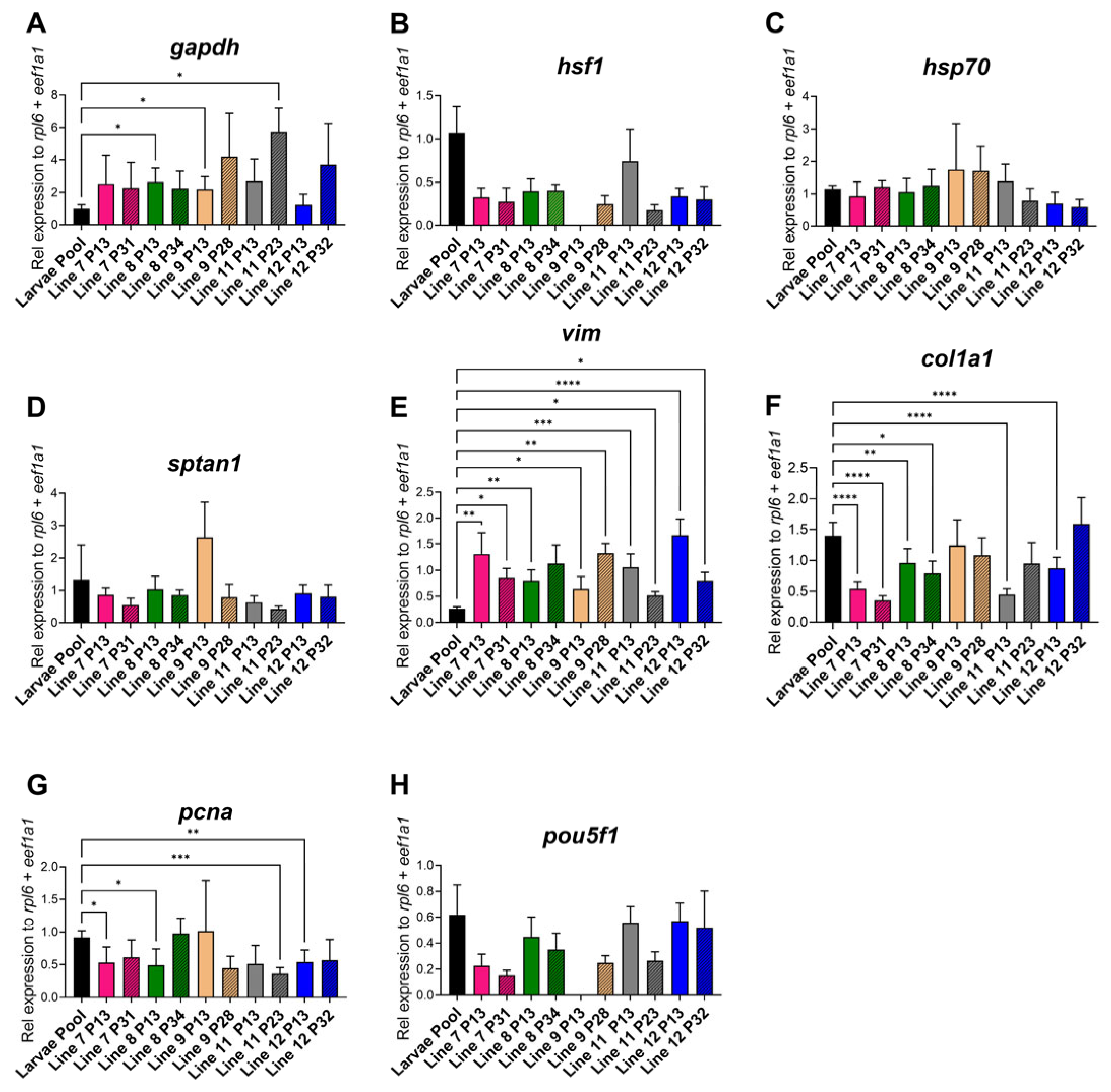
- The lines exhibited biological variation in cell morphology, mitochondrial activity, and extracellular acidification rates while the gene expression profiles closely resembled those of whole larvae with in vitro adaptations observed for gapdh, vim, col1a1, and pcna.
- First establishment of larval cell lines from the critically endangered species Siberian sturgeon (Acipenser baerii).
- These new larval cell lines provide a powerful in vitro model to explore developmental biology and stress responses in endangered sturgeons.
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Isolation
2.3. Cell Line Analysis
2.4. Analysis of Metabolic Function
2.5. Mitochondrial Membrane Potential (MMP) Analysis
2.6. Analysis of Gene Expression
2.6.1. Selection of Genes and Primer Design
2.6.2. RNA Extraction and Fluidigm PCR
2.7. Statistical Analysis
3. Results
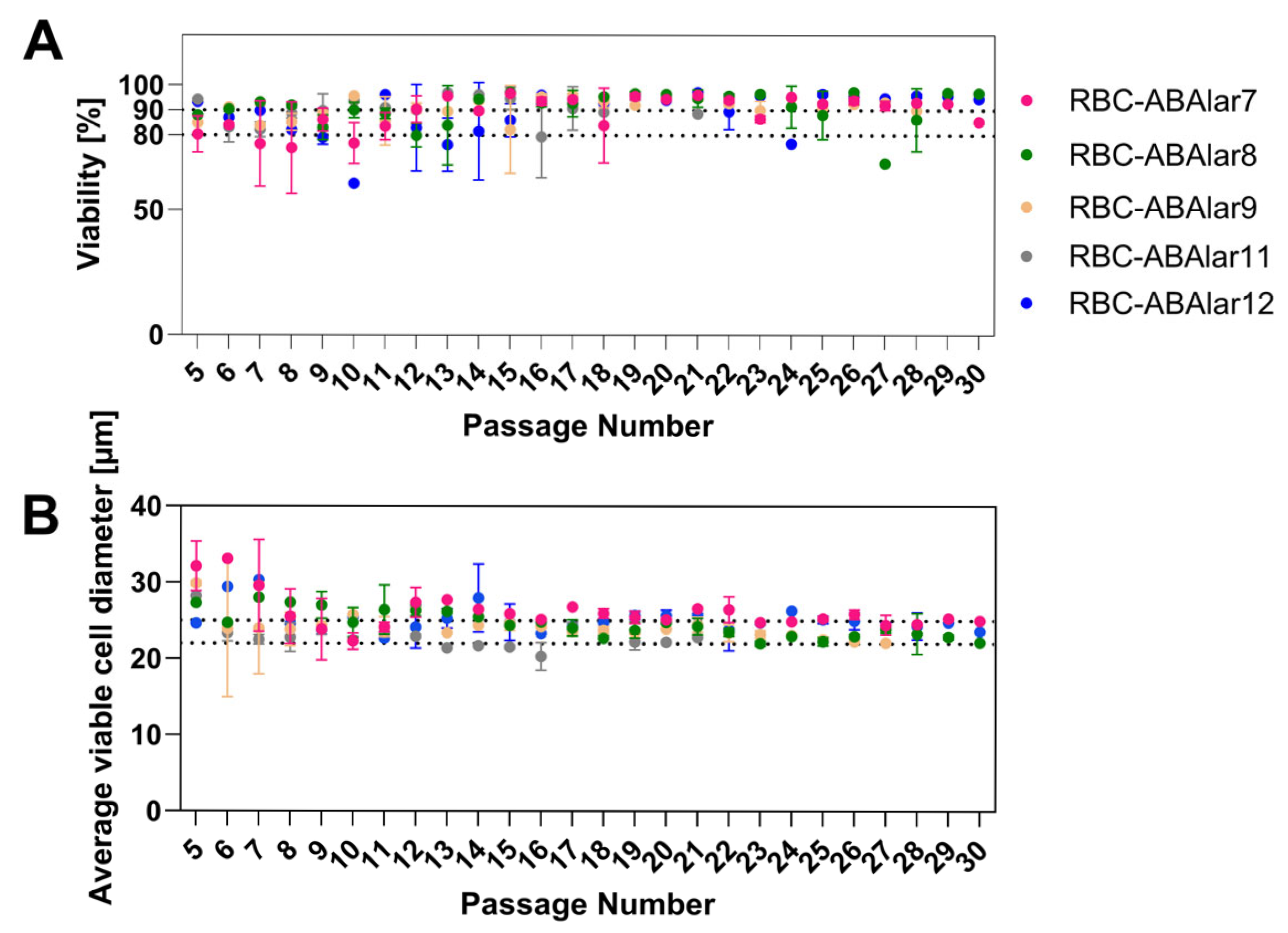
3.1. Cell Culture Development
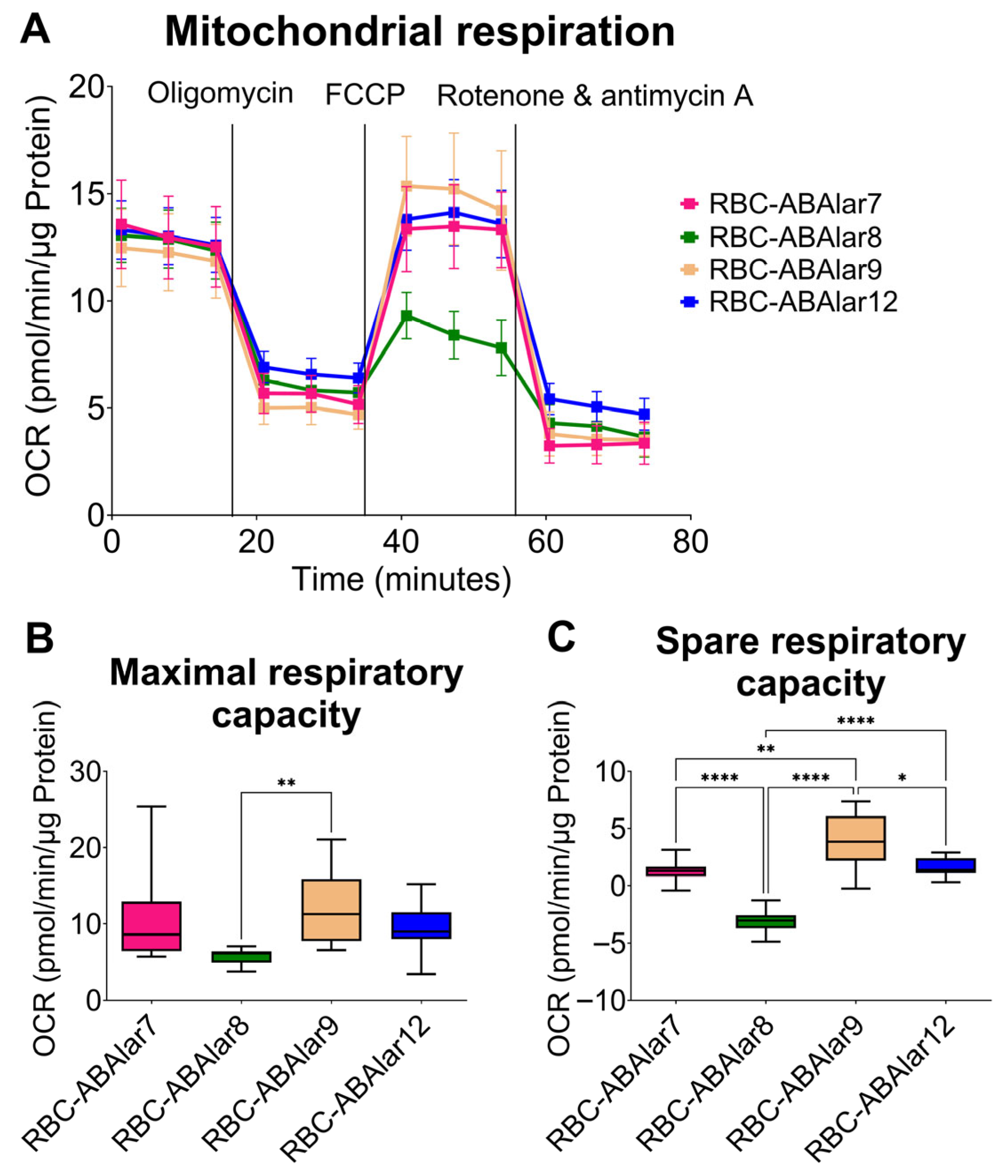
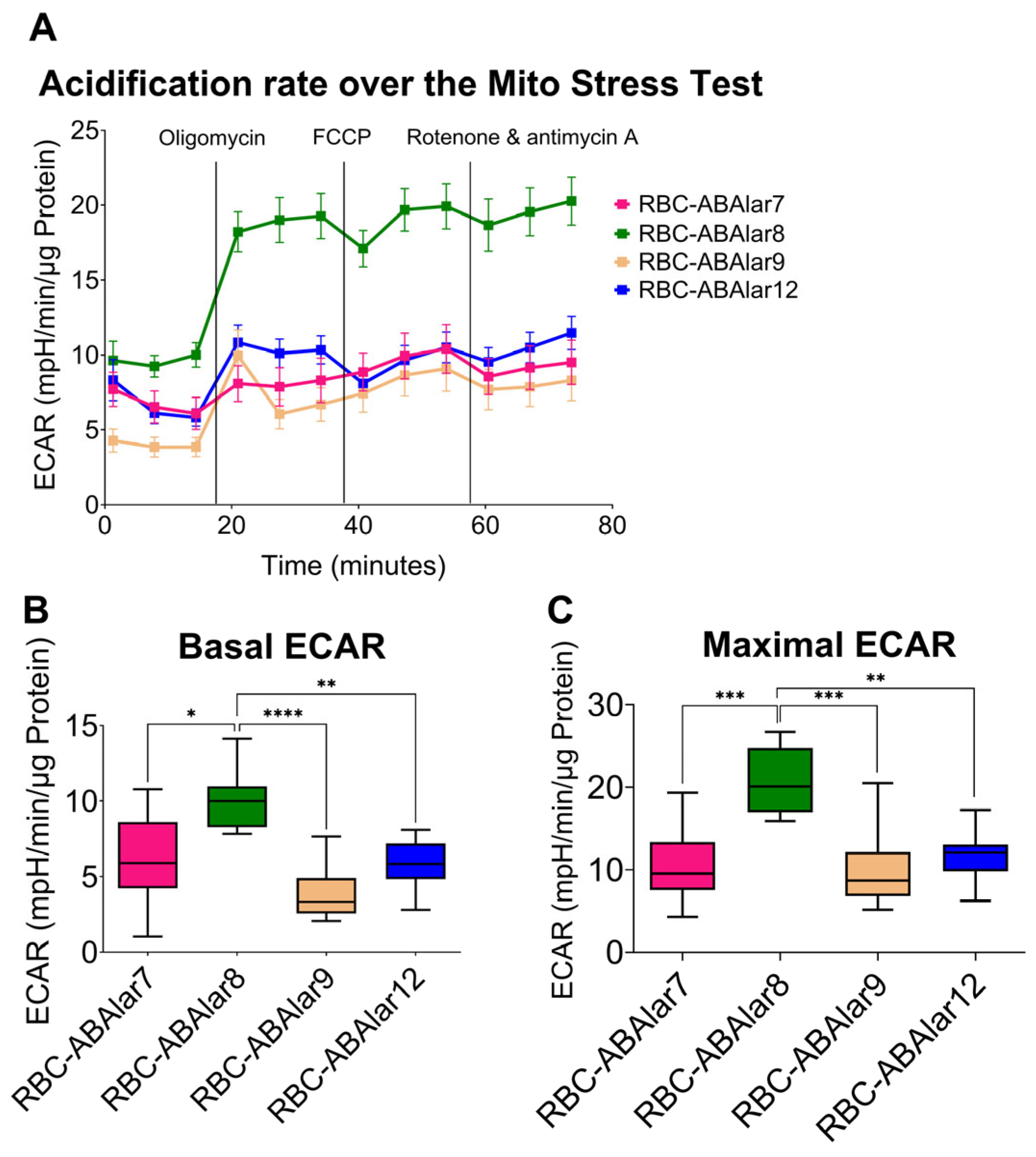
3.2. Metabolic Function of the Cells
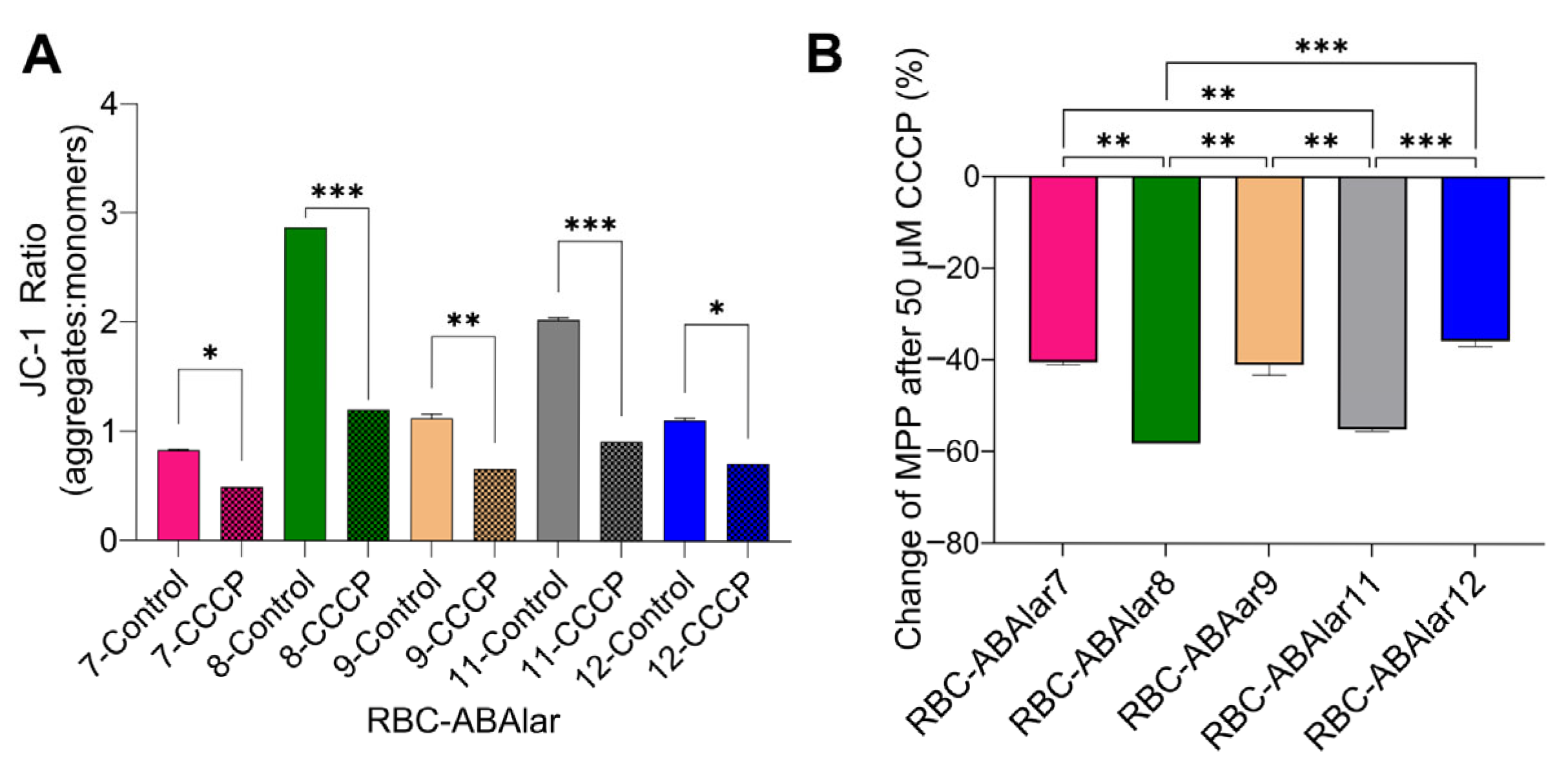
3.3. Flow Cytometry and MMP
3.4. Gene Expression Analysis
4. Discussion
4.1. Cell Culture Development and Gene Expression
4.2. Metabolic and MMP Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, L.; Zhao, C.; Xiao, Q.; Zhao, J.; Liu, H.; Jiang, J.; Cao, Q. Profiling the Physiological Roles in Fish Primary Cell Culture. Biology 2023, 12, 1454. [Google Scholar] [CrossRef] [PubMed]
- Ronald, F.; Eschmeyer, W.N.; Van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References; California Academy of Sciences: San Francisco, CA, USA, 2025; Available online: https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 12 December 2025).
- Scholz, S.; Sela, E.; Blaha, L.; Braunbeck, T.; Galay-Burgos, M.; García-Franco, M.; Guinea, J.; Klüver, N.; Schirmer, K.; Tanneberger, K.; et al. A European Perspective on Alternatives to Animal Testing for Environmental Hazard Identification and Risk Assessment. Regul. Toxicol. Pharmacol. 2013, 67, 506–530. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, R.; Martini, A.; Martinoli, M.; Newton, R.W.; Pulcini, D.; Tonachella, N.; Capoccioni, F. Environmental Performance of Sturgeon Caviar Production. An LCA Study on Luxury Food. Aquaculture 2025, 608, 742766. [Google Scholar] [CrossRef]
- Congiu, L.; Joern, G.; Arne, L. IUCN Red List Reassessment Reveals Further Decline of Sturgeons and Paddlefishes. Oryx 2023, 57, 9–10. [Google Scholar] [CrossRef]
- Bairoch, A. The Cellosaurus, a Cell-Line Knowledge Resource. J. Biomol. Tech. 2018, 29, 25–38. [Google Scholar] [CrossRef]
- Grunow, B.; Noglick, S.; Kruse, C.; Gebert, M. Isolation of Cells from Atlantic Sturgeon (Acipenser Oxyrinchus Oxyrinchus) and Optimization of Culture Conditions. Aquat. Biol. 2011, 14, 67–75. [Google Scholar] [CrossRef]
- Grunow, B.; Franz, G.P.; Tönißen, K. In Vitro Fish Models for the Analysis of Ecotoxins and Temperature Increase in the Context of Global Warming. Toxics 2021, 9, 286. [Google Scholar] [CrossRef]
- Lutze, P.; Brenmoehl, J.; Tesenvitz, S.; Ohde, D.; Wanka, H.; Meyer, Z.; Grunow, B. Effects of Temperature Adaptation on the Metabolism and Physiological Properties of Sturgeon Fish Larvae Cell Line. Cells 2024, 13, 269. [Google Scholar] [CrossRef]
- Downie, A.; Illing, B.; Faria, A.; Rummer, J. Swimming Performance of Marine Fish Larvae: Review of a Universal Trait under Ecological and Environmental Pressure. Rev. Fish Biol. Fish. 2020, 30, 93–108. [Google Scholar] [CrossRef]
- Garrido, S.; Ben-Hamadou, R.; Santos, A.M.P.; Ferreira, S.; Teodósio, M.A.; Cotano, U.; Irigoien, X.; Peck, M.A.; Saiz, E.; Ré, P. Born Small, Die Young: Intrinsic, Size-Selective Mortality in Marine Larval Fish. Sci. Rep. 2015, 5, 17065. [Google Scholar] [CrossRef]
- Goodrich, H.; Clark, T. Why Do Some Fish Grow Faster Than Others? Fish Fish 2023, 24, 796–811. [Google Scholar] [CrossRef]
- Salin, K.; Villasevil, E.M.; Anderson, G.J.; Lamarre, S.G.; Melanson, C.A.; McCarthy, I.; Selman, C.; Metcalfe, N.B. Differences in Mitochondrial Efficiency Explain Individual Variation in Growth Performance. Proc. Biol. Sci. 2019, 286, 20191466. [Google Scholar] [CrossRef] [PubMed]
- Swirplies, F.; Wuertz, S.; Baßmann, B.; Orban, A.; Schäfer, N.; Brunner, R.M.; Hadlich, F.; Goldammer, T.; Rebl, A. Identification of Molecular Stress Indicators in Pikeperch Sander Lucioperca Correlating with Rising Water Temperatures. Aquaculture 2019, 501, 260–271. [Google Scholar] [CrossRef]
- Rebl, A.; Rebl, H.; Verleih, M.; Haupt, S.; Köbis, J.M.; Goldammer, T.; Seyfert, H.-M. At Least Two Genes Encode Many Variants of Irak3 in Rainbow Trout, but Neither the Full-Length Factor nor Its Variants Interfere Directly with the TLR-Mediated Stimulation of Inflammation. Front. Immunol. 2019, 10, 2246. [Google Scholar] [CrossRef]
- Ballester, M.; Cordón, R.; Folch, J.M. DAG Expression: High-Throughput Gene Expression Analysis of Real-Time PCR Data Using Standard Curves for Relative Quantification. PLoS One 2013, 8, e80385. [Google Scholar] [CrossRef]
- Tudor, E.P.; Lewandrowski, W.; Tomlinson, S. Integrating Animal Physiology into the Adaptive Management of Restored Landscapes. Environ. Manag. 2023, 72, 519–528. [Google Scholar] [CrossRef]
- Seebacher, F.; Narayan, E.; Rummer, J.L.; Tomlinson, S.; Cooke, S.J. How Can Physiology Best Contribute to Wildlife Conservation in a Warming World? Conserv. Physiol. 2023, 11, coad038. [Google Scholar] [CrossRef]
- Bronzi, P.; Chebanov, M.; Michaels, J.; Wei, Q.; Rosenthal, H.; Gessner, J. Sturgeon Meat and Caviar Production: Global Update 2017. J. Appl. Ichthyol. 2019, 35, 257–266. [Google Scholar] [CrossRef]
- Dubuc, A.; Burns, C.M.; Debaere, S.F.; Dobszewicz, C.; Gayford, J.H.; Hoffecker, L.J.; Marshall, I.T.; Zanforlin, M.D.; Rummer, J.L. Harnessing Physiological Research for Smarter Environmental Policy. J. Exp. Biol. 2025, 228, jeb249867. [Google Scholar] [CrossRef]
- Nam, A.; Mohanty, A.; Bhattacharya, S.; Kotnala, S.; Achuthan, S.; Hari, K.; Srivastava, S.; Guo, L.; Nathan, A.; Chatterjee, R.; et al. Dynamic Phenotypic Switching and Group Behavior Help Non-Small Cell Lung Cancer Cells Evade Chemotherapy. Biomolecules 2022, 12, 8. [Google Scholar] [CrossRef]
- Grassivaro, F.; Martino, G.; Farina, C. The Phenotypic Convergence between Microglia and Peripheral Macrophages During Development and Neuroinflammation Paves the Way for New Therapeutic Perspectives. Neural Regen Res 2021, 16, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Stuart, J.A.; Fonseca, J.; Moradi, F.; Cunningham, C.; Seliman, B.; Worsfold, C.R.; Dolan, S.; Abando, J.; Maddalena, L.A. How Supraphysiological Oxygen Levels in Standard Cell Culture Affect Oxygen-Consuming Reactions. Oxidative Med. Cell. Longev. 2018, 1, 8238459. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Virtue, S.; Norris, D.M.; Conway, O.J.; Yang, M.; Bidault, G.; Gribben, C.; Lugtu, F.; Kamzolas, I.; Krycer, J.R.; et al. Limited Oxygen in Standard Cell Culture Alters Metabolism and Function of Differentiated Cells. EMBO J. 2024, 43, 2127–2165. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, F.; Peterson, M.K.; Caldeira Araújo, H.; Lautenschläger, F.; Gad, A.K. Vimentin Diversity in Health and Disease. Cells 2018, 7, 147. [Google Scholar] [CrossRef]
- Rossert, J.; Terraz, C.; Dupont, S. Regulation of Type I Collagen Genes Expression. Nephrol. Dial. Transpl. 2000, 15, 66–68. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Strzalka, W.; Ziemienowicz, A. Proliferating Cell Nuclear Antigen (PCNA): A Key Factor in DNA Replication and Cell Cycle Regulation. Ann. Bot. 2011, 107, 1127–1140. [Google Scholar] [CrossRef]
- Cerulus, B.; New, A.M.; Pougach, K.; Verstrepen, K.J. Noise and Epigenetic Inheritance of Single-Cell Division Times Influence Population Fitness. Curr. Biol. 2016, 26, 1138–1147. [Google Scholar] [CrossRef]
- Huang, S. Non-Genetic Heterogeneity of Cells in Development: More Than Just Noise. Development 2009, 136, 3853–3862. [Google Scholar] [CrossRef]
- Kwon, D.; Kim, J.-S.; Cha, B.-H.; Park, K.-S.; Han, I.; Park, K.-S.; Bae, H.; Han, M.-K.; Kim, K.-S.; Lee, S.-H. The Effect of Fetal Bovine Serum (FBS) on Efficacy of Cellular Reprogramming for Induced Pluripotent Stem Cell (iPSC) Generation. Cell Transplant. 2016, 25, 1025–1042. [Google Scholar] [CrossRef]
- van de Pol, I.; Flik, G.; Gorissen, M. Comparative Physiology of Energy Metabolism: Fishing for Endocrine Signals in the Early Vertebrate Pool. Front. Endocrinol. 2017, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.D.; Wang, Z.H.; Yan, B. Strategies for Hypoxia Adaptation in Fish Species: A Review. J. Comp. Physiol. B 2013, 183, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Torres, N.; Tobón-Cornejo, S.; Velazquez-Villegas, L.A.; Noriega, L.G.; Alemán-Escondrillas, G.; Tovar, A.R. Amino Acid Catabolism: An Overlooked Area of Metabolism. Nutrients 2023, 15, 3378. [Google Scholar] [CrossRef]
- Metallo, C.M.; Vander Heiden, M.G. Understanding Metabolic Regulation and Its Influence on Cell Physiology. Mol. Cell 2013, 49, 388–398. [Google Scholar] [CrossRef]
- Espinosa-Ruiz, C.; Mayor-Lafuente, J.; Esteban, M.Á. Mitochondrial Metabolism Characterization of Four Different Fish Cell Lines. Fishes 2022, 7, 354. [Google Scholar] [CrossRef]
- Symersky, J.; Osowski, D.; Walters, D.E.; Mueller, D.M. Oligomycin Frames a Common Drug-Binding Site in the ATP Synthase. Proc. Natl. Acad. Sci. USA 2012, 109, 13961–13965. [Google Scholar] [CrossRef]
- Divakaruni, A.S.; Paradyse, A.; Ferrick, D.A.; Murphy, A.N.; Jastroch, M. Chapter Sixteen—Analysis and Interpretation of Microplate-Based Oxygen Consumption and PH Data. In Methods in Enzymology; Murphy, A.N., Chan, D.C., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 547, pp. 309–354. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine Reliance in Cell Metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef]
- Kim, C.S.; Ding, X.; Allmeroth, K.; Biggs, L.C.; Kolenc, O.I.; L’Hoest, N.; Chacón-Martínez, C.A.; Edlich-Muth, C.; Giavalisco, P.; Quinn, K.P.; et al. Glutamine Metabolism Controls Stem Cell Fate Reversibility and Long-Term Maintenance in the Hair Follicle. Cell Metab. 2020, 32, 629–642.e8. [Google Scholar] [CrossRef]
- Choudhury, D.; Rong, N.; Ikhapoh, I.; Rajabian, N.; Tseropoulos, G.; Wu, Y.; Mehrotra, P.; Thiyagarajan, R.; Shahini, A.; Seldeen, K.L.; et al. Inhibition of Glutaminolysis Restores Mitochondrial Function in Senescent Stem Cells. Cell Rep. 2022, 41, 111744. [Google Scholar] [CrossRef]
- Zimmer, A.M.; Wright, P.A.; Wood, C.M. Ammonia and Urea Handling by Early Life Stages of Fishes. J. Exp. Biol. 2017, 220, 3843–3855. [Google Scholar] [CrossRef]
- Marchetti, P.; Fovez, Q.; Germain, N.; Khamari, R.; Kluza, J. Mitochondrial Spare Respiratory Capacity: Mechanisms, Regulation, and Significance in Non-Transformed and Cancer Cells. FASEB J. 2020, 34, 13106–13124. [Google Scholar] [CrossRef]
- Aguer, C.; Gambarotta, D.; Mailloux, R.J.; Moffat, C.; Dent, R.; McPherson, R.; Harper, M.-E. Galactose Enhances Oxidative Metabolism and Reveals Mitochondrial Dysfunction in Human Primary Muscle Cells. PLoS One 2011, 6, e28536. [Google Scholar] [CrossRef]
- Ruas, J.S.; Siqueira-Santos, E.S.; Amigo, I.; Rodrigues-Silva, E.; Kowaltowski, A.J.; Castilho, R.F. Underestimation of the Maximal Capacity of the Mitochondrial Electron Transport System in Oligomycin-Treated Cells. PLoS One 2016, 11, e0150967. [Google Scholar] [CrossRef]
- Ruas, J.S.; Siqueira-Santos, E.S.; Rodrigues-Silva, E.; Castilho, R.F. High Glycolytic Activity of Tumor Cells Leads to Underestimation of Electron Transport System Capacity When Mitochondrial ATP Synthase Is Inhibited. Sci. Rep. 2018, 8, 17383. [Google Scholar] [CrossRef]


| Gene Symbol | Official Name | NCBI Reference Sequence | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Product Size (bp) |
|---|---|---|---|---|---|
| Target Gene | |||||
| Regulation of glycolysis | |||||
| gapdh | Glyceraldehyde-3-phosphate dehydrogenase | XM_034042013.3 | ACACCCGCTCAT-CAATCTTT | AGGTCCACGACTCTGTTGCT | 80 |
| Proliferation | |||||
| pcna | Proliferating cell nuclear antigen | XM_059013649.1 | GCTGTGACGATCGAGATGAA | AACCAGAGCACACATGCTG | 215 |
| Pluripotency | |||||
| pou5f1 | POU domain, class 5, transcription factor 1 | XM_058986684.1 | GAGTCCCCT-CGTGATACAGG | CAGCACAGCCC-CTTTGATAC | 150 |
| Cytoskeleton | |||||
| sptan1 | Spectrin alpha chain, non-erythrocytic 1 | XM_034927705.2 | AGGGACACTTC-TCATCCGACAT | TGCAGCAGCCGCACACCCTT | 106 |
| vim | Vimentin | XM_034058632.3 | GATTTCGCCTTGTCCGATGC | TTGGTGGTGCGT-TTTCCCTT | 350 |
| col1a1 | Collagen Type I Alpha 1 Chain | EU241879.1 | CACCGAGGACGGTTACACAA | GTGCAATGTCG-ATGATGGGC | 101 |
| Stress-related genes | |||||
| hsp70 | Heat shock protein 70 | XM_033996031.3 | CCCGTGGAGAAGTCC | CCCGTTGAAGAAATCCTG | 123 |
| hsf1 | Heat Shock Transcription Factor 1 | MH917287.1 | CCAAGATTTGCTCGCACAGG | ACCAGCTGT-TTCCCAGTGTC | 100 |
| Reference genes | |||||
| eef1a1 | Eukaryotic Translation Elongation Factor 1 Alpha 1 | XM_034004589.3 | GGACTCCACTGAGCCACCT | GGGTTGTAGC-CGATCTTCTTG | 90 |
| rpl6 | Ribosomal Protein L6 | HQ449564.1 | GTGGTCAAACTC-CGCAAGA | GCCAGTAAG-GAGGATGAGGA | 177 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Leonardo, V.; Tönißen, K.; Brenmoehl, J.; Ohde, D.; Wanka, H.; Benning, K.; Grunow, B. Sibling-Derived Cell Lines of Whole Larval Siberian Sturgeon as an In Vitro Model System for Studying Inter-Individual Differences Within the Same Genomic Heritage. Cells 2025, 14, 2004. https://doi.org/10.3390/cells14242004
Di Leonardo V, Tönißen K, Brenmoehl J, Ohde D, Wanka H, Benning K, Grunow B. Sibling-Derived Cell Lines of Whole Larval Siberian Sturgeon as an In Vitro Model System for Studying Inter-Individual Differences Within the Same Genomic Heritage. Cells. 2025; 14(24):2004. https://doi.org/10.3390/cells14242004
Chicago/Turabian StyleDi Leonardo, Valeria, Katrin Tönißen, Julia Brenmoehl, Daniela Ohde, Heike Wanka, Kenneth Benning, and Bianka Grunow. 2025. "Sibling-Derived Cell Lines of Whole Larval Siberian Sturgeon as an In Vitro Model System for Studying Inter-Individual Differences Within the Same Genomic Heritage" Cells 14, no. 24: 2004. https://doi.org/10.3390/cells14242004
APA StyleDi Leonardo, V., Tönißen, K., Brenmoehl, J., Ohde, D., Wanka, H., Benning, K., & Grunow, B. (2025). Sibling-Derived Cell Lines of Whole Larval Siberian Sturgeon as an In Vitro Model System for Studying Inter-Individual Differences Within the Same Genomic Heritage. Cells, 14(24), 2004. https://doi.org/10.3390/cells14242004







